Abstract
In the process of diagnosing a brain tumor, the most prominent task is the analysis of the MRI images, and so it is important to precisely assess the images. Despite advancements in the field of medical science and research, there are very few methodologies that provide us with accurate brain tumor segmentation. The segmentation of the images, which is done manually becomes a laborious, tedious task; moreover, the 3D nature of the data imposes several challenges in the segmentation of images, which is done in an automatic manner. Our research focuses on the approach for tackling the task of the segmentation by training a network architecture encouraged by U-Net. Worked on 2018 dataset of brain tumor segmentation challenge (BraTS), we got better results with our projected system than other state-of-the-art architectures like native U-Net architecture.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Artificial intelligence (AI) is the training of machines to enable them to perform tasks that are performed by humans. AI is significant since it can help comprehend troublesome issues in different ventures, for example, entertainment, education, health, commerce, transport, and utilities. The utilization of computerized reasoning and, specifically, AI is getting progressively prominent in the field of research. These frameworks exceed expectations at rapid analysis of data, interpretation, and in certain research tasks such as image assessment and segmentation. Presently, researchers have started to utilize AI for brain tumor analysis. AI strategies are applied in the field of medication to improve the precision of analytic and diagnostic techniques.
Computer vision takes a shot at empowering computers to examine, distinguish and analyze pictures just like humans visualize and then provide acceptable output. The field is linked with artificial intelligence, as the computer must examine or act like humans. This interdisciplinary subject simulates and automates those elements of human imaginative and prescient structures with the usage of sensors, computer systems, and machine learning algorithms. Recently, deep learning algorithms perform a significant role in the computer vision discipline. Its application is that it is presently utilized in comfort stores, driverless car testing, every day therapeutic diagnostics, and in observing the well-being of crops and livestock.
Identifying with computer vision, ongoing journals show wide utilization of one of the sorts of deep artificial neural network, which is known as convolutional neural networks (CNN), especially for division and grouping of brain tumors. Brain tumor is a genuine infection wherein an irregular development of tissue inside the mind can upset appropriate cerebrum work and it has also been recorded as a major cause for increase in death rate cases. This paper (Menze et al., 2015) proposes that the main aim of BraTS challenge is empowering the improvement of present day strategies and methodologies for tumor segmentation by giving an enormous dataset of clarified low-grade gliomas (LGG) and high-grade glioblastomas (HGG). In general, tumor developed in the brain is analyzed using magnetic resonance imaging (MRI). After confirmed brain tumor detection using MRI, a biopsy or surgical operation is undertaken to decide the brain tumor type and to examine the outcomes from a pattern of tissue. Brain tumor detection helps in preliminary clinical trials. Use of the advanced technology can be extended for the educational purpose giving the new generation medical students as well as high school students, better understanding and insights of the assessment of brain tumors, its patterns and diagnosis.
2 Related Work
Brain tumor detection has received significant attention from the researchers due to the discovery of a few detection techniques during recent decades. Initially, CNN was used to feature image into a vector which can be further utilized in classification. With CNN, we learned such feature mapping, but the same feature maps can be used to extend the vector to a segmented image. This is the main idea behind U-Net, and this reverse process would reduce the resulting distortions in the image considerably. A fusion intelligent machine learning technique which engages computational methods such as segmentation of images using feedback pulse coupled neural network, extricating specifications utilizing discrete wavelet transform method, lessening the dimensionality of the wavelet coefficients utilizing principal component analysis, and the feedforward backpropagation neural network to classify inputs into typical or irregular, and for computer-aided detection system for brain tumor detection through magnetic resonance images is proposed by El-Dahshan et al. (2014). Abd-Ellah et al. (2016) anticipated a method in which classification of magnetic resonance images (MRIs) was done into normal and abnormal by using combination of morphological filters, extraction of features utilizing the discrete wavelet transform (DWT) approach, and dimensionality decrease of the parameters by applying the principal component analysis (PCA) technique. For the usage of the order of MRI pictures, a kernel support vector machine (KSVM) was utilized.
Mahmoud Khaled Abd-Ellah et al. (2018) proposed a two-phase multi-model automatic diagnosis system. This research addresses the development of computer-aided diagnosis (CAD) system for tumor detection and magnetic resonance images (MRIs) were used for locating the tumor. The first phase is of tumor detection where the system consolidates a CNN for include extraction and feature classification is finished with an error correcting output codes support vector machine (ECOC-SVM). In the second phase, a five-layer completely developed region-based convolutional neural network (RCNN) is employed to locate the position of the tumor. Rączkowski et al. (2019) implemented accurate, reliable, and active (ARA) framework with CNN, which is a Bayesian deep learning model, called ARA-CNN for histopathological image classification.
Isensee et al. (2017) present a modified algorithm of segmentation in the form of CNN, to obtain best results of segmentation of brain tumor. The system design is based on the popular U-Net which is trained on the BraTS 2017 validation set and modifications are done with caution. This network architecture presented potential scores. The resulting mean dice coefficients were 0.858 for whole tumor (WT), 0.775 for tumor core (TC), and 0.647 for the contrast enhancing tumor (ET). Dong et al. (2017) proposed a completely programmed technique using U-Net-based deep convolutional networks for performing brain tumor segmentation. The creators evaluated their technique utilizing multimodal brain tumor image segmentation (BraTS 2015) datasets which involved information of various numbers of cases for high-grade brain tumor and low-grade tumor. This method provided with promising segmentation of brain tumor proficiently. Clinical data was utilized to develop a way to segment three sub-regions of glioma by Tuan (2018). Their proposed strategy aims at producing more images using bit-plane which assembles the most and least significant bits. Thereafter, U-Net is implemented with numerous kernels, to section the entirety of glioma areas which provides progressively precise outcomes. They assessed this method with the BraTS challenge database of 2018 (Menze et al., 2015), and the same process accomplishes efficacy of 82, 68, and 70% dice scores on validation data.
For the purpose of performing easy automatic tumor detection and segmentation, Rajan and Sundar (2019) proposed a fresh hybrid energy-efficient technique called K-means clustering, unified with fuzzy C-means (KMFCM) and active contour by level set, for effectively performing tumor segmentation, detection of edges, and intensity improvement.
The BraTS Dataset 2018 provided by The Center for Biomedical Image Computing and Analytics (CBICA) essentially is the MRI scans of Glioblastoma (HGG) and Lower Grade Glioma (LGG), with a clinically affirmed prognosis. The CBICA provides a separate set of 66 MRI scans without ground truth, which was used for testing by Menze et al. (2015); 2017), Bakas et al. (2018).
Zikic et al. (2014) utilized max-pooling and separated two convolutional layers of a shallow CNN with stride 3, and in this way utilized one fully connected (FC) layer and a softmax layer. This research shows that CNNs are more promising than machine learning algorithms such as randomized forests (RF). For the brain segmentation task, Urban et al. (2014) used the 3D CNN. Voxel-wise classifier is used on the data, and the cubes of voxels are fed into the classifier, which helps in the prediction of voxel(s) in the center of the cube. Pereira et al. (2016) explored small 3 × 3 kernels while using automatic segmentation method on CNN. On stacking up multiple convolutional layers, the author examined that as the depth increased, the extracted features became abstract. The authors also experimented with the use of the larger kernels/filter and concluded that even after using a larger number of feature maps, shallow architectures exhibited lower performance. Cireşan et al. (2012) made use of the deep neural network as the pixel classifier to perform the segmentation of neuronal structures shown as electron microscopy (EM) images automatically. The author uses two-dimensional CNN as a pixel classifier on every slice in the sliding window fashion to segment an entire stack of 512 × 512 × 30 electron microscopy images. This approach causes redundancy in the computation leading to time inefficiency and the neural network cannot learn the global features in it.
The design of the U-Net is inspired from the fully connected layers mentioned in Long et al. (2015). A fully connected layer learns features from all the combinations of the features of the yield of the previous convolution or pooling layer. This fully connected layer flattens this output into a single vector of values, each representing a probability of a class. Image Segmentation involves classification of each image pixel of the input and later creating a map of all identified object areas on the image. Long et al. (2015) later projected the replacement of these detected areas with convolutions using 1 × 1 sized filter. Converting the fully connected layer into an equivalent convolution layer facilitates the usage of the neural net as a convolution to image larger than the original training image and spatially dense output can be obtained. With the purpose of increasing the receptive field and avoiding the input size reduction and information loss from the boundaries of the image, they used padding in the CNN.
Build upon the “fully convolution network” anticipated by Long et al. (2015), Ronneberger et al. (2015) attempts using a 2D completely convolutional neural network for segmentation of stacks of EM pictures. The network consists of a “contracting” or convolution stage, where the input size decreases, and the count of feature maps increases and an “expansive” or deconvolution stage in which the reverse process of the convolution happens. In the up-sampling part, the architecture gains U shape because of numerous feature channels, which allow the propagation of background information to higher resolution layers through the network, due to which the expansive path is to some extent conforming to the contracting path, altogether the network has 23 convolutional layers. In the last layer, the desired number of classes were achieved mapping the feature vector and is performed using a 1X1 convolution. The extension of the 2D network to 3D network is done in paper (Çiçek et al. 2016; Milletari et al. 2016).
3 Proposed Method
3.1 Architecture
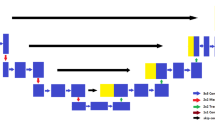
Our architecture consists of the contracting and growing stages found in different works utilizing similar networks (Çiçek et al. 2016; Milletari et al. 2016; Ronneberger et al. 2015), where long skip connections are made between feature maps from the principle stage and feature maps from the consequent stage. Concatenation is the method that has been used frequently for concatenating feature maps in many different works. Inspired by Long et al. (2015), where the authors perform combination of layers of the feature hierarchy and enhance the spatial precision of the output to connect coarseness of final segmentation, segmentations created at different stages in the network are combined in our approach. Normalizing over every channel in every training sample, for instance, normalization is put in practice at test time unlike batch normalization which normalizes across input features and does not apply due to interdependency of small batches at the time of testing. We have implemented multi-class adaptation of dice-loss (Milletari et al. 2016) (Fig. 1).
Network Architecture. Our architecture, which is U-Net influenced, consists of the contracting pathways (on the left-hand side), where the features are extracted and input size reduces, and an expanding pathway (on the right-hand side) where the up-sampling is performed so as to scale up the activation size to the same image size. The skip connections connect feature maps from the principal stage to feature maps in the consequent stage
3.2 Data
In this paper, our emphasis is on brain tumor segmentation task with DNN. For the same task, we are utilizing the BraTS 2018 dataset for the purpose of training and assessment. The dataset includes multimodal MRI sweeps of HGG and LGG obtained in differing organization with distinct conventions. These MRI scans contain native T1, post-contrast T1-weighted, T2-weighted, and T2 fluid attenuated inversion recovery (FLAIR) volumes and are co-enrolled to a similar structural format interjected to a similar resolution (1 mm) and are skull stripped. One to four raters performed manual annotation on these MRI scans and even acceptance was achieved from veteran radiologists. Various segmentation labels were used to designate different glioma sub-regions such as TC, ET, and WT. Altogether, the dataset incorporates 285 MRIs for training (210 HGG and 75 LGG images), 67 validations and 192 testing MRIs.
3.3 Preprocessing
The BraTS challenge dataset is now given preprocessed which included skull stripping, and the data were co-enlisted and re-sampled to a resolution of 1 mm × 1 mm × 1 mm. The dimensions of every volume were 240 × 240 × 155. The data is first preprocessed by image-wise normalization, and correction is performed on it. Bias field correction is performed to reduce the signal of certain modalities, caused by the magnetic fields while collecting the data. Then, all the nonzero-pixel, foreground locations from all the images are combined. The resultant image still has blank space background in every direction. To tackle this, the image is cropped and the smallest image that still contains foreground is obtained. Then using the same cropping values all the images in the input data are cropped. Later, those images are re-sliced to match the input shape of (128, 128, 128).
3.4 Training
The architecture was trained with the patches of the input data due to computational limitations. After mining 3D voxels with size 64 × 64 × 64 from the training data, they are provided as an input to the model, with the batch size of 2. The model was trained for 300 training steps, with each step involving iterating over 100 batches on NVIDIA GTX 1080ti GPU. The preliminary rate of learning was set to 5 × 10−4, and ADAM optimizer was used. We made use of the dice-loss function to cope up with the issue of class imbalance,
where a is the prediction, i.e., network’s softmax output and b being the ground truth. Both a and b have shape j by c where j is the training patch’s pixel quantity and i ∈ I, being the classes, so the scoring is repeated over all the classes and averaged (Fig. 2).
We additionally use augmentation techniques, for example, gamma correction augmentation and mirroring with random notations, scaling, elastic deformations, to avoid overfitting for images (Fig. 3).
4 Result Analysis
Our network is trained and assessed on the dataset of BraTS 2018 via fivefold cross-validation. Utilizing the BraTS 2018 validation set, we accomplished dice scores for WT = 0.89, TC = 0.79, and ET = 0.73 (Fig. 4).
It was observed that the revised approach introduced by an amalgamation of several segmentation maps generated at diverse scales implied in this paper gave better results than the native U-Net architecture and other conventional neural network approaches such as deep neural networks (DNN), fully convolution neural network (FCNN), and 3D CNN combined with random fields (Table 1).
All T1, T1C, T2, FLAIR images are skull stripped. Results in the T1 images highlight fat tissue within the brain and fat and fluid within the brain is highlighted in T2 images. FLAIR images appear like T2 weighted images where gray matter appears brighter than white matter. The abnormal glioblastoma in FLAIR weighted image are represented by a high signal (brighter/ white portion in the image) and similarly in T1 weighted image is represented by a low signal (darker portion in image). The ground truth is manually segmented, and the output through model is segmented with large and fine-grained regions accurately (Figs. 5, 6, 7, 8, 9 and 10).
5 Discussion
Thus, to putting everything in a nutshell, our paper centers around the segmentation task and post going through different model approaches to the segmentation of the medical images, we trained the BraTS 2018 data on the Modified U-Net Architecture (Tuan 2018) which includes residual weights, deep supervision, and equally weighted dice coefficient. Using this architecture, we were able to achieve more accuracy than the original U-Net Architecture. We first normalized the image to rule out bias fields/gain fields, after which the data is alienated into 85% of training and 25% validation. The training data was fed into the architecture. We applied various data augmentation methods, for example, random notations, random scaling, and random mirroring. With the above-mentioned methods, we achieved a dice coefficient of 0.89 (80.3% average).
In order to make the model more accurate, we wish to train the model on more data with more variation in the architecture and data augmentation for better results. We can also append the pipeline with classifiers such as efficient net to make the result more accurate and robust.
References
Abd-Ellah, M. K., Awad, A. I., Khalaf, A. A., & Hamed, H. F. (2016, September). Classification of brain tumor MRIs using a kernel support vector machine. In International Conference on Well-Being in the Information Society (pp. 151–160). Cham: Springer.
Abd-Ellah, M. K., Awad, A. I., Khalaf, A. A., & Hamed, H. F. (2018). Two-phase multi-model automatic brain tumour diagnosis system from magnetic resonance images using convolutional neural networks. EURASIP Journal on Image and Video Processing, 2018(1), 97.
Bakas, S., Akbari, H., Sotiras, A., Bilello, M., Rozycki, M., Kirby, J. S., & Davatzikos, C. (2017). Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Scientific Data, 4, 170117.
Bakas, S., Reyes, M., Jakab, A., Bauer, S., Rempfler, M., Crimi, A., & Prastawa, M. (2018). Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. arXiv:1811.02629
Chen, W., Liu, B., Peng, S., Sun, J., & Qiao, X. (2018, September). S3D-UNet: Separable 3D U-Net for brain tumor segmentation. In International MICCAI Brainlesion Workshop (pp. 358–368). Cham: Springer.
Çiçek, Ö., Abdulkadir, A., Lienkamp, S. S., Brox, T., & Ronneberger, O. (2016, October). 3D U-Net: Learning dense volumetric segmentation from sparse annotation. In International Conference on Medical Image Computing and Computer-assisted Intervention (pp. 424–432). Cham: Springer.
Ciresan, D., Giusti, A., Gambardella, L. M., & Schmidhuber, J. (2012). Deep neural networks segment neuronal membranes in electron microscopy images. In Advances in neural information processing systems (pp. 2843–2851).
Dong, H., Yang, G., Liu, F., Mo, Y., & Guo, Y. (2017, July). Automatic brain tumor detection and segmentation using U-Net based fully convolutional networks. In Annual Conference on Medical Image Understanding and Analysis (pp. 506–517). Cham: Springer.
El-Dahshan, E. S. A., Mohsen, H. M., Revett, K., & Salem, A. B. M. (2014). Computer-aided diagnosis of human brain tumor through MRI: A survey and a new algorithm. Expert Systems with Applications, 41(11), 5526–5545.
Havaei, M., Davy, A., Warde-Farley, D., Biard, A., Courville, A., Bengio, Y., & Larochelle, H. (2017). Brain tumor segmentation with deep neural networks. Medical Image Analysis, 35, 18–31.
Isensee, F., Kickingereder, P., Wick, W., Bendszus, M., & Maier-Hein, K. H. (2017, September). Brain tumor segmentation and radiomics survival prediction: Contribution to the brats 2017 challenge. In International MICCAI Brainlesion Workshop (pp. 287–297). Cham: Springer.
Kamnitsas, K., Bai, W., Ferrante, E., McDonagh, S., Sinclair, M., Pawlowski, N., & Glocker, B. (2017b, September). Ensembles of multiple models and architectures for robust brain tumour segmentation. In International MICCAI Brainlesion Workshop (pp. 450–462). Cham: Springer.
Kamnitsas, K., Ledig, C., Newcombe, V. F., Simpson, J. P., Kane, A. D., Menon, D. K., & Glocker, B. (2017a). Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Medical Image Analysis, 36, 61–78.
Kayalibay, B., Jensen, G., & van der Smagt, P. (2017). CNN-based segmentation of medical imaging data. arXiv:1701.03056
Long, J., Shelhamer, E., & Darrell, T. (2015). Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (pp. 3431–3440).
Menze, B. H., Jakab, A., Bauer, S., Kalpathy-Cramer, J., Farahani, K., Kirby, J., Burren, Y., Porz, N., Slotboom, J., Wiest, R., et al (2015). The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Transactions on Medical Imaging, 34(10), 1993–2024.
Milletari, F., Navab, N., & Ahmadi, S. A. (2016, October). V-net: Fully convolutional neural networks for volumetric medical image segmentation. In 2016 Fourth International Conference on 3D Vision (3DV) (pp. 565–571). IEEE.
Myronenko, A. (2018, September). 3D MRI brain tumor segmentation using autoencoder regularization. In International MICCAI Brainlesion Workshop (pp. 311–320). Cham: Springer.
Pereira, S., Pinto, A., Alves, V., & Silva, C. A. (2016). Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Transactions on Medical Imaging, 35(5), 1240–1251.
Rączkowski, Ł, Możejko, M., Zambonelli, J., & Szczurek, E. (2019). ARA: Accurate, reliable and active histopathological image classification framework with Bayesian deep learning. Scientific Reports, 9(1), 1–12.
Rajan, P. G., & Sundar, C. (2019). Brain tumor detection and segmentation by intensity adjustment. Journal of Medical Systems, 43(8), 282.
Ronneberger, O., Fischer, P., & Brox, T. (2015, October). U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention (pp. 234–241). Cham: Springer.
Singh, P. K., Kar, A. K., Singh, Y., Kolekar, M. H., & Tanwar, S. (Eds.). (2019a). In Proceedings of ICRIC 2019: Recent Innovations in Computing (Vol. 597). Springer.
Singh, P. K., Panigrahi, B. K., Suryadevara, N. K., Sharma, S. K., & Singh, A. P. (Eds.). (2019b). In Proceedings of ICETIT 2019: Emerging Trends in Information Technology (Vol. 605). Springer.
Tuan, T. A. (2018, September). Brain tumor segmentation using bit-plane and UNET. In International MICCAI Brainlesion Workshop (pp. 466–475). Cham: Springer.
Urban, G., Bendszus, M., Hamprecht, F., & Kleesiek, J. (2014). Multi-modal brain tumor segmentation using deep convolutional neural networks. In MICCAI BraTS (Brain Tumor Segmentation) Challenge. Proceedings, Winning Contribution (pp. 31–35)
Zhao, X., Wu, Y., Song, G., Li, Z., Zhang, Y., & Fan, Y. (2018). A deep learning model integrating FCNNs and CRFs for brain tumor segmentation. Medical Image Analysis, 43, 98–111.
Zikic, D., Ioannou, Y., Brown, M., & Criminisi, A. (2014). Segmentation of brain tumor tissues with convolutional neural networks. In Proceedings MICCAI-BRATS (pp. 36–39).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Trivedi, H., Thumar, K., Ghelani, K., Gandhi, D. (2021). An Analysis of Brain Tumor Segmentation Using Modified U-Net Architecture. In: Singh, P.K., Polkowski, Z., Tanwar, S., Pandey, S.K., Matei, G., Pirvu, D. (eds) Innovations in Information and Communication Technologies (IICT-2020). Advances in Science, Technology & Innovation. Springer, Cham. https://doi.org/10.1007/978-3-030-66218-9_16
Download citation
DOI: https://doi.org/10.1007/978-3-030-66218-9_16
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-66217-2
Online ISBN: 978-3-030-66218-9
eBook Packages: Earth and Environmental ScienceEarth and Environmental Science (R0)