Abstract
Labeled data is scarce in clinical practice, and labeling 3D medical data is time-consuming. The study aims to develop a deep learning network with a few labeled data and investigate its segmentation performance of lumbosacral structures on thin-layer computed tomography (CT). In this work, semi-cGAN and fewshot-GAN were developed for automatic segmentation of nerve, bone, and disc, compared with 3D U-Net. For evaluation, dice score and average symmetric surface distance are used to assess the segmentation performance of lumbosacral structures. Another dataset from SpineWeb was also included to test the generalization ability of the two trained networks. Research results show that the segmentation performance of semi-cGAN and fewshot-GAN is slightly superior to 3D U-Net for automatic segmenting lumbosacral structures on thin-layer CT with fewer labeled data.
H. Liu and H. Xiao—These two authors equally contribute to the study.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Three-dimensional (3D) radiographic image is beneficial for preoperative assessment of spinal surgery [1]. Computed tomography (CT) excels at demonstrating bony structures, while magnetic resonance imaging (MRI) is good at differentiating soft tissues. However, the combined evaluation of lumbosacral structures on multimodal radiographic images for surgical planning is time-consuming and costly. Thin-layer CT is an ideal candidate for constructing a 3D model with bony structures and soft tissue for surgical planning. However, manual segmentation is cumbersome for 3D reconstruction.
Deep learning is a promising technology to achieve the automatic segmentation of medical images. However, labeled data is scarce in clinical practice, and labeling 3D medical data is time-consuming. In 2015, a concise neural network named U-Net was developed for the semantic segmentation of two-dimensional biomedical images with a few training data [2]. One year later, the same group developed 3D U-Net for volumetric segmentation [3]. U-Net or 3D U-Net has been validated as a successful supervised neural network for segmenting medical images [4,5,6,7,8]. Generative adversarial networks (GAN) can integrate unlabeled data to achieve precise segmentation [9].
The study aims to develop a conditioned GAN for 3D images (semi-cGAN and fewshot-GAN) with a few labeled data and investigate its segmentation performance of lumbosacral structures on thin-layer CT.
2 Methods
The local institutional ethical committee approved this retrospective study before data extraction. All the CT data was obtained from Shanghai Tenth People’s Hospital, and algorithms were mainly developed and tested using Tensorflow on a personal computer (GPU: An Nvidia Tesla M40 GPU with 11180 MB memory CPU: An Intel(R) Xeon(R) CPU E5-2682 v4 @ 2.50 GHz CPU with eight cores).
2.1 Manual Annotations
Thirty-one cases of thin-layer CT were manually segmented with Slicer 4.8. Lumbosacral nerves, bones and nerves were meticulously segmented and labeled (see Fig. 1). These manual annotations were regarded as the ground truth.
2.2 Data Preprocessing
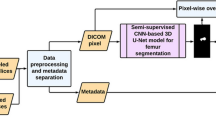
All thin-layer CT were preprocessed using the following steps: resampling, cropping, and intensity normalization.
-
1.
Resampling: we standardized the voxel size of the original CT and manually labeled 3D masked images into 1 mm × 0.5 mm × 0.5 mm nearest-neighbor interpolation.
-
2.
Cropping: we cropped the resampled CT and manually-labeled 3D masked images into 32 × 64 × 64 patches, which were used as input data.
-
3.
Intensity normalization: For more accurate semantic segmentation, the average brightness and contrast fluctuations of CT images of different samples should have a degree of consistency. For this reason, the CT images in the dataset are standardized so that the pixel values of the CT images have zero mean and unit variance. The scale and bias were obtained by statistic computing from the training dataset, and they were used for whitening during all phases, including training, validation and testing.
-
4.
Data augmentation: we conducted data augmentation with the following methods:
-
Adding a small amount of white noise to patch which will be input to the neural network
-
Performing vertical flipping and horizontal flipping at a certain probability
-
Voxel size is randomly disturbed within the range of ±0.2 mm to introduce a degree of size variation
-
Performing random rotation around the Height axis with angles within ±10°.
-
2.3 Network Architecture
As Fig. 2 shown, generator G of fewshot-GAN is a volume generator proposed by Wu et al. [10] The input for generator G is a noise vector z and the output of generator G is a fake patch. In association with a labeled patch and an unlabeled patch, they are input into Discriminator D. Discriminator D is a modified 3D U-Net with leaky ReLUs, weight-normalization (rather than batch normalization) and average pooling (instead of Max pooling). Discriminator D serves as a segmentation network to gain corresponding masks. Features of generated images match features in an intermediate layer of discriminator by feature matching loss.
Generator G of semi-cGAN is a 3D U-Net. The input for generator G is CT patch x (labeled CT or unlabeled CT), and the output of generator G is the corresponding masks of automatic segmentation. G(x) has four channels (4 classes: nerve, bone, disc, background), which is the same to manually labeled masks y (unlabeled CT has no corresponding masks y). G(x) or y concatenate to x, which generates (x, G(x)) or (x, y) and is input to Discriminator D. Discriminator D is a 3D convolutional neural network (3D-CNN). The last two layers are full connection layer, and the last layer generates the probability of (x, G(x)) or (x, y) as a real sample.
2.4 Material
Thirty-one labeled cases and 54 unlabeled cases of thin-layer CT were included for semi-supervised semantic segmentation. The ratio of training labeled cases and validation labeled cases for both semi-cGAN and fewshot-GAN is 20:6. The remaining 5 cases serve as a testing function. As a comparison, the same data used for training 3D U-Net except unlabeled cases. Besides, another ten labeled cases from SpineWeb [11] were also included to test the generalization ability of the two trained networks.
2.5 Training
Fewshot-GAN.
This method consists of a generator and a discriminator.
Generator Loss.
We adopted the Feature Matching (FM) loss for the generator. This method is aimed at matching the expected value of features \( f\left( x \right) \) in an intermediate layer of the discriminator.
Discriminator Loss.
The discriminator loss function is the sum of three losses incorporating a weighted loss for labeled images.
The loss for labeled images is weighted cross-entropy loss. Different classes have unique values. In terms of unlabeled data, we constraint the output to correspond to one of the K classes of real data. Similarly, to calculate the loss for fake images, we impose each pixel of an input image to be predicted as fake.
where \( {\text{F}}\left( {\text{x}} \right) = \mathop \sum \limits_{k = 0}^{N} { \exp }\left( {a_{k} \left( x \right)} \right) \), \( a_{k} \left( x \right) \) is the activation value at the last layer of the discriminator of voxel point \( x \) in channel k; \( a_{l} \left( x \right) \) is the activation value at the last layer of voxel point \( x \) in the ground truth channel; \( w\left( x \right) \) is the cross entropy weight of voxel x.
Semi-cGAN.
This method also includes a generator and a discriminator.
Generator Loss.
The generator is aimed to fool the discriminator and to be near the ground truth in a weighted softmax cross-entropy sense. We made two main modifications to the standard Generator Loss. The first one is including unlabeled data to improve the generalization, and the other is adding a weighted softmax cross-entropy loss for labeled data. Therefore, the final formula is as follow:
Among which:
In the above function, \( x \in \varOmega \) is voxel point of labeled CT; \( a_{k} \left( x \right) \) is the activation value at the last layer before softmax of voxel point \( x \) in channel k; \( a_{l} \left( x \right) \) is the activation value at the last layer before softmax of voxel point \( x \) in the ground truth channel; \( w\left( x \right) \) is the cross-entropy weight of voxel x.
Discriminator Loss.
The loss function of the discriminator is as follows:
Among which:
Training Process.
We adopted the Adam algorithm to optimize generator loss and discriminator loss. During training, the batch size of labeled data and unlabeled data is 4. It took 48.01 h to complete the training with 100 thousand iterations for semi-cGAN and 70.52 h for fewshot-GAN with the same number of iterations. To compared with state-of-art algorithms, we also trained a 3D U-Net with 20 labeled cases. During the training process, we preserve the best model according to the highest dice score on the testing dataset.
2.6 Testing
A total of 5 labeled cases were used to test the trained semi-cGAN, fewshot-GAN and 3D U-Net. Another 10 cases from SpineWeb were also included to test the generalization ability of the two trained networks. During testing, any case in the test dataset will be selected and undergo standard processing, and then a sliding window of size 32 × 64 × 64 is used to traverse the case with stride = (20 × 40 × 40) to obtain the patch \( x_{i} \). The patch \( x_{i} \) will be input to the trained model \( M \), and then the model will generate the corresponding probability mask \( y_{i} \). Finally, the automatically segmented mask \( S \) will be obtained with the above-mentioned combined algorithm combining \( y_{i} \) based on the location of \( x_{i} \) on \( Y \).
3 Results
3.1 Own Dataset
Final segmentation performances on five cases are compared to each other (see Fig. 3) Confusion matrixes are also calculated to display a summary of prediction results (see Fig. 4). The Dice score of semi-cGAN and fewshot-GAN is 91.5117% and 90.0431%, respectively, compared with 89.8137% of 3D U-Net (see Table 1). The average ASD of semi-cGAN and fewshot-GAN is 1.2726 and 1.5188, compared with 1.4747 of 3D U-Net (see Table 2).
3.2 Cross Dataset
Similarly, ten cases from the cross dataset are applied to assess segmentation performances (see Fig. 5), and confusion matrixes will be shown in Fig. 6. The Dice score of semi-cGAN and fewshot-GAN is 89.0644% and 88.3881% respectively, compared with 88.6382% of 3D U-Net (see Table 3). The average ASD of semi-cGAN and fewshot-GAN is 0.6869 and 0.6954, compared with 1.3109 of 3D U-Net (see Table 4).
4 Discussion
Based on enormous raw data, models trained by deep learning network are able to extract pertinent features and accomplish complex tasks. Therefore, relevant clinical data is essential for this promising method to train a prospective model. [12] However, specific task entails corresponding data, and public datasets are usually hard to meet particular research requirements. Besides, labeled data is scarce in clinical practice, and labeling 3D medical data is time-consuming. Apart from these problems, manual segmentation is not always consistent or entirely precise due to inter-expert variance. [13] It is also time-consuming to examine the accuracy of collected data. All in all, accurate and satisfying clinical data is rare, which is a bottleneck of this technology.
Transfer learning is one of the possible methods to remedy the scarce data problem. It applies knowledge learned from other similar data into target domain, which means little target data is required. However, this technology is not a simple task to implement, as the potential similarities between source data distribution and target data distribution need to be identified. This issue may result in negative transfer, which means reduced performance. [14] Because of little publicly related clinical data, this method can not perfectly tackle our problems.
Unsupervised learning is another sensible approach, making its inferences based on unlabeled data. However, unsupervised learning demands more training unlabeled data than supervised learning. It is also time-consuming to obtain relevant data and clean them. Meanwhile, unsupervised learning may show a poor performance in segmentation with lower accuracy because of unlabeled data. Also, given the subjectivity of unsupervised learning and its lack of gold standard, this method can not fix the existing challenges.
Semi-supervised learning subtly utilizes a combination of labeled data and unlabeled data. In this case, few labeled data and relatively more unlabeled data are required. One semi-supervised method proposed by Wenjia Bai et al. [15] has applied to MR Image, which proved the potentiality of semi-supervised learning. Other successful applications like Li et al. [16] and Papandreou et al. [17] showed a promising prospect. Therefore, we adopted semi-supervised learning to tackle the scarcity of clinical data.
U-NET [2] is a well-known convolutional neural network applied to medical image segmentation. This network creatively combined contraction path and symmetric expanding path, which eventually facilitated both segmentation speed and performance. Besides, it has been widely applied to various specific issues and proved its excellent segmentation ability. GANs are a semi-supervised method which has obtained great achievement in medical image processing despite lack of adequate labeled data. By synthesizing realistic images to alleviate the data limitation, this efficient network has an outstanding performance in segmentation. Other than this merit, GANs are highly effective in extracting meaningful features from training data compared with conventional approaches [18].
Both our methods try to combine the benefits of U-NET and GANs. In the fewshot-GAN, the discriminator is a modified 3D U-NET, and the generator is a volume generator. Semi-cGAN consists of a generator based on 3D U-NET and a 3D CNN discriminator. 3D U-NET acts as a different role in two models, and one of our goals is trying to explore which combination has better performance. Recently, there is also a novel U-NET-GAN proposed by Dong et al. However, in their proposed model, three-label-based segmentations are included and trained each of them separately, which might need massive computational powers. Our trained models are more concise with promising results, which automatically segment multiple lumbosacral structures.
5 Conclusions
Segmentation performance of semi-cGAN or fewshot-GAN is slightly superior to 3D U-Net for automatic segmenting lumbosacral structures on thin-layer CT with fewer labeled data.
References
Kochanski, R.B., Lombardi, J.M., Laratta, J.L., Lehman, R.A., O’Toole, J.E.: Image-guided navigation and robotics in spine surgery. Neurosurgery 84, 1179–1189 (2019)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, William M., Frangi, Alejandro F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Çiçek, Ö., Abdulkadir, A., Lienkamp, S.S., Brox, T., Ronneberger, O.: 3D U-Net: learning dense volumetric segmentation from sparse annotation. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) MICCAI 2016. LNCS, vol. 9901, pp. 424–432. Springer, Cham (2016). https://doi.org/10.1007/978-3-319-46723-8_49
Wang, C., Macgillivray, T., Macnaught, G., Yang, G., Newby, D.: A two-stage 3D Unet framework for multi-class segmentation on full resolution image (2018)
Funke, J., et al.: Large scale image segmentation with structured loss based deep learning for connectome reconstruction. IEEE Trans. Pattern Anal. Mach. Intell. 41, 1669–1680 (2018)
Norman, B., Pedoia, V., Majumdar, S.: Use of 2D U-Net convolutional neural networks for automated cartilage and meniscus segmentation of knee MR imaging data to determine relaxometry and morphometry. Radiology 288, 177–185 (2018)
Weston, A.D., et al.: Automated abdominal segmentation of CT scans for body composition analysis using deep learning. Radiology 290, 669–679 (2019)
Huang, Q., Sun, J., Ding, H., Wang, X., Wang, G.: Robust liver vessel extraction using 3D U-Net with variant dice loss function. Comput. Biol. Med. 101, 153–162 (2018)
Dong, X., et al.: Automatic multi-organ segmentation in thorax CT images using U-Net-GAN. Med. Phys. 46, 2157–2168 (2019)
Wu, J.J., Zhang, C.K., Xue, T.F., Freeman, W.T., Tenenbaum, J.B.: Learning a probabilistic latent space of object shapes via 3D generative-adversarial modeling. In: Advances in Neural Information Processing Systems, vol. 29 (2016)
Ibragimov, B., Likar, B., Pernuš, F., Vrtovec, T.: Shape representation for efficient landmark-based segmentation in 3-D. IEEE Trans. Med. Imaging 33, 861–874 (2014)
Chen, D., et al.: Deep learning and alternative learning strategies for retrospective real-world clinical data. Npj Digit. Med. 2, 43 (2019)
Retter, F., Plant, C., Burgeth, B., Botella, G., Schlossbauer, T., Meyer-Bäse, A.: Computer-aided diagnosis for diagnostically challenging breast lesions in DCE-MRI based on image registration and integration of morphologic and dynamic characteristics. EURASIP J. Adv. Sig. Process. 2013, 157 (2013)
Pan, S.J., Yang, Q.A.: A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 22, 1345–1359 (2010)
Bai, W., et al.: Semi-supervised learning for network-based cardiac MR image segmentation. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S. (eds.) MICCAI 2017. LNCS, vol. 10434, pp. 253–260. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-66185-8_29
Li, X., Yu, L., Chen, H., Fu, C.-W., Heng, P.-A.: Transformation consistent self-ensembling model for semi-supervised medical image segmentation (2019)
Papandreou, G., Chen, L.C., Murphy, K.P., Yuille, A.L.: Weakly- and semi-supervised learning of a deep convolutional network for semantic image segmentation. In: IEEE International Conference on Computer Vision, pp. 1742–1750 (2015)
Kazeminia, S., et al.: GANs for medical image analysis (2018)
Acknowledgement
We would like to thanks William M. Wells in Brigham and Women’s Hospital, Harvard Medical School for the guidance of this project.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this paper
Cite this paper
Liu, H. et al. (2020). Semi-supervised Semantic Segmentation of Multiple Lumbosacral Structures on CT. In: Cai, Y., Wang, L., Audette, M., Zheng, G., Li, S. (eds) Computational Methods and Clinical Applications for Spine Imaging. CSI 2019. Lecture Notes in Computer Science(), vol 11963. Springer, Cham. https://doi.org/10.1007/978-3-030-39752-4_5
Download citation
DOI: https://doi.org/10.1007/978-3-030-39752-4_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-39751-7
Online ISBN: 978-3-030-39752-4
eBook Packages: Computer ScienceComputer Science (R0)