Abstract
Chest radiography is the most common examination for a radiologist. This demands correct and immediate diagnosis of a patient’s thorax to avoid life threatening diseases. Not only certified radiologists are hard to find, stress, fatigue and experience contribute to the quality of an examination. It is ideal that a chest X-ray can be interpreted by an automated deep learning algorithm. In this paper, we proposed a stage-wise model that is founded on a ResNet-50 based deep convolutional neural networks architecture to detect the presence and absence of twelve thorax diseases. This novel model has incorporated various recent techniques such as transfer learning, fine tuning, fit one cycle function and discriminative learning rates. The experiments were performed on 10% of the largest collection of chest X-rays to date, the MIMIC-CXR dataset. The model was trained for eight epochs using a subset of the available multi-view chest X-rays. The absolute labelling performance has achieved an encouraging average AUC of 0.779.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Currently, analyzing chest x-rays depends on the availability of professional radiologist. In some regions, access to such radiologists is limited [1]. Additionally, clinicians in emergency department and intensive care unit needs fast and accurate interpretations of medical images [2]. Globally, chest X-ray is the most common radiological examinations that required correct and fast analysis [1]. An automated and precise system that can flag potentially life-threatening diseases could allow care providers to handle emergency cases efficiently.
However, interpreting X-rays to detect thoracic diseases is still a challenging job. This is due to the highly diverse appearance of lesion areas on chest X-rays. Unlike the traditional computer-aided detection (CAD) systems that interpret medical images automatically to offer an objective diagnosis that assist radiologists [3], deep learning is able to learn useful features which are beyond the limit of radiology detection [4]. For example, deep learning has been applied on Mammography to discriminate breast cancer with microcalcification [5], on ultrasound to differentiate breast lesions and on CT lung scans to classify pulmonary [6]. Researchers [5, 6] showed a significant performance boost by their deep learning based models over the conventional CAD systems.
In this study, we present a supervised deep learning model using convolutional neural network to detect twelve thoracic diseases by reading a given chest X-ray. Residual network (ResNet-50) [7] is the backbone network for our model because it has clearly shown its outstanding performance on computer vision.
2 Related Work
Recently, several deep learning models that classify thorax diseases have been proposed as a result of the public release of a collection of large datasets namely Indiana Chest X-Ray [8], ChestX-ray14 [9], CheXpert [1], PadChest [10] and MIMIC-CXR [9]. For example, CheXNet [11], text-image embedding network (TieNet) [12], attention guided convolutional neural network (AG-CNN) [13], learning to diagnose from scratch network [14] classify thorax diseases from frontal chest x-rays using ChestX-ray14. However, [15] suggest that using lateral view enhances the performance for certain prediction tasks such as pleural effusion. Further, [2] proposed DualNet model to prove that simultaneous processing of both frontal and lateral chest X-ray inputs results in better classification performance. Unlike ChestX-ray14 [9] that only presents the frontal view of chest X-ray, MIMIC-CXR is a multi-view version of radiographs dataset. DualNet employed a limited released version of the MIMIC-CXR dataset to automate reading of frontal and lateral chest X-rays.
Convolutional neural network (CNN) which is a supervised deep learning model is the most common used deep learning technique for thoracic disease classification. It has also seen the widest variety in architectures, such as AlexNet [16], VGG-16 [17], DenseNet [18] and ResNet [7]. CNN-based classification model [19], for instance, adopt VGG-16 and ResNet-101 to classify X-rays based on nine chest diseases like emphysema and bronchitis. ResNet won the ImageNet large scale visual recognition challenge (ILSVRC) in 2015 with 3.6% top five error rate, which enables automated image classification to beat human brains with 5% error for the first time. ResNet is a feed forward network that contains several basic residual blocks, refer to Fig. 1, to handle the vanishing gradients [20] and the degradation issue.
Consistent with recent proposed CNN models on automated chest x-rays classification [2, 11, 19], we focus on training CNN models to detect 12 common thoracic diseases namely enlarged cardiomediastinum, cardiomegaly, airspace opacity, lung lesion, edema, consolidation, pneumonia, atelectasis, pneumothorax, pleural effusion, pleural other and fracture (Fig. 2). Unique from past works, we propose a novel stage wise training approach to observe the model’s performance and hence reduce training time and increase accuracy. We adopt a combination of recent techniques on multi-view chest X-rays including ResNet-50, transfer learning, fine tuning, fit one cycle function [21] and discriminative learning rates [22].
3 Proposed Model
3.1 Structure Overview
The task of detecting thorax diseases in chest x-rays is divided into 12 sub-tasks, where each task considers the presence and absence of a specific disease. Among the proposed variations of ResNet layers (i.e. 34, 50, 101, 152 and 1202), we adopt the popular ResNet-50 network which consists of 49 convolution layers and ends with 1 fully connected layer. Equation 1 defines the last output of residual unit \( x_{l} \), where \( F\left( {x_{l - 1} } \right) \) is the generated output after performing the convolution operations, batch normalization and activation function on \( x_{l - 1} \). Importantly, we use cyclical learning rates to enhance performance by decreasing the number of epochs required to accomplish the accuracy threshold. For each binary label problem, ResNet is used as the baseline CNN architecture in three main training stages (Fig. 3).
3.2 Training Stages
In the first stage, the pre-trained ResNet-50 with the default fastai [23] hyperparameter values is trained for three epochs. That is setting all layers to frozen, excluding the final dense layer and examining each X-ray three times. In other words, the first stage embraces transfer learning approach to train faster with a model that is already trained to recognize 1000 categories of things in ImageNet. At the end of stage-1, model’s weights were saved.
In the second stage, the whole model is trained again for one epoch by unfreezing the layers and calling the fit-one-cycle method. The objective of this stage is to observe the model’s performance to reduce training time and increase accuracy. If the AUC is decreased at the end of this training stage, stage-1 weights are re-loaded.
In the third stage, the whole model is trained again for four epochs using the optimal learning rate finder. The learning rate is set by default to about 1e−3 at stage-1 and changed manually to a range of lower learning rates (1e−6 to 1e−4) at stage 3. Figure 4 illustrates the plotted learning rate after the first and second stages of the model, where the red dots on the graphs indicate the steepest gradient point. Using different learning rates for each layer at this stage is in line with the discriminate fine-tuning technique to tune each layer with various learning rates. In this case, the model’s parameters \( \theta \) and the learning rate \( \eta \) are split into \( \left\{ {\theta^{1} , \ldots , \theta^{L} } \right\} \) at time step \( t \) and \( \left\{ {\eta^{1} , \ldots , \eta^{L} } \right\} \) respectively, where \( L \) is the number of layers. This updated version of the regular stochastic gradient descent (SGD) with discriminative fine-tuning is defined in Eq. 2, where \( \nabla_{{\theta^{l} }} J \) is the gradient of the model’s objective function.
4 Experiment
4.1 Dataset
MIMIC-CXR is the largest dataset of chest x-rays to date that consist of 371,920 images and relevant 227,943 studies derived from Beth Israel Deaconess Center [24]. Images are annotated with 14 labels, which overlap with those of the popular ChestX-ray14 dataset and match the co-released CheXpert dataset. Labels are extracted from the associated free-text radiology reports using the CheXpert labeler tool. The training labels for each observation are 0 for negative, 1 for positive, −1 for uncertain and blank for unknown. We organized a subset of 10% of the MIMIC-CXR v1.0.0 into training and validation sets that contains 33,195 and 3,688 images respectively. The validation set was selected at random. During training, the uncertain and unknown labels were ignored. Table 1 shows the positive and negative cases for each observation.
4.2 Pre-processing
Prior to models training, we employ several augmentation strategies (refer to Table 2) as data augmentation is a critical step of deep CNNs in medical imaging [25]. We crop each x-ray in both the training and validation sets to 224 by 224 pixels to reduces training time while maintaining robust model’s performance. For example, training the model to diagnose cardiomegaly using 299 by 299 pixels would increase training time without improving the AUC per epoch (refer to Table 3). We perform a horizontal flip only for each image in the training set, since vertical flips often do not reflect chest x-rays (i.e. an upside-down chest x-ray may not improve training). The maximum lighting of the image is set to 0.3 with applying probability of 0.5. Note that no vertical flips, rotations, zooms or wraps were done on the images. In addition, uncertain and unknown labels were dropped.
4.3 Training
The training algorithms were evaluated in twelve pathologies: enlarged cardiomediastinum, cardiomegaly, airspace opacity, lung lesion, edema, consolidation, pneumonia, atelectasis, pneumothorax, pleural effusion, pleural other and fracture. We used PyTorch software [26], fastai library, n1-highmem-8 (8 vCPUs, 52 GB memory) machine and 4 x NVIDIA Tesla P4 GPUs. This is in accordance with [27] work that demonstrate how time-per-epoch for the ResNet-50 architecture scale much better when training it on multiple GPUs. Table 4 records the time per epoch for training ResNet-50 based model to detect cardiomegaly using different number of GPUs, where parallel training on 4 GPUs reduce training time by around 20 min.
4.4 Results
Table 5 shows the Area Under Curve (AUC) results of each pathology computed on the validation set for each of the eight training epochs. It can be seen that detection performance for each pathology fluctuate over epochs. For individual training epochs, the eighth unfrozen epoch accomplish a higher average AUC (0.777), compared to the first (0.670), second (0.704), third (0.718), forth (0.711), fifth (0.753), sixth (0.765) and seventh (0.776). Compared with stage-1 (epoch 1–3) and stage 2 (epoch 4), stage 3 (epoch 5–8) results in larger AUC values for all pathologies. This difference is likely due to the discriminative learning rates at the third stage of training.
Table 6 compares the per pathology AUC results between our proposed model and DualNet architecture using MIMIC-CXR dataset. We employed 10% of the dataset using all available frontal and lateral views of the chest X-rays. DualNet, on the other hand, considered a combination of posteroanterior (PA) and lateral as well as a composite of anteroposterior (AP) and lateral. In 5 out of 7 overlap pathologies, our model performs better than both DualNet models. Overall, it can be seen that average AUC is higher for our multi-view classifiers (0.779), compared to both PA-lateral (0.722) and AP-lateral (0.677).
4.5 Analysis
In DualNet model, chest X-rays labels were extracted from the associated radiology reports using an open source tool developed by the National Institute of Health (NIH), the NegBio labelerFootnote 1 [28]. This tool was used to annotate the popular ChestX-ray14 dataset. In contrast, our model follows the public released labels by [24] that utilized a different open source tool created by Stanford machine learning group, the CheXpert labelerFootnote 2. Although the labeling algorithm of CheXpert is built upon the work of NegBio, it achieves a higher F1 score. Hence, our model is trained on a better annotated chest X-rays than DualNet. Interestingly, we reach improved results over those achieved by DualNet using small image sizes 224 by 224 pixels instead of 512 by 512 pixels.
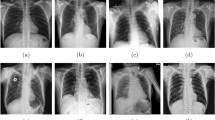
Nevertheless MIMIC-CXR is the largest open source X-ray images to date, the class labels in the training set are noisy because they were mined by natural language processing tool, rather than by experienced radiologist. Figure 5 visualizes the most incorrect predicted X-rays by our model with heatmaps, using the activations of the wrongly predicted class. In addition, the positive-negative subsets ratio was highly imbalanced in the enlarged cardiomediastinum, lung lesion, consolidation, pneumothorax, pleural other and fracture sets (Table 1). Yet, our model’s AUC for each of these pathologies is above 0.7 (Table 6).
5 Conclusion
In this paper, ResNet-50 CNN based stage wise models have been proposed to detect twelve thorax diseases on 10% of the largest chest X-rays dataset to date, the MIMIC-CXR dataset. The absolute labelling performance with an average weighted AUC of 0.779 is encouraging, since we used only a subset of the available chest X-rays. In future work, we plan to improve our CNN model performance through utilizing common image-based classification techniques, in particular data augmentation. Importantly, we will incorporate useful information from the free-text radiology reports such as patient’s history and clinical records to accurately recognize the presence and absence of thorax diseases.
References
Irvin, J., et al.: CheXpert: a large chest radiograph dataset with uncertainty labels and expert comparison. arXiv preprint arXiv:1901.07031 (2019)
Rubin, J., Sanghavi, D., Zhao, C., Lee, K., Qadir, A., Xu-Wilson, M.: Large scale automated reading of frontal and lateral chest x-rays using dual convolutional neural networks. arXiv preprint arXiv:1804.07839 (2018)
van Ginneken, B., Schaefer-Prokop, C.M., Prokop, M.: Computer-aided diagnosis: how to move from the laboratory to the clinic. Radiology 261, 719–732 (2011)
Kohli, M., Prevedello, L.M., Filice, R.W., Geis, J.R.: Implementing machine learning in radiology practice and research. Am. J. Roentgenol. 208, 754–760 (2017)
Wang, J., Yang, X., Cai, H., Tan, W., Jin, C., Li, L.: Discrimination of breast cancer with microcalcifications on mammography by deep learning. Sci. Rep. 6, 27327 (2016)
Cheng, J.-Z., et al.: Computer-aided diagnosis with deep learning architecture: applications to breast lesions in US images and pulmonary nodules in CT scans. Sci. Rep. 6, 24454 (2016)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016)
Demner-Fushman, D., et al.: Preparing a collection of radiology examinations for distribution and retrieval. J. Am. Med. Inform. Assoc. 23, 304–310 (2015)
Wang, X., Peng, Y., Lu, L., Lu, Z., Bagheri, M., Summers, R.M.: Chestx-ray8: hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 3462–3471. IEEE (2017)
Bustos, A., Pertusa, A., Salinas, J.-M., de la Iglesia-Vayá, M.: PadChest: a large chest x-ray image dataset with multi-label annotated reports. arXiv preprint arXiv:1901.07441 (2019)
Rajpurkar, P., et al.: Chexnet: radiologist-level pneumonia detection on chest x-rays with deep learning. arXiv preprint arXiv:1711.05225 (2017)
Wang, X., Peng, Y., Lu, L., Lu, Z., Summers, R.M.: TieNet: Text-image embedding network for common thorax disease classification and reporting in chest x-rays. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 9049–9058 (2018)
Guan, Q., Huang, Y., Zhong, Z., Zheng, Z., Zheng, L., Yang, Y.: Diagnose like a radiologist: attention guided convolutional neural network for thorax disease classification. arXiv preprint arXiv:1801.09927 (2018)
Yao, L., Poblenz, E., Dagunts, D., Covington, B., Bernard, D., Lyman, K.: Learning to diagnose from scratch by exploiting dependencies among labels. arXiv preprint arXiv:1710.10501 (2017)
Bertrand, H., Hashir, M., Cohen, J.P.: Do lateral views help automated chest X-ray predictions? arXiv preprint arXiv:1904.08534 (2019)
Krizhevsky, A., Sutskever, I., Hinton, G.E.: ImageNet classification with deep convolutional neural networks. In: Advances in Neural Information Processing Systems, pp. 1097–1105 (2012)
Simonyan, K., Zisserman, A.: Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556 (2014)
Huang, G., Liu, Z., Van Der Maaten, L., Weinberger, K.Q.: Densely connected convolutional networks. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 2261–2269. IEEE (2017)
Dong, Y., Pan, Y., Zhang, J., Xu, W.: Learning to read chest X-ray images from 16000+ examples using CNN. In: Proceedings of the Second IEEE/ACM International Conference on Connected Health: Applications, Systems and Engineering Technologies, pp. 51–57. IEEE Press (2017)
Glorot, X., Bengio, Y.: Understanding the difficulty of training deep feedforward neural networks. In: Proceedings of the Thirteenth International Conference on Artificial Intelligence and Statistics, pp. 249–256 (2010)
Smith, L.N.: A disciplined approach to neural network hyper-parameters: part 1–learning rate, batch size, momentum, and weight decay. arXiv preprint arXiv:1803.09820 (2018)
Howard, J., Ruder, S.: Universal language model fine-tuning for text classification. arXiv preprint arXiv:1801.06146 (2018)
Johnson, A.E., et al.: MIMIC-CXR: a large publicly available database of labeled chest radiographs. arXiv preprint arXiv:1901.07042 (2019)
Hussain, Z., Gimenez, F., Yi, D., Rubin, D.: Differential data augmentation techniques for medical imaging classification tasks. In: AMIA Annual Symposium Proceedings, p. 979. American Medical Informatics Association (2017)
Ketkar, N.: Introduction to PyTorch. Deep Learning with Python: A Hands-on Introduction, pp. 195–208. Apress, Berkeley (2017)
Coleman, C., et al.: Analysis of dawnbench, a time-to-accuracy machine learning performance benchmark. arXiv preprint arXiv:1806.01427 (2018)
Peng, Y., Wang, X., Lu, L., Bagheri, M., Summers, R., Lu, Z.: NegBio: a high-performance tool for negation and uncertainty detection in radiology reports. In: AMIA Summits on Translational Science Proceedings 2017, p. 188 (2018)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Monshi, M.M.A., Poon, J., Chung, V. (2019). Convolutional Neural Network to Detect Thorax Diseases from Multi-view Chest X-Rays. In: Gedeon, T., Wong, K., Lee, M. (eds) Neural Information Processing. ICONIP 2019. Communications in Computer and Information Science, vol 1142. Springer, Cham. https://doi.org/10.1007/978-3-030-36808-1_17
Download citation
DOI: https://doi.org/10.1007/978-3-030-36808-1_17
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-36807-4
Online ISBN: 978-3-030-36808-1
eBook Packages: Computer ScienceComputer Science (R0)