Abstract
Deep Convolutional Neural Networks (CNN) are at the backbone of the state–of–the art methods to automatically analyze Whole Slide Images (WSIs) of digital tissue slides. One challenge to train fully-supervised CNN models with WSIs is providing the required amount of costly, manually annotated data. This paper presents a semi-weakly supervised model for classifying prostate cancer tissue. The approach follows a teacher-student learning paradigm that allows combining a small amount of annotated data (tissue microarrays with regions of interest traced by pathologists) with a large amount of weakly-annotated data (whole slide images with labels extracted from the diagnostic reports). The task of the teacher model is to annotate the weakly-annotated images. The student is trained with the pseudo-labeled images annotated by the teacher and fine-tuned with the small amount of strongly annotated data. The evaluation of the methods is in the task of classification of four Gleason patterns and the Gleason score in prostate cancer images. Results show that the teacher-student approach improves significatively the performance of the fully-supervised CNN, both at the Gleason pattern level in tissue microarrays (respectively \(\kappa = 0.594 \pm 0.022\) and \(\kappa = 0.559 \pm 0.034\)) and at the Gleason score level in WSIs (respectively \(\kappa = 0.403 \pm 0.046\) and \(\kappa = 0.273 \pm 0.12\)). Our approach opens the possibility of transforming large weakly–annotated (and unlabeled) datasets into valuable sources of supervision for training robust CNN models in computational pathology.
S. Otálora and N. Marini—Equal contribution.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Prostate cancer (PCa) is the fourth most common cancer worldwide, with 1.2 million new cases in 2018, and it has the second-highest incidence of all cancers in men. The gold standard for the diagnosis of PCa is the visual inspection of needle biopsies or tissue samples such as prostatectomies. Currently, the Gleason score (GS) is the standard grading system used to determine the aggressiveness of PCa. The GS system is based on the architectural patterns shown in prostate tissue samples that describe tumor appearance and the presence of alterations in the glands. The Gleason score results from the sum of the two patterns (Gleason patterns from 1 to 5) most present in the tissue slide producing a final grade in the range of 2 to 10. Typical scores range from 6 to 10, where cases with higher values are more likely to grow and spread faster. The Gleason score system has been revised in 2016 [5] to propose a simpler system by having a smaller number of grades (five-groups) with the most significant prognostic differences, Nevertheless, GS is still commonly used in pathology reports, in conjunction with the new five-groups classes. Thanks to the recent improvements in digital microscopy, the diagnosis is increasingly made through the visual inspection of high-resolution scans of a tissue sample or a Whole-Slide Image (WSI).
One of the current challenges in medical imaging and particularly in computational pathology (CP), is the lack of datasets with copious region annotations for training robust supervised deep convolutional neural networks (CNN) [4]. For example, to train the deep learning models in Nagpal et al. [9], the authors collected 112 million image patches derived from 912 slides, which required approximately 900 pathologist hours to annotate. Such efforts raise the question of investigating models that minimize this costly labeling effort and reuse publicly available data to train CNN-based models.
While there is an increasing amount of available raw data, it is well known that finding reliable annotations accompanying the WSI, which are made of up to \(100000^{2}\) pixels, is a problem in this field. Examples of valuable, publicly available datasets are the Camelyon dataset for breast cancer [8] and The Cancer Genome Atlas datasets, containing up to 500 Whole slide images for individual organs, including the prostate (TCGA-PRAD)Footnote 1. The main drawback of the TCGA datasets is that the repository does not provide region annotations for the images. The lack of strong labels poses a challenge to use the dataset to train state–of–the–art supervised CNN models for CP tasks such as the classification and segmentation of tissue subtypes of PCa. The available strongly annotated datasets in CP usually contain few images annotated or small regions of larger images [2], since the annotation of such large slides is a costly process that takes a considerable amount of time from highly-specialized personnel. In machine learning and computer vision, the use of semi-supervised and semi–weakly supervised learning has recently shown the potential of leveraging on large unlabeled and weakly–labeled datasets, reaching better performance than state–of–the–art supervised models in the classification of the ImageNet dataset [14]. Also, combining few strongly labeled and many weakly labeled images has been proposed in [11], achieving competitive results on natural image datasets, while requiring significantly less annotation effort.
Recently in CP, deep CNN approaches using weakly supervision have reached good performance for automatic Gleason scoring in WSI [10]. Obtaining pseudo-labeled data that is automatically annotated and that can improve the robustness against dataset heterogeneity and performance of CNN models is highly valuable, given a large amount of unlabeled (and weakly annotated) datasets that are publicly available and the improvement that it can bring to the results.
In this paper, the simple, yet effective, teacher-student approach of fine-tuning very large pre-trained models to generate pseudo-labeled examples is explored for the first time in the task of classifying prostate cancer tissue. Our approach employs a high-capacity (22 million parameters) ResNext-based model as a teacher. The teacher is pre-trained with a dataset of nearly one billion natural images retrieved from Instagram and its hashtags, and fine-tuned with both, weakly–annotated images from TCGA-PRAD, and annotated tissue microarrays. The smaller student model, a DenseNet-BC-121 with 7 million parameters, is then trained with the TCGA-PRAD pseudo-labeled regions annotated by the teacher and fine-tuned with the tissue microarray strong pixel-wise labels. Experimental results show that the teacher-student approach improves with statistical significance the performance of the fully-supervised CNN, both at the Gleason pattern level in tissue microarrays (respectively \(\kappa = 0.594 \pm 0.022\) and \(\kappa = 0.559 \pm 0.034\)) and at the Gleason score level in WSI (respectively \(\kappa = 0.403 \pm 0.046\) and \(\kappa = 0.273 \pm 0.12\)).
2 Experimental Setup
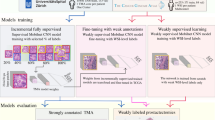
The teacher-student approach: The teacher model is involved in the steps 1 to 4 (yellow background, top) and the student model is steps 5 and 6 (green background at bottom). The teacher model is first fine-tuned (from the trained model of [14]) to predict the weak labels of the TCGA-PRAD patches (primary GP) and then fine-tuned with the strongly-annotated patches from the TMA dataset. The teacher then pseudo-annotate the TCGA-PRAD patches, and the student is pre-trained using the top-\(\rho \) ranked patches. Finally, the student is fine-tuned with the strongly annotated patches from the TMA dataset. (Color figure online)
The overall workflow of the proposed semi-weakly supervised approach for classifying PCa images is summarized in Fig. 1. The details of each step involved in the training of the models are further explained in Sect. 2.2. The cardinality and characteristics of the datasets used in the article are described in Sect. 2.1.
2.1 Datasets
The two datasets of prostate images are gathered from two different sources. The TCGA-PRAD WSI repository and Tissue Microarrays (TMA). TCGA-PRAD includes WSIs from 19 different medical centers. It implies visual heterogeneity between dataset content, even though the tissues are stained in both datasets with the same reagents: hematoxylin and eosin (H&E). The dataset is comprised of pairs of WSIs, up to \(100000^{2}\) pixels, scanned at 40x resolution and the corresponding weak labels (one label per WSI) from the diagnostic report of prostate cancer cases with Gleason scores between 6 and 10.
The WSIs are available from The Cancer Genome Atlas (TCGA), which is an extensive publicly available collection of data including digital pathology images that contains 500 cases of prostate adenocarcinoma (TCGA-PRAD). The used WSIs are a subset of the data containing only images used for diagnostic purposes (no frozen sections). The division of the dataset is the same as in baseline sets for cross-validation: 171 cases for training, 84 for validation, and 46 for testing. Each WSI is paired with its global Gleason score. For the task of Gleason pattern prediction at the patch level, the reported primary Gleason pattern of the WSI is used as a weak label. The patches are densely extracted only from tissue-regions of the WSI. For this, the HistoQC tool [7] is used first to generate tissue masks of the WSIs. Then, the blue-ratio mapping described in Chang et al. [3] is used to prevent selecting areas without nuclei such as those containing fat, connective tissue, or background.
The TMA dataset includes pixel-wise annotations, made by pathologists, of 886 prostate TMA cores. Each core is \(3100^{2}\) pixels, scanned at 40x resolution (0.23 microns per pixel). The training, validation and test sets as well as the patches are the same as in the study of Arvaniti et al. [2]. The total number of microarrays, WSIs and patches extracted from them is shown in Table 1.
2.2 Weakly Semi-supervised Teacher-Student Approach
The hypothesis in the semi-supervised setting is that if one has a dataset with labeled data and another without, it is possible to train a model that can use both sources, of which the performance is higher than the one obtained using only used the labeled samples [15].
The teacher-student paradigm is a semi-supervised strategy where the teacher’s role is to transform the labels from the relevant examples of the weakly–annotated (or unlabeled) data. The teacher model output is pseudo–labels for the unlabeled data (resembling the strong labels) for training the student model with both sources of supervision, the strong annotations, and the pseudo-annotated dataset. Formally, if we denote the loss of a model M trained with a dataset X by \(\mathcal {L_M(X)}\), then ideally, \(\mathcal {L}_M(S) > \mathcal {L}_M(S \cup T(U))\), where S stand for the strongly-annotated, and T(U) for a pseudo-labeled set transformed using a mapping T of the unlabeled (or weakly labeled) dataset U.
The six-steps setup presented bellow resembles the best-performing configuration from the weakly-supervised teacher-student setup originally presented by Yalniz et al. [14]. In the weakly-supervised setup, the authors exploit the weak labels and characteristics of the datasets resembling the characteristics in our application to computational pathology, where it is feasible to use image-level labels as a weak form of supervision. Our main methodological novelties are the use of very high resolution and highly heterogeneous images with weak labels and the student variants, which are specifically designed for the prostate cancer image classification problem and not presented in the baseline paper [14]. While our approach might resemble commonly used bootstrapping techniques, our method differs from them because there is no random sampling involved since the teacher makes a non-trivial selection of unlabeled samples, and the models do not use subsets of the same training set to estimate the performance measures.
1) Weakly Supervised Teacher Fine-tuning: In this first step, the model is fine-tuned with the TCGA-PRAD dataset to predict the primary Gleason pattern label extracted from the reports. The teacher model weights are initialized from the trained model of [14]. The pre-trained model from Instagram, a ResNext-50 is a high-capacity model with 22 million parameters, that better fits with noisy labels [6]. TCGA-PRAD can be considered a noisy dataset since only a subset of patches actually contains the relevant pattern reported as primary Gleason pattern. In this step, the model is trained for ten epochs with a categorical-cross entropy loss to predict the primary Gleason pattern and stopped if convergence is reached early.
2) Fine-tuning of the Teacher with Strong Annotations: In this step, the weights of the model are refined to classify the TMA patches with ground-truth data. In this case the teacher is also presented with samples from the benign class. Ten models (with different initialization) are trained for 15 epochs, as the TMA dataset is not as large as TCGA-PRAD. Then, the model with the best average performance in the validation TMA partition and validation TCGA partition is kept to pseudo-annotate the patches in the next step. The performance of the teacher up to this step is reported in the results section.
3) & 4) Pseudo-labeling and Patch Selection of TCGA-PRAD: In this step, the previously selected teacher model is used to infer the class-wise probabilities of all the TCGA-PRAD patches. For each class, the \(\rho \) highest-ranked patches per class are selected according to the softmax probability of the output of the last fully connected layer. The trade-off between performance and \(\rho \) is shown in the results section.
5) Pre-training of the Student Model with Pseudo-labeled Data: The student model is trained in a supervised fashion using the pseudo-labeled images annotated by the teacher. The distillation procedure aims at training the student in such a way that it best reproduces the output of the teacher. This strategy was shown to be successful for several image recognition tasks [14]. The student has a smaller architecture than the teacher model because it is more efficient for evaluation: the student model is the one for which the hyper-parameter selection and test set evaluations are made. Therefore, it is better to have a smaller, faster inference architecture. In the fifth step, the student model is pre-trained with the \(\rho \) patches per Gleason pattern that are pseudo-labeled by the teacher. Ten models are trained in this step for 15 epochs. The best student model is then selected (i.e., the one that has the best performance in \(\kappa \)-score in the TMA validation partition).
6) Training of the Student and Variants: In the last step, the best student is trained with the strongly-annotated TMA patches. Ten model runs are trained for 15 epochs, selecting the best (the best average run) and reporting the final performance in the \(\kappa \)-score, both in the TMA and in the TCGA-PRAD test sets.
Four training variants of the student are evaluated. A) Fully-supervised training: here, only the TMA annotated patches are used for training the student; the training scheme is similar to the one described in [2]. B) Using only the pseudo-labeled images: in this case, the student never sees any patch with ground-truth data from the pathologist annotations, just the pseudo-labeled patches from the teacher model. C) Pre-training with pseudo-labeled samples and then fine-tuning with the strong annotations. D) Combining the pseudo-labeled and strongly annotated patches in one single training set: this variant is similar to C), with the difference that all the TMA and TCGA-PRAD patches are mixed at training, instead of having two training stages. These three ablation experiments results for the student model, are reported in the results sections Table 2.
2.3 Implementation, Architectures and Hyperparameter Selection
The implementation of all models was done in PyTorch, initialized with the Instagram/ImageNet pre–trained weights for the teacher and student models, respectively. Batch sizes of 128 samples were used for the first weakly supervised pre-training of the teacher (step 1), and the fine-tuning of the teacher was done with a batch size of 32 TMA patches (step 2). Several CNN models, namely, DenseNet121, DenseNet161, MobileNet, MobileNetV2, were tested for the student. Among these, the one that showed the best performance in the validation TMA set was DenseNet121. Therefore, this architecture was chosen to train the four variants of the student. The choice of a pre–trained network is done for speeding up the convergence of the model, as described for the teacher model. The CNN parameters were selected using a grid search over the validation sets of both TCGA and TMA. The best values found on the validation set are the ones used for training the ten repetitions. Specifically, the values explored for the learning rate are in the set \(\{10^{-5},10^{-4},10^{-3}, 10^{-2}\}\). In each of the student training variants, the Adam optimizer is used with a learning rate of 0.001 and a decay rate of \(10^{-6}\).
3 Results and Analysis
There are two evaluation criteria: patch-level Gleason pattern classification and image-level GS classification. For the GS classification, the models are evaluated using the revised Gleason score as defined by the International Society of Urological Pathology. All model performances are measured as the inter-rater agreement and pathologist ground-truth. A performance measure that is often used [1, 13] is Cohen’s kappa, that is defined as \( \kappa = 1 - \frac{\sum _{i,j}w_{i,j}O_{i,j}}{\sum _{i,j}w_{i,j}E_{i,j}}, w_{i,j} = \frac{(i-j)^{2}}{(N-1)^{2}}\) Where i, j are the ordered scores, \(N=5\) is the total number of Gleason scores (or \(N=4\) Gleason pattern classes). \(O_{i,j}\), is the number of images that were classified with a score of i by the first rater and j by the second. \(E_{i,j}\) denotes the expected number of images receiving rating i by the first expert and rating j by the second. The quadratic term \(w_{i,j}\) penalizes the ratings that are not close. When the predicted Gleason score is far from the ground-truth class, \(w_{i,j}\) gets closer to 1. For obtaining the GS using the patch probabilities, all the predicted probabilities are combined and a majority voting decides the GS, as in [1].
In Table 2 the test set performance for the four variants of the student models is shown. The best model is variant four, where both TMA and pseudo-labeled patches from TCGA-PRAD are mixed in one single training set. The teacher-student approach improves the performance of the fully-supervised CNN, both at the Gleason pattern level in tissue microarrays (respectively \(\kappa = 0.594 \pm 0.022\) and \(\kappa = 0.559 \pm 0.034\)) as well as in the Gleason score level performance in WSI (respectively \(\kappa = 0.403 \pm 0.046\) and \(\kappa = 0.273 \pm 0.12\)). The results entries with ‘*’ also show that the only student variant performs significantly better than the baseline in both test sets is the combination of pseudo-labeled and strongly-annotated samples, despite the other variants showing relative improvements.
4 Discussion
An analysis of the optimal \(\rho \) for the number of examples presented to the student is shown in Figure 2. The performance of two of the student variants for Gleason pattern classification remains flat with respect to the number of pseudo-labeled patches, likely because the student saturates with few pseudo-labeled patches. Similar behavior was shown in the baseline method of Yalniz et al. [11] where the student reaches a maximum performance with \({\sim }10\%\) of the pseudo-labeled data and then starts decreasing, probably due to the introduction of many noisy samples.
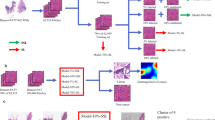
Example of TCGA-PRAD patches pseudo-labeled by the teacher model: each class-box has five uniformly sampled patches from the top hundred ranked samples by the teacher and in the second row five from the hundred lowest ranked for that class. The probability of each patch belonging to the class is shown on top (first row) and in the bottom (second row). The Xe-Y is shorthand for \(X\times 10^{-Y}\).
In Figure 3, a set of pseudo-labeled patches from the teacher are shown. Most of the top-ranked patches match the tissue morphology from the strongly-annotated data. There are a few noisy patches at the lowest probabilities, suggesting that the model is also lowering the relevance of artifacts and other sources of noise. The top-ranked patches for GP3, GP4, and GP5 are similar and typical for the class morphology.
The code and datasets generated during the current study are available from the corresponding author on request. Also, a supplemental document accompanying this paper, details the training of the teacher and each of the three student variants.
Concurrently to the publication of this work, Shaw et al. [12] extended the teacher-student model by generating a chain of student models for the application of classifying colon cancer regions. The results obtained by the authors showed that with the chain of students, using only \(0.5\%\) of the original labeled data, is possible to obtain the same performance as using \(100\%\) of the annotations, showing the potential for use of this approach in other computational pathology tasks.
5 Conclusion
We present a simple yet effective approach for increasing the training dataset size by obtaining pseudo–labeled regions in the task of prostate cancer classification. The evaluation of the proposed semi-weakly supervised teacher-student approach yielded better quantitative results than a fully supervised approach in two highly heterogeneous datasets of PCa. A qualitative assessment also shows how the annotated images by the teacher follow the same gland morphology patterns of the strongly annotated data. The assessment of the trade-off between performance and the amount of pseudo-labeled data shows that increasing the number of patches can deteriorate the student performance by introducing noise in training. We are now working on the semi-supervised approach only, i.e., without using any weak label, as well as the evaluation of the approach in classification tasks for other tissues, validating the pseudo-labeled images with pathologists.
Notes
- 1.
https://portal.gdc.cancer.gov/projects/TCGA-PRAD Retrieved 1st of July, 2020.
References
Arvaniti, E., Claassen, M.: Coupling weak and strong supervision for classification of prostate cancer histopathology images. In: Medical Imaging Meets NIPS Workshop, NIPS 2018 (2018)
Arvaniti, E., et al.: Automated Gleason grading of prostate cancer tissue microarrays via deep learning. Sci. Rep. 8, 1–11 (2018)
Chang, H., Loss, L.A., Parvin, B.: Nuclear segmentation in H&E sections via multi-reference graph cut (MRGC). In: International Symposium Biomedical Imaging (2012)
Cheplygina, V., de Bruijne, M., Pluim, J.P.: Not-so-supervised: a survey of semi-supervised, multi-instance, and transfer learning in medical image analysis. Med. Image Anal. 54, 280–296 (2019)
Epstein, J.I., et al.: A contemporary prostate cancer grading system: a validated alternative to the Gleason score. Eur. Urol. 69(3), 428–435 (2016)
Han, B., et al.: Co-teaching: robust training of deep neural networks with extremely noisy labels. In: Advances in Neural Information Processing Systems, pp. 8527–8537 (2018)
Janowczyk, A., Zuo, R., Gilmore, H., Feldman, M., Madabhushi, A.: HistoQC:: an open-source quality control tool for digital pathology slides. JCO Clin. Cancer Inf. 3, 1–7 (2019)
Litjens, G., et al.: 1399 H&E-stained sentinel lymph node sections of breast cancer patients: the Camelyon dataset. GigaScience 7(6), giy065 (2018)
Luo, F., Nagesh, A., Sharp, R., Surdeanu, M.: Semi-supervised teacher-student architecture for relation extraction. In: Proceedings of the Third Workshop on Structured Prediction for NLP, pp. 29–37. Association for Computational Linguistics, Minneapolis, Jun 2019. https://doi.org/10.18653/v1/W19-1505, https://www.aclweb.org/anthology/W19-1505
Otálora, S., Atzori, M., Khan, A., Jimenez-del Toro, O., Andrearczyk, V., Müller, H.: A systematic comparison of deep learning strategies for weakly supervised Gleason grading. In: Medical Imaging 2020: Digital Pathology, vol. 11320, p. 113200L. International Society for Optics and Photonics (2020)
Papandreou, G., Chen, L.C., Murphy, K.P., Yuille, A.L.: Weakly-and semi-supervised learning of a deep convolutional network for semantic image segmentation. In: Proceedings of the IEEE International Conference on Computer Vision, pp. 1742–1750 (2015)
Shaw, S., Pajak, M., Lisowska, A., Tsaftaris, S.A., O’Neil, A.Q.: Teacher-student chain for efficient semi-supervised histology image classification. arXiv preprint arXiv:1911.04252 (2020)
Ström, P., et al.: Artificial intelligence for diagnosis and grading of prostate cancer in biopsies: a population-based, diagnostic study. Lancet Oncol. 21(2), 222–232 (2020). https://doi.org/10.1016/S1470-2045(19)30738-7
Yalniz, I.Z., Jégou, H., Chen, K., Paluri, M., Mahajan, D.: Billion-scale semi-supervised learning for image classification. arXiv preprint arXiv:1905.00546 (2019)
Zhu, X., Goldberg, A.B.: Introduction to Semi-supervised Learning. Synthesis Lectures on Artificial Intelligence and Machine Learning, pp. 1–130. Morgan Kaufmann Publishers, San Francisco (2009)
Acknowledgements.
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No. 825292 (ExaMode, https://www.examode.eu). Infrastructure from the SURFsara HPC center was used to train the CNN models in parallel. Otálora thanks Minciencias through the call 756 for Ph.D. studies.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
1 Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Otálora, S., Marini, N., Müller, H., Atzori, M. (2020). Semi-weakly Supervised Learning for Prostate Cancer Image Classification with Teacher-Student Deep Convolutional Networks. In: Cardoso, J., et al. Interpretable and Annotation-Efficient Learning for Medical Image Computing. IMIMIC MIL3ID LABELS 2020 2020 2020. Lecture Notes in Computer Science(), vol 12446. Springer, Cham. https://doi.org/10.1007/978-3-030-61166-8_21
Download citation
DOI: https://doi.org/10.1007/978-3-030-61166-8_21
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-61165-1
Online ISBN: 978-3-030-61166-8
eBook Packages: Computer ScienceComputer Science (R0)