Abstract
Segmentation is one of the key enabling technologies in medical image analysis with a great variety of methods proposed (Histace et al. 2009; Zhang et al. 2010, 2013; Matuszewski et al. 2011). Methods based on deep learning, with the features learned directly from data rather than handcrafted, showed significant improvement in the quality of the segmentation including the analysis of colonoscopy images.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
1 Motivation
Segmentation is one of the key enabling technologies in medical image analysis with a great variety of methods proposed (Histace et al. 2009; Zhang et al. 2010, 2013; Matuszewski et al. 2011). Methods based on deep learning, with the features learned directly from data rather than handcrafted, showed significant improvement in the quality of the segmentation including the analysis of colonoscopy images. The recent advances in fully convolutional networks and in particular the dilation convolution and squeeze-and-excitation unit have inspired the two architectures proposed in this chapter. More specifically, the first proposed network can be seen as a specific example of an encoder-decoder architecture with the multi-channel encoder providing features operating at different spatial resolution of the input image. The dilation kernels in each channel facilitate a compromise between the capacity of the network and the size of the receptive fields. The second network combines the base U-net architecture with squeeze-and-excitation units, to take better advantage of the extracted features. Overall, the key motivation behind the proposed solutions is to strike a balance between network capacity and the size of the receptive field. The objective is to use a possibly large receptive field, without significantly increasing the network capacity. This way, the network is less prone to overfitting, particularly when trained on relatively small data sets with somewhat limited dimensionality of the underlying segmentation problem.
2 Introduction of the Base Structure
The fully convolutional network (FCN) architecture was the first type of end-to-end network to be successfully used for semantic image segmentation based on deep learning (Long et al. 2015). FCN can process images of any size and obtain a full-size segmentation result without the need for additional pre-processing. The structure of an FCN can be divided into two parts, an encoder and a decoder. The former is used to extract low resolution, high-level features from the input image. The latter fuses these features and converts them into low-resolution segmentation results, then restores their size by means of up-sampling and cropping layers. The loss in the backward direction is determined by processing the full-scale segmentation result and ground truth. Then, the errors are propagated to each hidden layer that needs to be trained. This method not only simplifies the steps of image segmentation, but also is more accurate than the traditional methods.
Encoder
The encoder can be any CNN whose fully connected layer has been removed. It can be one of the existing CNN architectures or a custom built one. When designing an FCN, the choice of the encoder is usually determined by the complexity of the images and the performance of the hardware, with the goal of avoiding unnecessary calculations. It should be noted that when using an FCN model, the final feature map is required to be of a certain size, otherwise, some smaller objects of interest could be missed. Therefore, the rate of down-sampling should be chosen based on the characteristics of the specific segmentation problem.
Decoder
The decoder consists of a pixel classifier, an up-sampling layer and a cropping layer. The pixel classifier is used to classify the pixels in the feature maps one by one. It is a convolutional layer rather than a fully connected layer. This is because the number of outputs of a fully connected layer is fixed, making it impossible to process images of different sizes. For general pixel classifiers, a 1 \(\times \) 1 convolution kernel is used to fuse the feature maps and generate low-resolution segmentation results. Large convolution kernels can also be used, but additional padding is needed to ensure that the size of the feature maps is not significantly reduced.
To reduce the loss of segmentation details caused by down-sampling, feature maps of different resolutions can be extracted from convolution layers at different depths in the encoder, and corresponding pixel classifiers can then be designed separately. After that, the results can be fused through up-sampling, creating the so-called skip structure (Long et al. 2015). As the fusion method, a direct addition could be used, or build structures are stacked and fused by using a 1 \(\times \) 1 convolution kernel. The up-sampling layer is a critical hidden layer in an FCN, and it serves as the basis for end-to-end training. The up-sampling layer is essentially a special convolutional layer controlled by a set of three parameters, namely, the size of the convolution kernel, the stride and the kernel weighs. The stride size corresponds to the scale of previous down-sampling operations. The kernel weights often are selected to correspond to bilinear interpolation, subsequently in some cases, they are adjusted during the network training. Finally, the up-sampled results are cropped to match the size of the ground truth.
The original architecture of FCN inducted three sub-architectures, namely, FCN32s, FCN16s and FCN8s (Long et al. 2015) (Fig. 8.1). In all three, VGG16 was used as an encoder. The difference in the sub-architecture is that the sizes of the skip structures are different. FCN8s performs classification after FC7, pool4 and pool3 and generates a corresponding segmentation result for each case. VGG16 contains a total of 5 down-sampling layers, and each output is reduced by a factor of 2. Therefore, the results of the last two down-sampling layers are required to be up-sampled and then merged with the result of the pool3 classifier to obtain the final segmentation result. Since the output of pool3 is only 1/8th the size of the original image, the fusion result needs to be enlarged by a factor of 8 (hence the name FCN8s). In FCN16s classifiers are included only after pool4 and FC7, and their outputs are fused. The output of pool4 is 1/16th the size of the original image, so the segmentation result needs to be enlarged by a factor of 16. FCN32s uses only the output of FC7 as the segmentation result.
One of the important concepts in the design of FCN is the receptive field, which refers to the size of the area in the input image to which each unit in the output layer corresponds. A larger receptive field allows the output to contain more global features, which helps to improve the accuracy of the segmentation results. However, reducing the stride for down-sampling to improve local spatial segmentation accuracy will make the receptive field smaller.
In the example shown in Fig. 8.2, when the pooling stride is 2 (Fig. 8.2a), the receptive field is 6 (a single output unit is connected to 6 input units). When the pooling stride becomes 1 (Fig. 8.2b), the output size is increased to 7, but only 4 input units are connected to a single output unit. In this case, there is no doubt that the output size is improved, but the amount of information contained in each output unit is reduced.
Regular convolution (a)–(c) and atrous convolution (d). a Regular convolution, with pooling stride 2 and 1 \(\times \) 3 kernel. b Regular convolution, with pooling stride 1 and 1 \(\times \) 3 kernel. c Regular convolution, with pooling stride 1 and 1 \(\times \) 5 kernel. d Atrous convolution, with pooling stride 1, 1 \(\times \) 5 kernel and dilation 2; kernel size is 5 but only 3 weights are trainable
Using a larger convolution kernel can solve this problem, but it will increase the computational cost and the number of parameters to be estimated, as shown in Fig. 8.2c. To solve this problem more efficiently, the so-called dilated convolution (Yu and Koltun 2016) (also known as atrous convolution) was proposed. The underlying idea is to increase the size of the convolution kernel by adding 0s between the weights without changing the number of weights, as shown in Fig. 8.2d. The definition of the dilated convolution is given as
where y[i] is the output, x[i] is an 1-D input signal, w[k] represents the weight in a kernel. The parameter r is called dilation rate and it controls the stride between each weight in an atrous kernel.
3 Methodology Explanation
Based on analysis of existing machine learning and polyp image segmentation techniques, a novel hybrid deep learning segmentation method (Guo and Matuszewski 2019, 2020; Guo 2019) has been proposed for both SD and HD GIANA polyp segmentation problems. The method consists of two fully convolutional networks. The first network named “Dilated ResFCN” takes advantage of dilation convolution layers (Chen et al. 2017) to increase receptive fields, and therefore, makes the algorithm aware of various multi-scale relationships between the polyps and their surroundings. The second network “SE-Unet” is designed to segment small and flat polyps which have been missed by the Dilated ResFCN, however, overall it tends to produce more false positive pixels.
3.1 Dilated ResFCN
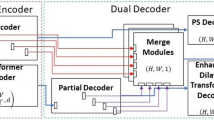
The architecture of the first proposed network, Dilated ResFCN, is shown in Fig. 8.3. This architecture is inspired by Long et al. (2015), Chen et al. (2017), and the Global Convolutional Network (Peng et al. 2017). The proposed FCN consists of three sub-networks performing specific tasks: feature extraction, multi-resolution classification and fusion. The feature extraction sub-network is based on the ResNet-50 model (He et al. 2016).
The classification sub-network consists of four parallel paths. Each such path includes a dilation convolutional layer, which is used to increase the receptive field without increasing computational complexity. The larger receptive fields are needed to access contextual information about polyp neighbourhood areas. The dilation rate is determined by the number of active kernel weights (Guo and Matuszewski 2020). The dilation rates for sub-nets connected to Res5-Res2 are 2, 4, 8, 16 and the corresponding kernel sizes are 5, 9, 17 and 33. The fusion sub-network corresponds to the deconvolution layers of the FCN model. The segmentation results from each classification sub-network are up-sampled and fused by a bilinear interpolation.
The feature extraction sub-network weights are initialized by a publicly available ResNet-50 (Deep residual networks 2017). The convolutional layers in the four parallel classification paths are initialized by the Xavier method (Glorot and Bengio 2010).
3.2 SE-Unet
The Dilated ResFCN focuses on learning features using a larger receptive field. However, smaller polyps may be ignored by networks with a large receptive field, this is because smaller polyps may not excite lower resolution feature maps strongly enough. To solve this problem, the SE-Unet network has been proposed. It has been designed specifically for the detection and segmentation of small polyps missed by the Dilated ResFCN network.
The SE-Unet, shown in Fig. 8.4, is based on the classic U-net architecture (Ronneberger et al. 2015). In the encoder, the original architecture of U-net is replaced by the VGG16 network (Simonyan and Zisserman 2015), and a modified atrous spatial pyramid pooling (ASPP) module (Chen et al. 2017). The ASPP module has four kernels, with respective sizes of \(1 \times 1, 3 \times 3, 5 \times 5, 7 \times 7\). The last two are dilated kernels, with corresponding dilation rates of 2 and 4. These two modifications have the purpose of improving the performance of image feature extraction. The decoder can be regarded as a mirrored VGG16 network where the down-sampling layers are replaced by up-sampling layers. The original U-net fuses different level feature maps after each up-sampling layer to provide more features to the pixel level classifier. In SE-Unet, this is further reinforced with the squeeze-and-excitation (SE) module (Hu et al. 2018) added between the up-sampling and fusion layer. The SE module aims to assign higher weights to the high importance features and lower weights to minor importance features and therefore the network is expected to focus more on important features in the decoder. The parameter “r in the SE module is set to 16. The SE-Unet training consists of two stages. In the first stage, the SE modules are removed from SE-Unet. In the second stage, the SE modules are added and the whole network is re-trained. Both Dilated ResFCN and SE-Unet are trained using the Adam algorithm.
3.3 Training-Time Data Augmentation
One of the key advantages of deep learning is that features are learned directly from data rather than been designed/handcrafted. Therefore, in many cases, these features inherently better represent complex data. However, for this to work, the training data should adequately represent data variabilities, including size, pose, shape, texture, colour, etc. From that perspective, the training data available for the GIANA polyp segmentation challenge was rather small. Therefore, available data were heavily augmented with random rotation, translation, scale changes as well as colour and contrast jitter. In total, after augmentation, the training dataset consisted of more than 90,000 images.
3.4 Test-Time Data Augmentation
Since the convolutional neural networks are not inherently rotation invariant, a possible option to improve segmentation results is to perform the data augmentation during the test time (Simonyan and Zisserman 2015). For this, rotated versions of the original test image are also presented to the network and the corresponding outputs, after restoring to the original image reference frame, are averaged to take advantage of the network generalization capabilities. The adopted test-time augmentation process is explained in Fig. 8.5. The implemented augmentation uses 24 images derived from the original test image rotated in 15\(^{\circ }\) intervals.
Visualization of the test-time data augmentation. The image on the left shows an input test image. Images in the middle represent rotated, in 15\(^{\circ }\) intervals, versions of the original image; the corresponding results of the binary segmentation in the rotated image reference frame; and the results after restoration to the original image reference frame. The image on the right shows final segmentation results, superimposed on the original image, with (in red) and without (in blue) test-time augmentation
4 Example of Results
This section presents validation results of the proposed methods using GIANA SD training images, with a standard 4-fold cross-validation scheme. Frames extracted from the same video are always in the same validation sub-set, i.e. they are not used for training and validation at the same time. The three main configurations have been tested: Dilated ResFCN, SE-Unet and the hybrid method. The hybrid method uses Dilated ResFCN as the base network and switches to the SE-Unet when the base network does not detect any polyps. These three architectures have been compared against the FCN8s and simplified version of the Dilated ResFCN, called here ResFCN. The ResFCN has the same architecture like the one shown in Fig. 8.3, but without the dilation kernels. This network has been included to demonstrate the significance of the dilation kernels on the segmentation performance.
Figure 8.6 shows a sample of segmentation results for typical small, medium and large polyps. The polyp occurrence confidence maps show that FCN8s can determine the approximate position of a polyp, but it generates a large number of false positives and false negatives with diffused network response and irregular shape of the segmented polyps. For the large polyp, FCN8s generate many strong responses outside of the polyp. For the Dilated ResFCN, the confidence maps are more accurate than those of the other methods with a clear boundary defining polyp edges.
Typical results obtained for the SD images using FCN8s, ResFCN, Dilated ResFCN and SE-Unet networks (Guo 2019). For each image: the left column shows the polyp occurrence confidence maps with the red colour representing the high confidence and blue colour representing the low confidence of a polyp presence; the right column shows the original images with superimposed red and blue contours representing the ground truth and segmentation results, respectively
Table 8.1 presents the corresponding results for the Dice Index, Precision, Recall and the Hausdorff distance metrics. It can be seen that overall all best results are provided by the hybrid method closely followed by the Dilated ResFCN, indeed the latter outperforms the former with respect to the Hausdorff distance.
Figure 8.7 shows a more detailed representation of the mean Dice index results achieved by the tested methods. For each method, the results are shown as histograms of a number of polyps calculated as a function of the Dice index. It can be observed that Dilated ResFCN segments the largest number of polyps within the top Dice index range (i.e. with the Dice index between 0.9 and 1). The Hybrid method produces very similar results within the top range, but improving (reducing the number of polyps) within the bottom range (i.e. with the Dice index between 0 and 0.4), due to improvement in segmentation of small polyps.
References
Chen, L.-C., Papandreou, G., Kokkinos, I., Murphy, K., & Yuille, A. L. (2017). DeepLab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. IEEE Transactions on Pattern Analysis and Machine Intelligence, 40(4), 834–848.
Deep residual networks. https://github.com/KaimingHe/deep-residual-networks. Retrieved 03 August 2017.
Glorot, X., & Bengio, Y. (2010). Understanding the difficulty of training deep feedforward neural networks. In Proceedings of the Thirteenth International Conference on Artificial Intelligence and Statistics (pp. 249–256).
Guo, Y., & Matuszewski, B. J. (2020). Polyp segmentation with fully convolutional deep dilation neural network. In Zheng, Y., Williams, B., Chen, K. (Eds.) Medical Image Understanding and Analysis. MIUA (2019). Communications in Computer and Information Science, vol 1065. Springer, Cham
Guo, Y. B. (2019). Polyp segmentation in colonoscopy images with convolutional neural networks. Ph.D. thesis, University of Central Lancashire.
Guo, Y. B., & Matuszewski, B. (2019). GIANA polyp segmentation with fully convolutional dilation neural networks. In Proceedings of the 14th International Joint Conference on Computer Vision, Imaging and Computer Graphics Theory and Applications (pp. 632–641). SCITEPRESS-Science and Technology Publications.
He, K., Zhang, X., Ren, S., & Sun, J. (2016). Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (pp. 770–778).
Histace, A., Matuszewski, B. J., & Zhang, Y. (2009). Segmentation of myocardial boundaries in tagged cardiac MRI using active contours: A gradient-based approach integrating texture analysis. International Journal of Biomedical Imaging, 2009, 983794:1–983794:8.
Hu, J., Shen, L., & Sun, G. (2018). Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (pp. 7132–7141).
Long, J., Shelhamer, E., & Darrell, T. (2015). Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (pp. 3431–3440).
Matuszewski, B. J., Murphy, M. F., Burton, D. R., Marchant, T. E., Moore, C. J., Histace, A., et al. (2011). Segmentation of cellular structures in actin tagged fluorescence confocal microscopy images. In 2011 18th IEEE International Conference on Image Processing (pp. 3081–3084).
Peng, C., Zhang, X., Yu, G., Luo, G., & Sun, J. (2017). Large kernel matters–improve semantic segmentation by global convolutional network. In IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA (pp. 1743–1751). IEEE.
Ronneberger, O., Fischer, P., & Brox, T. (2015). U-Net: Convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention (pp. 234–241). Springer.
Simonyan, K., & Zisserman, A. (2015). Very deep convolutional networks for large-scale image recognition. In 3rd International Conference on Learning Representations, (ICLR), San Diego, CA, USA. May 7–9arXiv:1409.1556.
Yu, F., & Koltun, V. (2016). Multi-scale context aggregation by dilated convolutions. In /textit4th International Conference on Learning Representations, (ICLR), San Juan, Puerto Rico. May 2–4. arXiv:1511.07122.
Zhang, Y., Matuszewski, B. J., Histace, A., Precioso, F., Kilgallon, J., & Moore, C. (2010). Boundary delineation in prostate imaging using active contour segmentation method with interactively defined object regions. In International Workshop on Prostate Cancer Imaging (pp. 131–142). Springer.
Zhang, Y., Matuszewski, B. J., Histace, A., & Precioso, F. (2013). Statistical model of shape moments with active contour evolution for shape detection and segmentation. Journal of Mathematical Imaging and Vision, 47(1–2), 35–47.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Guo, Y., Matuszewski, B.J. (2021). Dilated ResFCN and SE-Unet for Polyp Segmentation. In: Bernal, J., Histace, A. (eds) Computer-Aided Analysis of Gastrointestinal Videos. Springer, Cham. https://doi.org/10.1007/978-3-030-64340-9_8
Download citation
DOI: https://doi.org/10.1007/978-3-030-64340-9_8
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-64339-3
Online ISBN: 978-3-030-64340-9
eBook Packages: Computer ScienceComputer Science (R0)