Abstract
RNA-binding proteins (RBPs) constitute a diverse group of proteins that control the fate and expression of the transcriptome via events collectively termed post-transcriptional gene regulation. They are a relatively understudied class of regulators, where historically the focus has been on gene regulators such as transcription factors and small RNAs. This has been due partly to the inability to globally identify the RNA-binding portion of the proteome. However, this has recently changed with the development of “mRNA-interactome capture”; the UV cross-linking of RNAs to proteins that are in direct contact, followed by the isolation of these protein-RNA complexes and subsequent identification of the RNA-bound proteins by mass spectrometry. In plants, this methodology has now confirmed the RNA-binding nature of 100s of bioinformatically predicted RBPs, as well as the identification of many proteins that were not previously known to bind RNA. Characterizing these RBPs will begin to elucidate the true scope of post-transcriptional gene regulation in plants, revealing novel regulatory mechanisms and biotechnological opportunities for improvement of crop species. We highlight three areas of immediate interest to which this UV cross-linking method can contribute; gene silencing, translational control of protein synthesis during abiotic stress, and the epitranscriptome.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
5.1 Introduction: RNA-Binding Proteins Execute Post-Transcriptional Regulation
Gene regulation is fundamental to life, being coordinated via a myriad of molecular interactions that enables the execution of differential gene expression programs that underpin development and responses to environmental cues (Briggs 2016; Hicks 2001). Gene expression commences with the production of RNA via transcription. These RNA molecules are simply carriers of genetic information, needing to interact with cellular factors and machinery in order for them to perform their genetic function. This not only applies to precursor mRNAs (pre-mRNAs) that corresponds to the coding portion of the transcriptome, but also to the non-coding portion, for example, primary-microRNAs (pri-miRNAs). The majority of these cellular factors and machinery correspond to RNA-binding proteins (RBPs), whose complex interaction with the transcriptome determines its fate (Hentze et al. 2018). RBPs not only mediate the processing and modification of RNAs resulting in their maturation, but they also determine expression (translation), localization, and stability (Fig. 5.1) (Obernosterer et al. 2006; Floris et al. 2009; Maldonado-Bonilla 2014; Schwartz 2016). For instance, in the nucleus RBPs mediate the capping of pre-mRNAs at their 5` end, and polyadenylation at their 3` end. Most RNAs are decorated with chemical modifications that are added by RBPs referred to as “epitranscriptomic writers” (Li and Mason 2014; Vandivier and Gregory 2018). Ubiquitously, eukaryotic mRNAs contain introns that are processed by the spliceosome, a complex composed of small nuclear RNAs and proteins including RBPs (Fig. 5.1) (Xiao et al. 2016). Once exported to cytoplasm, mRNAs may be translated or transported to organelles or subcellular foci such as processing bodies (where mRNAs is degraded) or stress granule (where they are protected from translation and decay) (Fig. 5.1) (Chantarachot and Bailey-Serres 2018; Maldonado-Bonilla 2014). “Epitranscriptome readers” and “erasers” also interact with the chemical modifications, controlling RNA fate (Shen et al. 2019). Together these processes are considered post-transcriptional regulation, which ultimately controls the genomic output from a cell, a process that underpins life.
5.2 RBPs Are an Understudied Class of Gene Regulators
Despite this central role in controlling gene expression, RBPs have remained a relatively understudied cohort of gene regulators. One contributing factor to this, is that determining mRNA-protein interaction has remained challenging largely due to limiting technology. Historically, this was in contrast to methods that were available to study other classes of regulators. For instance, RNA-seq makes it relatively easy to identify the global cohort of small RNAs (sRNAs). Supporting these analyses are simple sequence complementary-based programs that predict their targets giving insights into their function (Li et al. 2014). Similarly, methodologies have long existed for the study of transcription factors and their targets. For example Chromatin—immunoprecipitation (ChIP-seq) methodology has been well developed and widely utilized, which again gives functional insight of these regulatory genes.
Consequently, regarding gene regulation, the focus has remained on transcription factors and sRNAs of which many have been functionally characterized. By contrast, for the vast majority of RBPs, little is known about their function, their targets, or even when they are actively binding RNA (Silverman et al. 2013). Confounding this challenge is their heterogeneity. RBPs correspond to a biochemically diverse and complex collection of proteins that interact with RNA via multiple mechanisms, be it RNA sequence motifs, RNA structures, or to the vast array different post-transcriptional chemical epitranscriptome marks decorated on RNA. Defining the cohort of RBPs in a cell, the RNAs to which they bind, and to what structural features they recognize, are all challenging experiments. Consequently, despite eukaryotic genomes contain hundreds of different RBPs (being similar to the number of genes encoding transcription factors), currently our knowledge on the function of vast majority of these RBPs, or the mechanism by which they operate, remain unknown (Wheeler et al. 2018; Lee and Kang 2016).
Of the few RBPs that have been characterized in plants, they have been shown to play crucial roles in development, including flowering (Lim et al. 2004), senescence (Wu et al. 2016), and environmental responses, including circadian rhythms (Staiger and Green 2011), stresses (Marondedze et al. 2019; Frei dit Frey et al. 2010) and hormones [for reviews see (Bazin et al. 2018; Silverman et al. 2013)]. For example, the FLOWERING CONTROL LOCUS A (FCA) protein harbors an RNA-Recognition Motif (RRM) domain that regulates RNA splicing to suppress target gene expression and promote flowering-time (Lim et al. 2004; Lee et al. 2015). Other RBPs have been shown to play a role in stress response, such as the GLYCINE RICH PROTEINs (GRPs). Their expression is regulated by ABA and they mediate a number of different physiological responses to counter stress (Czolpinska and Rurek 2018). Nevertheless, in plants, much of what is known regarding RBPs is rudimentary and comes via bioinformatic extrapolation from other kingdoms (Silverman et al. 2013).
5.3 The Global Identification of RBPs with mRNA-Interactome Capture
Until recently, our knowledge on which proteins bind RNA came mainly from targeted studies on individual proteins or from bioinformatic predictions of proteins containing known canonical RNA-binding domains (RBDs), as there were no global methods for their determination (Silverman et al. 2013). Attempts to solve this problem included the use of protein micro-arrays (Tsvetanova et al. 2010) or stable isotope labeling by amino acids in cell culture (SILAC) to identify peptides bound to RNA probes (Butter et al. 2009). However, these in vitro approaches are limited and may not reflect biologically significant interactions that occur in vivo.
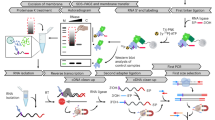
Solving this technical limitation has been the landmark development of mRNA-interactome capture, which was pioneered in animal cell lines (Castello et al. 2012; Baltz et al. 2012). Here 254 nm UV light is irradiated onto live cells which covalently cross-links proteins that are directly bound to RNAs in vivo, thereby “freezing” mRNA-protein interactions. The advantage of using UV light for cross-linking is that only proteins in direct contact with RNA will form covalent bonds with RNA, and unlike formaldehyde, no protein-protein cross-links will occur, therefore only genuine RBPs are captured (Castello et al. 2012; Baltz et al. 2012). Following cross-linking, mRNA-protein complexes are isolated using oligo(dT) beads. These complexes are stringently washed to remove non-cross-linked proteins. The mRNA-protein complexes are then eluted from the oligo(dT) beads, and then RNA is degraded via RNase treatment, leaving the RNA-bound protein fraction. These proteins are then digested with trypsin and then analysed by quantitative mass spectrometry (MS). Multiple large scale biological replicates are performed on both UV treated [cross-linked (CL)] or non-UV [non cross-linked (nCL)] samples. Proteins that are enriched in the CL sample compared to the nCL sample with strong statistical significance [e.g., a false discovery rate (FDR) of below 1%] are considered strong candidates for being RBPs.
Such an approach captures RBPs in a largely unbiased, systematic manner. Interactome capture experiments have been completed for human HeLa and human embryonic kidney HEK293 cells (Castello et al. 2012; Baltz et al. 2012). mouse embryonic stem cells (Kwon et al. 2013), liver cells, and yeast (Beckmann et al. 2015). Additionally, the approach has been used on whole organisms, such as Caenorhabditis elegans (Matia-Gonzalez et al. 2015) and Drosophila (Wessels et al. 2016). Together, these experiments have provided experimental evidence of RNA-binding for hundreds of predicted RBPs, which have classical RNA-binding domains (RBDs). In addition, a multitude of other potential RBPs has been identified, that neither have a classical RBD, nor any known association with RNA (Hentze et al. 2018). Therefore, like other unbiased “omics” approaches, the unexpected findings are leading to a paradigm shift in our perception of what an RBP is and what their potential roles in the cell are (Hentze et al. 2018).
5.4 Arabidopsis in Planta mRNA-Interactome Capture
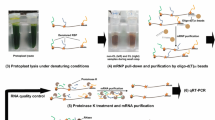
The method of mRNA-interactome capture has now been applied to Arabidopsis, including leaf mesophyll protoplasts (Zhang et al. 2016), cell suspension cultures (Marondedze et al. 2016), and an in planta study on intact etiolated seedlings (Fig. 5.2; Reichel et al. 2016). These studies have given insights into the portion of the proteome that is RNA-binding. They have provided the first experimental evidence of RNA-binding for 100s of bioinformatically predicted plant RBPs. Additionally, similar to the studies in animals, a large proportion of the captured proteins neither have a classical RBD nor any know association with RNA. This has raised the possibility of identifying many new RNA regulatory pathways and mechanisms that had not been previously considered (Koster et al. 2017; Bach-Pages et al. 2017).
For the Arabidopsis in planta seedling study, 737 proteins were captured, of which 300 were enriched in the CL compared to the nCL sample, with a false discovery rate of below 1%. This set of proteins was defined as “interactome RBPs.” The remainder of the proteins (437) did not meet these stringent criteria and were classified as “candidate RBPs,” which are of lower confidence but still likely to bind to RNA. Gene ontology (GO) analysis revealed that approximately 74% of the interactome RBPs and 46% of candidate RBPs had GO annotations linking their function to RNA, demonstrating that proteins associated with RNA have been preferentially captured (Reichel et al. 2016). Additionally, many of these proteins contained a known RNA-binding domain (RBD); this includes RNA-Recognition Motif (RRM) (80 proteins), K homology domain (12 proteins), DEAD-box helicase domain (12 proteins), pumilio repeats (six proteins), zinc finger types (19 proteins), or pentatricopeptide repeats (12 proteins). Well-known RBPs, such as COLD SHOCK PROTEINs (CSPs), GRPs and TUDOR-SN proteins were isolated, along with many housekeeping RBPs such as POLY(A) BINDING PROTEINs, splicing factors and proteins associated with gene silencing, including AGONAUTE family members (AGO1, AGO2, and AGO4) (Table 5.1). Additionally, a family of ten YTH (YT521B-Homology) domain-containing proteins were captured, also known as EVOLUTIONARY CONSERVED C-TERMINAL DOMAIN family proteins. These proteins are homologous to mammalian proteins that bind the most prevalent mRNA modification, adenosine 6 methylation (m6A), and are considered part of the epitranscriptome, with the ECT2 protein been shown to increase the stability of its target mRNAs (Wei et al. 2018). Additionally, ECT2 and ECT3 have now been demonstrated to recognize the m6A mRNA modifications in Arabidopsis, and functional analysis has shown that they control developmental timing and morphogenesis in Arabidopsis (Arribas-Hernandez et al. 2018). As these proteins are redundant with one another, it likely explains why they have not been previously identified with these phenotypes in mutant screens, an issue that is likely common among plant RBPs, as most belong to small to medium protein families (Arribas-Hernandez et al. 2018; Scutenaire et al. 2018).
5.5 The Use of mRNA-Interactome Capture to Address Key Areas of Plant Biology
Gene regulation at the translational level remains enigmatic. Given the ease at which mRNA levels are measured with RNA-seq, gene expression is predominantly quantified via transcriptomics, with the underlying assumption that transcript abundance acts as a proxy for protein levels. However, the plethora of post-transcriptional gene regulatory (PTGR) mechanisms means that the correlation between an mRNA’s abundance and its corresponding protein’s abundance is poor. In mammalian systems, mRNA levels only account for approximately 40% of the variability in protein levels, with translation efficiency the best predictor of protein expression (Schwanhausser et al. 2011). Moreover, although discrepancies between mRNA and protein levels are designated “translational control,” our understanding of the mechanisms behind such regulation is virtually non-existent. For instance, despite the intense focus on plant microRNAs (miRNAs), no unifying theme has as yet emerged of how they mediate repression of their targets via a translational mechanism (Axtell 2017).
Gene silencing. Firstly, ARGONAUTE (AGO) proteins, mediators of gene silencing, have been successfully cross-linked to mRNA (Reichel et al. 2016). For the in planta interactome, AGO1 and AGO2 were identified in the “interactome RBPs” (Table 5.1), and AGO4 in the candidate RBPs. In animals, miRNA target genes have been identified in numerous studies through cross-linking and immunoprecipitation of AGO complexes, followed by high-throughput sequencing of RNA (often referred to as HITS-CLIP or CLIP-seq) (Chi et al. 2009; Zisoulis et al. 2010). No such experiments have been achieved yet for plant systems, but this mRNA-interactome result implies this is possible, raising new opportunities to explore which mRNAs are being targeted by the different gene silencing effector proteins (pathways) in plants. Moreover, comparison of an AGO1 CLIP-seq to degradome data will give insights into silencing mechanisms by determining which targets are cleaved (present in degradome), compared to targets being translationally repressed (targets present in CLIP-seq data, but no degradome signature). Given the ongoing investigation into gene silencing, the mechanism by which it works, and the genes it targets, application of such methodology to plants would be highly significant to the field.
RBPs and selective translation during abiotic stress. Gene expression reprogramming during abiotic stress underpins a plant’s response and tolerance. This includes strong gene regulation at the translational level, which occurs during a wide range of stresses, including heat, cold, hypoxia (waterlogging), and water deficit (Merchante et al. 2017). Here, often two opposing translational regulatory events occur; a general decrease in global translation rates, coupled with increased translation efficiency of a select group of mRNAs required for stress survival (Merchante et al. 2017). This occurs as protein synthesis is potentially the most energy-expensive process in the cell; after translation, correct folding, modification, and transportation ensues (Roy and von Arnim 2013). Therefore, regulating what fraction of the transcriptome is translated is a key regulatory step enabling a rapid response to environmental perturbations while conserving energy (Matsuura et al. 2010). In the extreme cases of anaerobic or heat shock, the majority of cellular mRNA polyribosomes dissociate resulting in inhibition of general protein synthesis, while a small group of mRNAs required for stress survival are selectively translated (Minia et al. 2016). For anaerobiosis, enzymes involved in anaerobic metabolism are selectively translated, presumably to make enough ATP to survive the stress (Sachs et al. 1980). Thus, this post-transcriptional gene regulation not only couples a rapid response with energy conservation, but also focuses translation on a subset of proteins to maximize stress survival. Despite this hypoxic response being discovered over 35 years ago, the molecular mechanisms that underlie selective translation during hypoxia, or any other stress, remains unknown. These mechanisms are likely to be complex, but RBPs must be regarded as likely key players (Lorkovic 2009; Ambrosone et al. 2012; Marondedze et al. 2019). Identifying these regulatory RBPs will be central in understanding how these responses occur and may provide opportunities to manipulate them. Indeed, the RBP known as OLIGOURIDYLATE BINDING PROTEIN 1 (UBP1), identified from animal systems via homology, selectively sequesters non-stress-related mRNAs into stress granules during hypoxia to prevent their expression (Sorenson and Bailey-Serres 2014). Other RBPs that are known to play key roles in stress response have already been identified by mRNA-interactome capture during non-stress conditions (Reichel et al. 2016) (Table 1). This includes Tudor-SN proteins that are essential under stress where they stabilizes their targets (Frei dit Frey et al. 2010), and GRPs that are heavily involved in stress response (Czolpinska and Rurek 2018). Elucidating differential RNA-binders between control and stress conditions via mRNA-interactome capture will give the best chance of identifying RBPs that are key in coordinating abiotic stress responses.
The epitranscriptome. Relative to DNA methylation and epigenetics, the epitranscriptome has been poorly studied. This is despite there being over 100 known modifications, inferring there is huge regulatory potential via RNA modification. The most abundant modification is the methylation of adenosine, N6-methyladenosine (m6A) (Li and Mason 2014), and this modification is added by an RBP referred to as a “writer” (Fig. 5.3). These m6A modifications are essential for plants, as mutations in the writer, the RNA m6A methylase enzyme, are embryo lethal (Zhong et al. 2008). Recognition of m6A modified RNA is achieved by RBPs referred to as “readers” (Fig. 5.3). Their identity has been determined in animal cells as proteins containing an YTH domain, which binds to m6A modified mRNA facilitating their degradation (Wang et al. 2014), splicing (Xiao et al. 2016), or translation (Yang et al. 2018). In contrast to humans which only have five YTH domain-containing genes, Arabidopsis has 12 different YTH domain proteins (11 ECT proteins and CPSF30), ten of which were identified in the in planta mRNA-interactome (Reichel et al. 2016), confirming that these proteins are binding to mRNA in vivo. ECT2 and ECT3 have subsequently been demonstrated to regulate the branching of the trichomes, and together with ECT4, are required for leaf developmental timing and morphogenesis (Arribas-Hernandez et al. 2018; Scutenaire et al. 2018). ECT2 has been confirmed to stabilize the mRNAs related to trichome morphogenesis, and may also regulate the 3’UTR processing (Wei et al. 2018). Additionally, many ECT genes are strongly induced by stress, potentially linking the epitranscriptome to stress (Arribas-Hernandez et al. 2018; Scutenaire et al. 2018). However, the function of the majority of the m6A regulators in the plant kingdom is still unclear (Reichel et al. 2019) Therefore, it is likely that we are only beginning to understand the impact of the epitranscriptome, and how it controls genomic output during development and environmental response.
The mechanism of m6A function. Methylation to adenosine on the transcripts is mediated by writers [e.g., METTL3 (methyltransferase like 3) and METTL14 in mammal, MTA (mRNA adenosine methylase) in plant], and can be demethylased by erasers [e.g., FTO (fat mass and obesity-associated gene) in mammal and AtALKBH10B in plant]. Then the m6A is directly interacted by readers (YTH proteins) which lead the transcripts to different processes
5.6 Conclusions
Plant mRNA-interactomes will open up many new avenues of research that will likely elucidate post-transcriptional gene regulatory mechanisms not previously considered. Full development and exploitation of the methodology will serve as an exhaustive resource for the plant biology community, enabling researchers working on other plant (crop) species to adapt the methodology that has been pioneered in Arabidopsis. We believe interactome capture will be of great interest to the plant scientific community; as has the development of next-generation sequencing revolutionized the field of transcriptomics resulting in an intense focus on sRNA biology, we anticipate that enabling the global, unbiased analysis of the interactome will facilitate such a focus for plant RBPs.
References
Ambrosone A, Costa A, Leone A, Grillo S (2012) Beyond transcription: RNA-binding proteins as emerging regulators of plant response to environmental constraints. Plant Sci 182:12–18. https://doi.org/10.1016/j.plantsci.2011.02.004
Arribas-Hernandez L, Bressendorff S, Hansen MH, Poulsen C, Erdmann S, Brodersen P (2018) An m(6)A-YTH module controls developmental timing and morphogenesis in Arabidopsis. Plant Cell 30(5):952–967. https://doi.org/10.1105/tpc.17.00833
Axtell MJ (2017) Lost in translation? microRNAs at the rough ER. Trends Plant Sci 22(4):273–274. https://doi.org/10.1016/j.tplants.2017.03.002
Bach-Pages M, Castello A, Preston GM (2017) Plant RNA interactome capture: revealing the plant RBPome. Trends Plant Sci 22(6):449–451. https://doi.org/10.1016/j.tplants.2017.04.006
Baltz AG, Munschauer M, Schwanhausser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, Wyler E, Bonneau R, Selbach M, Dieterich C, Landthaler M (2012) The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell 46(5):674–690. https://doi.org/10.1016/j.molcel.2012.05.021
Bazin J, Romero N, Rigo R, Charon C, Blein T, Ariel F, Crespi M (2018) Nuclear speckle RNA binding proteins remodel alternative splicing and the non-coding Arabidopsis transcriptome to regulate a cross-talk between auxin and immune responses. Front Plant Sci 9:1209. https://doi.org/10.3389/fpls.2018.01209
Beckmann BM, Horos R, Fischer B, Castello A, Eichelbaum K, Alleaume AM, Schwarzl T, Curk T, Foehr S, Huber W, Krijgsveld J, Hentze MW (2015) The RNA-binding proteomes from yeast to man harbour conserved enigmRBPs. Nat Commun 6:10127. https://doi.org/10.1038/ncomms10127
Briggs WR (2016) Plant Biology: Seedling Emergence through Soil. Curr Biol 26(2):R68–R70. https://doi.org/10.1016/j.cub.2015.12.003
Butter F, Scheibe M, Morl M, Mann M (2009) Unbiased RNA-protein interaction screen by quantitative proteomics. Proc Natl Acad Sci USA 106(26):10626–10631. https://doi.org/10.1073/pnas.0812099106
Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, Krijgsveld J, Hentze MW (2012) Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149(6):1393–1406. https://doi.org/10.1016/j.cell.2012.04.031
Chantarachot T, Bailey-Serres J (2018) Polysomes, stress granules, and processing bodies: a dynamic triumvirate controlling cytoplasmic mRNA fate and function. Plant Physiol 176(1):254–269. https://doi.org/10.1104/pp.17.01468
Chi SW, Zang JB, Mele A, Darnell RB (2009) Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460(7254):479–486. https://doi.org/10.1038/nature08170
Czolpinska M, Rurek M (2018) Plant glycine-rich proteins in stress response: an emerging, still prospective story. Front Plant Sci 9:302. https://doi.org/10.3389/fpls.2018.00302
Floris M, Mahgoub H, Lanet E, Robaglia C, Menand B (2009) Post-transcriptional regulation of gene expression in plants during abiotic stress. Int J Mol Sci 10(7):3168–3185. https://doi.org/10.3390/ijms10073168
Frei dit Frey N, Muller P, Jammes F, Kizis D, Leung J, Perrot-Rechenmann C, Bianchi MW (2010) The RNA binding protein Tudor-SN is essential for stress tolerance and stabilizes levels of stress-responsive mRNAs encoding secreted proteins in Arabidopsis. Plant Cell 22(5):1575–1591. https://doi.org/10.1105/tpc.109.070680
Hentze MW, Castello A, Schwarzl T, Preiss T (2018) A brave new world of RNA-binding proteins. Nat Rev Mol Cell Biol 19(5):327–341. https://doi.org/10.1038/nrm.2017.130
Hicks GR (2001) Pathways in plant biology. Plant Physiol 127(3):704–706
Koster T, Marondedze C, Meyer K, Staiger D (2017) RNA-binding proteins revisited—the emerging Arabidopsis mRNA interactome. Trends Plant Sci 22(6):512–526. https://doi.org/10.1016/j.tplants.2017.03.009
Kwon SC, Yi H, Eichelbaum K, Fohr S, Fischer B, You KT, Castello A, Krijgsveld J, Hentze MW, Kim VN (2013) The RNA-binding protein repertoire of embryonic stem cells. Nat Struct Mol Biol 20(9):1122–1130. https://doi.org/10.1038/nsmb.2638
Lee K, Kang H (2016) Emerging roles of RNA-binding proteins in plant growth, development, and stress responses. Mol Cells 39(3):179–185. https://doi.org/10.14348/molcells.2016.2359
Lee S, Lee HJ, Jung JH, Park CM (2015) The Arabidopsis thaliana RNA-binding protein FCA regulates thermotolerance by modulating the detoxification of reactive oxygen species. New Phytol 205(2):555–569. https://doi.org/10.1111/nph.13079
Li J, Reichel M, Li Y, Millar AA (2014) The functional scope of plant microRNA-mediated silencing. Trends Plant Sci 19(12):750–756. https://doi.org/10.1016/j.tplants.2014.08.006
Li S, Mason CE (2014) The pivotal regulatory landscape of RNA modifications. Annu Rev Genomics Hum Genet 15:127–150. https://doi.org/10.1146/annurev-genom-090413-025405
Lim MH, Kim J, Kim YS, Chung KS, Seo YH, Lee I, Kim J, Hong CB, Kim HJ, Park CM (2004) A new Arabidopsis gene, FLK, encodes an RNA binding protein with K homology motifs and regulates flowering time via FLOWERING LOCUS C. Plant Cell 16(3):731–740. https://doi.org/10.1105/tpc.019331
Lorkovic ZJ (2009) Role of plant RNA-binding proteins in development, stress response and genome organization. Trends Plant Sci 14(4):229–236. https://doi.org/10.1016/j.tplants.2009.01.007
Maldonado-Bonilla LD (2014) Composition and function of P bodies in Arabidopsis thaliana. Front Plant Sci 5:201. https://doi.org/10.3389/fpls.2014.00201
Marondedze C, Thomas L, Gehring C, Lilley KS (2019) Changes in the Arabidopsis RNA-binding proteome reveal novel stress response mechanisms. BMC Plant Biol 19(1):139. https://doi.org/10.1186/s12870-019-1750-x
Marondedze C, Thomas L, Serrano NL, Lilley KS, Gehring C (2016) The RNA-binding protein repertoire of Arabidopsis thaliana. Sci Rep 6:29766. https://doi.org/10.1038/srep29766
Matia-Gonzalez AM, Laing EE, Gerber AP (2015) Conserved mRNA-binding proteomes in eukaryotic organisms. Nat Struct Mol Biol 22(12):1027–1033. https://doi.org/10.1038/nsmb.3128
Matsuura H, Ishibashi Y, Shinmyo A, Kanaya S, Kato K (2010) Genome-wide analyses of early translational responses to elevated temperature and high salinity in Arabidopsis thaliana. Plant Cell Physiol 51(3):448–462. https://doi.org/10.1093/pcp/pcq010
Merchante C, Stepanova AN, Alonso JM (2017) Translation regulation in plants: an interesting past, an exciting present and a promising future. Plant J 90(4):628–653. https://doi.org/10.1111/tpj.13520
Minia I, Merce C, Terrao M, Clayton C (2016) Translation regulation and RNA granule formation after heat shock of procyclic form trypanosoma brucei: many heat-induced mRNAs are also increased during differentiation to mammalian-infective forms. PLoS Negl Trop Dis 10(9):e0004982. https://doi.org/10.1371/journal.pntd.0004982
Obernosterer G, Leuschner PJ, Alenius M, Martinez J (2006) Post-transcriptional regulation of microRNA expression. RNA 12(7):1161–1167. https://doi.org/10.1261/rna.2322506
Reichel M, Koster T, Staiger D (2019) Marking RNA: m6A writers, readers, and functions in Arabidopsis. J Mol Cell Biol 11(10):899–910. https://doi.org/10.1093/jmcb/mjz085
Reichel M, Liao Y, Rettel M, Ragan C, Evers M, Alleaume AM, Horos R, Hentze MW, Preiss T, Millar AA (2016) In planta determination of the mRNA-binding proteome of Arabidopsis etiolated seedlings. Plant Cell 28(10):2435–2452. https://doi.org/10.1105/tpc.16.00562
Roy B, von Arnim AG (2013) Translational regulation of cytoplasmic mRNAs. Arabidopsis Book 11:e0165. https://doi.org/10.1199/tab.0165
Sachs MM, Freeling M, Okimoto R (1980) The anaerobic proteins of maize. Cell 20(3):761–767. https://doi.org/10.1016/0092-8674(80)90322-0
Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M (2011) Global quantification of mammalian gene expression control. Nature 473(7347):337–342. https://doi.org/10.1038/nature10098
Schwartz S (2016) Cracking the epitranscriptome. RNA 22(2):169–174. https://doi.org/10.1261/rna.054502.115
Scutenaire J, Deragon JM, Jean V, Benhamed M, Raynaud C, Favory JJ, Merret R, Bousquet-Antonelli C (2018) The YTH domain protein ECT2 is an m(6)A reader required for normal trichome branching in Arabidopsis. Plant Cell 30(5):986–1005. https://doi.org/10.1105/tpc.17.00854
Shen L, Liang Z, Wong CE, Yu H (2019) Messenger RNA modifications in plants. Trends Plant Sci 24(4):328–341. https://doi.org/10.1016/j.tplants.2019.01.005
Silverman IM, Li F, Gregory BD (2013) Genomic era analyses of RNA secondary structure and RNA-binding proteins reveal their significance to post-transcriptional regulation in plants. Plant Sci 205–206:55–62. https://doi.org/10.1016/j.plantsci.2013.01.009
Sorenson R, Bailey-Serres J (2014) Selective mRNA sequestration by OLIGOURIDYLATE-BINDING PROTEIN 1 contributes to translational control during hypoxia in Arabidopsis. Proc Natl Acad Sci U S A 111(6):2373–2378. https://doi.org/10.1073/pnas.1314851111
Staiger D, Green R (2011) RNA-based regulation in the plant circadian clock. Trends Plant Sci 16(10):517–523. https://doi.org/10.1016/j.tplants.2011.06.002
Tsvetanova NG, Klass DM, Salzman J, Brown PO (2010) Proteome-wide search reveals unexpected RNA-binding proteins in Saccharomyces cerevisiae. PLoS One 5(9). https://doi.org/10.1371/journal.pone.0012671
Vandivier LE, Gregory BD (2018) New insights into the plant epitranscriptome. J Exp Bot 69(20):4659–4665. https://doi.org/10.1093/jxb/ery262
Wang X, Lu Z, Gomez A, Hon G, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, Ren B, Pan T, He C (2014) N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505:117–120.
Wei LH, Song P, Wang Y, Lu Z, Tang Q, Yu Q, Xiao Y, Zhang X, Duan HC, Jia G (2018) The m(6)A reader ECT2 controls trichome morphology by affecting mRNA stability in Arabidopsis. Plant Cell 30(5):968–985. https://doi.org/10.1105/tpc.17.00934
Wessels HH, Imami K, Baltz AG, Kolinski M, Beldovskaya A, Selbach M, Small S, Ohler U, Landthaler M (2016) The mRNA-bound proteome of the early fly embryo. Genome Res 26(7):1000–1009. https://doi.org/10.1101/gr.200386.115
Wheeler EC, Van Nostrand EL, Yeo GW (2018) Advances and challenges in the detection of transcriptome-wide protein-RNA interactions. Wiley Interdiscip Rev RNA 9(1). https://doi.org/10.1002/wrna.1436
Wu Z, Zhu D, Lin X, Miao J, Gu L, Deng X, Yang Q, Sun K, Zhu D, Cao X, Tsuge T, Dean C, Aoyama T, Gu H, Qu LJ (2016) RNA binding proteins RZ-1B and RZ-1C play critical roles in regulating pre-mRNA splicing and gene expression during development in Arabidopsis. Plant Cell 28(1):55–73. https://doi.org/10.1105/tpc.15.00949
Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, Wang X, Ma HL, Huang CM, Yang Y, Huang N, Jiang GB, Wang HL, Zhou Q, Wang XJ, Zhao YL, Yang YG (2016) Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol Cell 61(4):507–519. https://doi.org/10.1016/j.molcel.2016.01.012
Yang Y, Hsu PJ, Chen YS, Yang YG (2018) Dynamic transcriptomic m(6)A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res 28(6):616–624. https://doi.org/10.1038/s41422-018-0040-8
Zhang Z, Boonen K, Ferrari P, Schoofs L, Janssens E, van Noort V, Rolland F, Geuten K (2016) UV crosslinked mRNA-binding proteins captured from leaf mesophyll protoplasts. Plant Methods 12:42. https://doi.org/10.1186/s13007-016-0142-6
Zhong S, Li H, Bodi Z, Button J, Vespa L, Herzog M, Fray RG (2008) MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell 20(5):1278–1288. https://doi.org/10.1105/tpc.108.058883
Zisoulis DG, Lovci MT, Wilbert ML, Hutt KR, Liang TY, Pasquinelli AE, Yeo GW (2010) Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat Struct Mol Biol 17(2):173–179. https://doi.org/10.1038/nsmb.1745
Funding
This project was supported from funds from the Research School of Biology (RSB), ANU.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Wang, N., Millar, A.A. (2021). Use of mRNA-Interactome Capture for Generating Novel Insights into Plant RNA Biology. In: Tang, G., Teotia, S., Tang, X., Singh, D. (eds) RNA-Based Technologies for Functional Genomics in Plants. Concepts and Strategies in Plant Sciences. Springer, Cham. https://doi.org/10.1007/978-3-030-64994-4_5
Download citation
DOI: https://doi.org/10.1007/978-3-030-64994-4_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-64993-7
Online ISBN: 978-3-030-64994-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)