Abstract
Human society has coevolved with our invisible, microbial neighbors. From their use in the processing of food and drink to the manufacture of commercial products and therapeutics, bacteria, yeast, and fungi are invaluable tools to many aspects of our existence. Despite our long history together, researchers are only recently beginning to understand the complexities of these relationships and how the microbial world can fully be exploited. In the last 50 years alone, researchers have shown that bacteria can be used to manufacture drugs to aid in the treatment of medical disorders such as diabetes, that environmental microbes can be used to clean up chemical spills and disasters, and that the microbial communities that reside within our bodies are capable of influencing our mental and physical health. With this greater understanding comes new avenues of research to utilize these microbes to advance human society. Here we discuss efforts to utilize both native and engineered membrane vesicles shed by bacteria and their potential applications to several areas of biotechnology. We highlight the use of bacterial membrane vesicles in vaccine research and as emerging therapeutics as well as exploring their potential commercial applications and benefits.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
10.1 Introduction
Society has a long and storied history with the microbial world. While most often thought of in the context of historical events of plague, disease, and infection, microbes such as bacteria, yeast, and fungi have benefited and advanced human society in countless ways. Microbes have become integral components in food and beverage production, in the manufacture of therapeutics, and as platforms for synthesis of commercial enzymes and other products. In recent years, the ability to engineer biology has made exponential leaps forward allowing researchers to explore alternatives to using natural, “wild-type” bacteria. Advances in DNA sequencing, the accessibility of molecular engineering technologies, improvements in omics’ instrumentation and analysis tools, and many other factors have opened new avenues of biological engineering and synthetic biology.
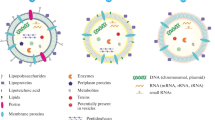
Since they were first observed under scanning electron microscopy, the vesicles shed by microbes have gone by many names: membrane vesicles, exosomes, outer membrane vesicles, microvesicles, and others (Knox et al. 1967). Often the nomenclature follows the characteristics of the parental organisms such as membrane vesicle (MV) for those particles originating from Gram-positive bacteria and outer membrane vesicle (OMV) for the vesicles produced by Gram-negative bacteria, though this is not always the case. Regardless of the nomenclature and the parental organism, these vesicles are shed from the outermost membrane of their parental microorganism (Fig. 10.1). The biomolecules’ composition is highly variable, as are many of their morphological properties. Most Gram-negative OMVs typically range in size from 50 to 300 nm, though particles of 85–125 nm are by far the most prevalent (Deatherage et al. 2009; Schwechheimer et al. 2013). In comparison, the MVs of Gram-positive bacteria often exhibit a bimodal distribution of particle sizes with a significant number of vesicles in the 10–30 nm range (Dean et al. 2019; Grande et al. 2017). The exosomes of eukaryotic cells show a distribution similar to the OMVs, while microvesicles of mammalian cells have been reported from 100 nm to 1 μm in range (Raposo and Stoorvogel 2013; Tricarico et al. 2017; Zomer et al. 2010). Typically, vesicles are released from the parental bacteria into the surrounding environment, however, researchers have observed some elongated tubules extending from bacterial surfaces that appear as chains of vesicles as seen with some Myxococcus species (Berleman et al. 2014).
Beyond simple morphological differences, the composition of biological vesicles is highly variable, as would be expected. Proteomic analysis of several bacterial species of both Gram-positive and Gram-negative classification has shown that membrane proteins are often the most prevalent components of the structures (Kroniger et al. 2018; Lee et al. 2007). However, in these studies, researchers have also shown that in many instances the protein composition of the vesicles does not always mimic that of the parental microbe. Early studies of Escherichia coli OMVs by Lee et al. highlighted this phenomenon, reporting that highly abundant membrane and periplasmic proteins were often not found at detectable levels in OMV preparations (Lee et al. 2007, 2008). Similar observations have been made with regard to toxins, small molecules, and peptides that appear to be enriched in vesicles by some microbes, suggesting a cellular mechanism or signal that facilitates loading (Dean et al. 2019; Jan 2017). Cell membranes are very heterogeneous by nature with specific proteins and lipids distributed across the exterior of the cell (Barak and Muchova 2013). For most microbes, the outermost membrane is a dynamic surface that not only changes through the various stages of the cells’ life cycle but also responds to environmental cues and conditions and changes in response to these stimuli. While there are many mechanisms proposed for vesicle biogenesis, evidence is mounting that formation and release may be a well-controlled cellular event in some bacteria (Elhenawy et al. 2016; Roier et al. 2016). The bacterial membrane vesicles therefore take on a variety of functions, serving as a defense mechanism from phages and antimicrobial peptides, facilitating toxin delivery in pathogenesis, and shuttling DNA and RNA between species allowing for horizontal gene transfer, among others.
In the subsequent sections, we will explore the potential biotechnology applications of both natural and engineered bacterial membrane vesicles. At present, membrane vesicles have seen their greatest acceptance in the area of vaccine development employing the OMVs of Gram-negative bacteria. Unlike many traditional strategies that require inactivation of the virus or toxin, OMV-based vaccines allow antigens to be maintained in their native state and delivered simultaneously with immunostimulatory epitopes that serve as the adjuvant. OMV-based vaccines offer an all-in-one vehicle that is relatively low cost and easy to manufacture. Beyond vaccines, the complexity of protein and other biomolecule composition opens numerous avenues of engineering bacterial membrane vesicles for specific applications that may have commercial value. Researchers have demonstrated that through careful design of molecular systems, specific proteins can be targeted to both the interior and exterior of these biological nanoparticles. These efforts have allowed for the development of OMV-based assays for antigen detection, enzyme-based systems for bioremediation, and tools for imaging cell–cell interactions. These early success in both health-related and commercial applications are foundational to ongoing efforts that may lead to new therapeutics, engineered probiotics, or tools for regulating complex microbial communities.
10.2 OMV Use in Vaccines
The design of vaccines is based on the concept that protective immunity against a given pathogen results from inducing the natural immune response against that pathogen without actually causing the associated disease. Therefore, the vaccine must resemble the pathogen enough to trigger the correct immune response, yet must not itself be infectious. Traditionally, this has been achieved through several strategies, including the use of attenuated pathogens (rendered nonpathogenic by various methods) or killed pathogens (inactivated by heat or chemical denaturation) (Zepp 2010). While numerous vaccines in use today are still produced by these strategies, there are drawbacks to both. Attenuated pathogens may potentially revert back to virulent forms or may pose a higher threat to individuals with compromised immune systems. On the other hand, killed pathogens may not stimulate the immune system enough to provide long-lasting protection, necessitating the use of adjuvants such as aluminum compounds that further stimulate the innate immune response (Coffman et al. 2010; Zepp 2010). Thus, modern vaccines are based on the use of carefully chosen antigens in combination with adjuvants to enhance the body’s response to the antigen (Fig. 10.2).
Current and emerging vaccine strategies. Current vaccines employ live and/or attenuated pathogens (upper left panel) or inactivated biomolecules such as proteins, carbohydrates, or peptides (lower left panel). DNA vaccines are based on isolated genetic sequences that encode an antigenic, neutralizing protein or peptide for a target pathogen. DNA is taken up by antigen-presenting cells, then transcribed and translated into a protein sequence that is presented on the cell surface to stimulate an immune response (middle panel). Membrane vesicles can be used in their natural form as they mimic the cell surface of pathogenic bacteria, thereby allowing the host immune system to respond to a wide range of antigenic features of the bacteria (upper right panel). Membrane vesicles can be engineered to contain recombinant proteins and peptides thus allowing for the development of vaccines for any number of bacterial or viral targets determined solely by the cargo within or on the surface of the vesicle itself (lower right panel)
OMVs possess a number of characteristics that make them ideal candidates as a vaccine platform. First, they are naturally produced by bacteria and have intrinsic immunostimulatory properties, as they contain species-specific antigens as well as pathogen-associated molecular patterns (PAMPs). Thus, OMVs induce both adaptive and innate immune responses, allowing them to function as antigen and adjuvant in one package (Ellis and Kuehn 2010; Kaparakis-Liaskos and Ferrero 2015). Second, OMVs are non-replicative and so do not need to be treated with inactivating agents, which preserves antigens and PAMPs in their native states. Third, while a number of nanoparticle-based vaccine delivery vehicles are in development today including OMVs, virus-like particles (VLPs), immunostimulatory complexes (ISCOMs), and inorganic nanoparticles (reviewed in Xiang et al. 2006), OMVs are uniquely suited to this purpose as their natural size range (20–250 nm) enables them to both drain freely into the lymph nodes to target immune cells residing there, as well as to be taken up by antigen-presenting cells such as dendritic cells (Fig. 10.3) (Gerritzen et al. 2017; Kulp and Kuehn 2010; Manolova et al. 2008). Finally, genetic engineering can be used in a number of ways to improve the production, immunostimulatory properties, and safety of OMV-based vaccines (discussed in detail below). As a result of their potential as a vaccine platform, a significant amount of research has been dedicated in recent decades to the engineering of OMV-based vaccines, including an FDA-approved vaccine against meningitis serogroup B.
Comparison of current vaccine platforms. Vaccine platforms vary in their size, methods of purification, and their mode of interaction with host immune cells. For each system, these properties have to be weighed to determine their feasibility in administration and commercialization. For example, the possibility of sterile filtrations simplifies the production design while the analysis of nanoparticle size distribution can increase the difficulty of standardizing the final product. Within this figure, the green box highlights the overall preferred size window for vaccine production. Used with permission from Gerritzen et al. Bioengineering bacterial outer membrane vesicles as vaccine platform. Biotechnology Advances, 2017. 35(5): p. 565–574
There are various factors to consider in the design and production of OMV-based vaccines. OMVs may be isolated directly from the target pathogen, though this often requires them to be extracted using detergents that remove certain toxic components from the OMV surface such as lipopolysaccharide (LPS) found on Gram-negative bacteria (Gnopo et al. 2017; van de Waterbeemd et al. 2010). Alternatively, mutant pathogen strains producing modified, less-toxic LPS can be used as the OMV source, or antigens can be heterologously expressed in engineered host species with modified LPS. In either case, this can be done in strains carrying mutations that increase vesicle formation to increase the OMV yield. Genetic engineering can also be used to introduce multiple antigens into a single OMV platform, thus increasing the strength or broadening the scope of the immune response to protect against additional strains of the pathogen. These considerations will be discussed in detail below in the context of the meningitis B vaccine as a primary case study, followed by several other examples of OMV-based vaccines against various diseases. This topic has also been extensively reviewed by other researchers and their work may be referred to for additional information (Gerritzen et al. 2017; Gnopo et al. 2017; Tan et al. 2018; van der Pol et al. 2015).
10.2.1 OMV Vaccines for Meningitis
To date, the development of OMV vaccines has seen the most attention for Meningitis type B (MenB), which remains the only disease for which they have been approved for human use. Neisseria meningitidis strains are classified into serogroups based on their capsular type, and effective vaccines containing a capsular polysaccharide coupled to a carrier protein have been developed against several serogroups. However, the serogroup B capsule bears structural similarity to a neural cell adhesion molecule in the human brain, which means that it is poorly immunogenic and also that its use in a vaccine has the potential to induce an autoimmune response (Finne et al. 1983; Rosenstein et al. 2001). Consequently, vaccine development for MenB has focused on other antigens, particularly the immunodominant outer membrane protein porin A (PorA) (Holst et al. 2009, 2013). As PorA is abundant in the outer membrane, it is naturally also present at high levels in OMVs isolated from N. meningitidis strains, making them an ideal candidate for vaccine development.
OMVs isolated from local serogroup B meningococcal strains proved effective as vaccines in Cuba, Norway, Chile, Brazil, and New Zealand, with the significant limitation that the immune response was strain-specific due to the high sequence variability of PorA (reviewed in Holst et al. 2009). To combat this limitation, an OMV vaccine was developed at the Netherlands Vaccine Institute that is produced from two genetically modified N. meningitidis strains that each express three PorA subtypes (Claassen et al. 1996; van der Ley et al. 1995). This hexavalent vaccine “HexaMen” was safe and effective in clinical trials and has been further improved to include a third trivalent OMV to provide coverage against the nine most frequently occurring subtypes of MenB in industrialized countries (de Kleijn et al. 2001; van den Dobbelsteen et al. 2007).
While OMV vaccines are safe and effective, their primary dependence on the PorA antigen necessitates that even the multivalent vaccines would still require periodic reformulation as the dominant subtypes of MenB change over time. Thus, work continued toward the identification of new, conserved antigens that would allow for the development of a “universal” vaccine for serogroup B meningococcal strains. The availability of whole genome sequencing technology gave rise to a new strategy of vaccine development termed “reverse vaccinology,” in which candidate antigens identified in silico are individually expressed in E. coli and tested for immunogenicity (Rappuoli 2000). This approach allowed researchers to identify a number of novel antigens that are conserved across multiple N. meningitidis strains (Pizza et al. 2000). A new vaccine was then developed that includes five of these novel antigens in the form of three recombinant proteins: a fragment of NadA, plus two fusion proteins consisting of NHBA–GNA1030 and GNA2091-fHbp (Giuliani et al. 2006). NadA is an adhesin, NHBA is a heparin binding protein, and fHbp is a lipoprotein that binds to human complement factor H; the functions of GNA1030 and GNA2091 remain unknown (Comanducci et al. 2002; Madico et al. 2006; Serruto et al. 2010). The final formulation of this vaccine, registered as Bexsero by Novartis Vaccines, includes the five recombinant antigens plus OMVs prepared from the New Zealand epidemic strain of N. meningitidis, which contribute both the PorA antigen to increase strain coverage and additional adjuvant activity (Gorringe and Pajón 2012).
In addition to the choice of antigenic component(s), other important factors to be considered in the development of OMV-based vaccines include the source organism and method of isolation for the OMVs. In the case of MenB vaccines, OMVs have traditionally been isolated from wild-type or recombinant strains of N. meningitidis, which requires them to be prepared by detergent extraction due to the highly toxic nature of lipopolysaccharide (LPS) found on the OMV surface (Holst et al. 2009). LPS (also known as endotoxin) is a potent adjuvant that stimulates the innate immune system through Toll-like receptor 4 (TLR4) found on macrophages, however, high levels of LPS can lead to fever, inflammation, and septic shock (Copeland et al. 2005; Raetz and Whitfield 2002). While detergent extraction of OMVs removes most of the LPS and can also increase OMV yield, there are also drawbacks to this process. Detergent use can result in aggregation of OMVs due to removal of negatively charged LPS and phospholipid molecules, which can increase size variability and decrease vaccine stability and shelf life (Holst et al. 2009; van de Waterbeemd et al. 2010). It also alters the proteomic profile of OMVs, through both contamination of OMVs with cytoplasmic proteins as a result of bacterial cell lysis and removal of components that contribute to immunogenicity and adjuvanticity (van de Waterbeemd et al. 2013, 2010; Zariri et al. 2016a). For example, detergent treatment removes the fHbp antigen, a surface-exposed lipoprotein that is highly immunogenic against various N. meningitidis strains and is naturally present on native OMVs (Masignani et al. 2003). The absence of antigens and PAMPs such as lipoproteins and phospholipids can reduce the immunogenic response to OMVs, requiring the use of additional adjuvants (Gnopo et al. 2017; Zariri et al. 2016a).
Recent efforts toward a second-generation OMV vaccine for MenB have focused on genetic engineering of N. meningitidis strains to reduce LPS toxicity and eliminate the need for detergent extraction of OMVs. The lpxL1 and lpxL2 deletion mutants produce modified lipid A, which is the major component of LPS responsible for its toxicity (Fig. 10.4) (van der Ley et al. 2001; Zariri et al. 2016b). The resulting lipid A is penta-acylated rather than hexa-acylated and is no longer toxic as it shows little to no stimulation of human TLR4 (Steeghs et al. 2008; van der Ley et al. 2001). Thus, OMVs isolated from lpxL1 and lpxL2 mutant strains do not require detergent treatment and are still immunogenic, though they may require additional adjuvants in order to be effective vaccines in humans due to their lack of stimulation of innate immunity through TLR4 (Fisseha et al. 2005; Koeberling et al. 2008; van de Waterbeemd et al. 2010). Further bioengineering of LPS biosynthesis or modification through the use of the lptA or lgtB mutations and heterologous expression of the pagL gene encoding lipid A 3-O-deacylase from Bordetella bronchiseptica has shown that the potential exists to produce OMVs that display a broad range of TLR4 activation, which show promise to be used as effective stand-alone vaccines (Geurtsen et al. 2006; Pupo et al. 2014; Zariri et al. 2016b).
Structural changes in Lipid A. Lipid A can be enzymatically modified by a number of enzymes as shown in the color panel (a). LpxL1 (green), LpxL2 (blue), LpxP (pink) and LptA (brown) all add the corresponding group to the molecule, whereas PagL (red) and LpxE (orange) remove the group. The ΔlptA and ΔlptL1-lpxP mutants produce modified LPS that is less toxic than LPS from wild-type E. coli. The abbreviation of the enzymes, organism source, and activity are presented (b). Used with permission from Zariri et al. Modulating endotoxin activity by combinatorial bioengineering of meningococcal lipopolysaccharide. Scientific Reports, 2016. 6: p. 36575
Finally, there are additional genetic mutations available that can increase overall OMV yield. A mutation in the RmpM protein, which links the outer membrane of the cell to the peptidoglycan layer, leads to a more loosely attached outer membrane and increased OMV release, but does not affect bacterial growth or OMV immunogenicity (Arigita et al. 2004; Klugman et al. 1989; van de Waterbeemd et al. 2010).
Research to date on MenB vaccines highlights both the advantages and challenges of using OMVs as a vaccine platform. Recent developments in bioengineering have done much to expand the potential in this area, and it is likely that the next generation MenB vaccine will make use of the various options in antigen choice and display, LPS detoxification, and OMV yield to result in improved effectiveness, safety, and ease of production.
10.2.2 OMV Vaccines for Gonorrhea
Gonorrhea is one of the most frequently reported communicable diseases in the USA, with a worldwide incidence estimated at 78 million new cases per year (Bolan et al. 2012; Newman et al. 2015). Despite more than a century of research, efforts to develop a vaccine against gonorrhea have been unsuccessful, while the need for such a vaccine has only increased as a number of antibiotic-resistant strains of gonorrhea have emerged (Bolan et al. 2012; Edwards et al. 2016). Interestingly, ecological data suggest a decline in gonorrhea during the time period immediately after the use of the OMV-based MenB vaccines in Cuba and Norway, indicating that the OMV vaccine may afford some protection against gonorrhea (Pérez et al. 2009; Whelan et al. 2016). Furthermore, a retrospective case-control study done in New Zealand estimated that the OMV vaccine used in that country, MeNZB, was 31% effective in preventing gonorrhea (Petousis-Harris et al. 2017). This landmark finding represents the first example of any vaccine being associated with protection against gonorrhea in humans.
Despite causing significantly different diseases, the causal pathogens of gonorrhea and meningitis, Neisseria gonorrhoeae and N.meningitidis, are closely related. The two are estimated to share 80–90% homology in DNA sequence, and several of the antigens found in the Bexsero MenB vaccine are also present in various N. gonorrhoeae strains at approximately 60–90% amino acid sequence identity to the N. meningitidis reference strain (Hadad et al. 2012; Semchenko et al. 2018; Tinsley and Nassif 1996). It was recently shown that both the OMV and recombinant protein antigen components of the Bexsero vaccine (which contains the same OMV component as the MeNZB vaccine) could elicit antibodies against N. gonorrhoeae in rabbits and humans (Semchenko et al. 2018). This provides some explanation of the cross-protection afforded by the MenB vaccines and suggests that development of a similar OMV-based strategy might be the key to an effective vaccine against gonorrhea.
The promising findings of Petousis-Harris et al. and Semchenko et al. with regard to vaccine cross-protection are very recent, and as such there have not yet been reports on OMV-based vaccines designed specifically against gonorrhea. However, some candidate antigens have been identified. For example, N. gonorrhoeae MetQ, a subunit of the methionine binding ABC transporter, was recently shown to be highly conserved, localize to the bacterial and OMV surface, and to play a role in adherence to cervical epithelial cells. Most importantly, antibodies against MetQ are bactericidal and can block adherence, indicating its potential as a candidate vaccine antigen (Semchenko et al. 2017). Additionally, it was recently demonstrated that a vaccine consisting of formalin-inactivated whole bacterial cells encapsulated in microparticles was effective in a mouse model (Gala et al. 2018), further supporting the hypothesis that a vaccine presenting surface-exposed antigens in their native state may be the most effective avenue for a successful gonorrhea vaccine.
10.2.3 OMV Vaccines for Influenza
While the above examples demonstrate the use of pathogen-derived OMVs as vaccines, other species of bacteria can be engineered to heterologously express and display antigens on their outer membrane to produce antibacterial or antiviral OMV vaccines. Despite the availability of seasonal vaccines, influenza infection remains an ongoing threat, particularly with the ability of influenza A viruses to form pandemic strains. Current influenza vaccines generally produce an immune response against the immunodominant glycoproteins hemagglutinin and neuraminidase, however, these proteins are extremely variable and thus strain-specific vaccines must be redeveloped annually (Sato et al. 2001; Treanor 2015). Variations in these surface epitopes and others also contribute to a low efficacy for influenza vaccinations (Osterholm et al. 2012). Therefore, there is significant interest in the development of a universal influenza vaccine that could reduce the need for annual redesign and revaccination and would provide protection should a new pandemic strain arise.
One of the most promising target antigens for a universal vaccine is M2e, an integral membrane protein of influenza A virus. Unlike hemagglutinin and neuraminidase, the M2e sequence is highly conserved across strains, however, it is not as immunogenic as the aforementioned antigens and requires adjuvants to be effective (Lamb et al. 1985). To produce a self-adjuvant, OMV-based vaccine, the probiotic E. coli strain Nissle 1917 was engineered to express a fusion protein consisting of M2e4xHet, a multimeric construct containing four M2e variants, as a C-terminal fusion to ClyA, an E. coli transmembrane protein that is known to be enriched in OMVs (Rappazzo et al. 2016). Mice vaccinated with the resulting recombinant OMVs showed a 100% survival rate after challenge with a lethal dose of a mouse-adapted H1N1 strain (Rappazzo et al. 2016). The OMV-based vaccine was further improved by production in ClearColi, an E. coli strain engineered to contain only the LPS precursor lipid IVa instead of full LPS, and these OMVs provided equal protection against influenza in mice and ferrets without LPS-based endotoxicity, which would otherwise hamper translation of this therapy to use in humans (Watkins et al. 2017).
10.2.4 OMV Vaccines for Cholera
Cholera, a secretory diarrheal disease caused by the Gram-negative bacterium Vibrio cholerae, is a major cause of mortality in developing countries, particularly for infants and young children. While cholera vaccines are currently available, they still suffer from drawbacks of high cost, short shelf life, and the need for cold storage, all of which limit their implementation in developing countries and highlight the need for new candidate vaccines (Bishop and Camilli 2011). V. cholerae OMVs could induce a specific, high-titer, and long-lasting immune response in mice, and immunization of female mice also resulted in protection of their neonatal offspring via the transfer of IgG and IgA antibodies in the mother’s milk (Schild et al. 2009, 2008). As was the case for the N. meningitidis and E. coli OMVs, genetic modification of lipid A resulted in reduced endotoxicity without diminishing the immunogenic potential of the vaccine (Leitner et al. 2013).
It has already been shown that there is a protective effect of breastfeeding against cholera due to the presence of IgA antibodies directed against V. cholerae surface structures and cholera toxin (Clemens et al. 1990; Glass et al. 1983; Hanson et al. 1985; Qureshi et al. 2006). The major protective antigen of the OMV vaccine is the O-antigen, which is present in high amounts on the OMV surface, and cholera toxin is known to be packaged into OMVs (Chatterjee and Chaudhuri 2011; Leitner et al. 2013). Thus, an OMV vaccine may be the ideal candidate to deliver effective antigens that can induce immunity via high IgA titers, and immunization of adult women would hopefully lead to significant protection of newborns and young children, often the population most affected during cholera epidemics (Leitner et al. 2013).
In addition to its effectiveness, the OMV vaccine is also a promising candidate to overcome the cost, stability, and transport limitations of the other available vaccines. Large-scale production of OMVs at reasonable cost has already been demonstrated for the meningitis vaccines. OMVs purified from V. cholerae were stable after 1 month at 37 °C, and could be easily administered for immunizations without accessory buffer solutions, indicating that cold storage and trained medical professionals may not be required for vaccine distribution (Leitner et al. 2013; Schild et al. 2009).
10.2.5 OMV and EV Vaccines for Other Diseases
In addition to the above examples, OMVs from various other pathogenic species have been tested as a vaccine platform against their associated diseases. These include Acinetobacter baumannii, Bordetella pertussis and B. parapertussis, Brucella melitensis, Burkholderia mallei and B. pseudomallei, Francisella novicida, Heliobacter pylori and H. felis, Klebsiella pneumoniae, Porphyromonas gingivalis, Salmonella enterica ssp. enterica ser. Typhimurium, and Shigella spp. (Alaniz et al. 2007; Asensio et al. 2011; Avila-Calderón et al. 2012; Bottero et al. 2013; Keenan et al. 1998; Kesavalu et al. 1992; Lee et al. 2012; McConnell et al. 2011; Mitra et al. 2013; Nieves et al. 2014; Pierson et al. 2011; Roberts et al. 2008).
While the above examples all pertain to the use of OMVs from Gram-negative organisms, the relatively recent discovery that Gram-positive bacteria also produce membrane vesicles has led several groups to test whether Gram-positive MVs might also be effectively used as vaccines (Brown et al. 2015; Lee et al. 2009; Liu et al. 2018b) (See Chap. 2). Promising results from vaccinations with MVs have been demonstrated for Bacillus anthracis, Clostridium perfringens, Mycobacterium tuberculosis, Staphylococcus aureus, and Streptococcus pneumoniae (Choi et al. 2015; Jiang et al. 2014; Olaya-Abril et al. 2014; Prados-Rosales et al. 2014; Rivera et al. 2010). One potential advantage of using MVs from Gram-positive bacteria is that they do not contain LPS and thus are unlikely to require detoxification; for example, Choi et al. reported that unmodified MVs from S. aureus were self-adjuvanting and could stimulate effective immunity against S. aureus with no observable toxic effects (Choi et al. 2015).
In each of the abovementioned cases, the administered OMVs or MVs induced an immune response in cell lines or animal models, and usually promoted survival or prolonged the time to death after challenge with the associated pathogen. As antibiotic-resistant strains are a serious problem for some of these pathogens, the potential for the development of OMV/MV vaccines that protect against these diseases is highly attractive.
10.2.6 OMV Vaccines Based on Recombinant Antigens
There are numerous reports of OMV vaccines based on heterologous expression of antigens. E. coli is often used as the host species for OMV production, as is the case for vaccine concepts against A. baumannii, Francisella tularensis, S. pneumoniae, Campylobacter jejuni, Plasmodium sp., Chlamydia sp., and Leishmania sp. (Bartolini et al. 2013; Chen et al. 2016; Fantappiè et al. 2014; Huang et al. 2016; Pritsch et al. 2016; Schroeder and Aebischer 2009).
Other host species have also been used for recombinant OMV production, particularly those whose OMVs have already been studied as vaccine candidates including N. meningitidis expressing antigens against Lyme disease or genital herpes, V. cholerae expressing enterotoxigenic E. coli antigens and S. enterica ssp. enterica ser. Typhimurium expressing protective antigens to prevent pneumococcal disease, tuberculosis, and chlamydia infections (Daleke-Schermerhorn et al. 2014; Del Campo et al. 2010; Kuipers et al. 2015; Leitner et al. 2015; Muralinath et al. 2011; Salverda et al. 2016).
As in the influenza vaccine described above, the target protein-based antigens are generally expressed as fusions with native membrane-localized proteins such as ClyA or OmpA in E. coli or fHbp in N. meningitidis. The E. coli autotransporter Hemoglobin protease (Hbp) was also recently engineered as a platform that can be used for simultaneous display of multiple heterologous antigens on a single, stable scaffold that localizes to the OMV surface at high densities (Daleke-Schermerhorn et al. 2014; Kuipers et al. 2015). Similarly, a system has been designed in which multiple proteins can be produced separately from OMVs, then assembled onto a scaffold located on the OMV surface (see the below section on biomass conversion for more detail) (Park et al. 2014). These developments open up the exciting possibility of creating multivalent OMV vaccines that present several antigens from the same pathogen or even antigens from several different pathogens.
10.2.7 OMV Vaccines Based on Bacterial Glycans
In some cases, the target vaccine antigens are not proteins but rather glycans. Many successful current vaccines, such as those for non-serogroup B meningitis, are based on the use of glycoconjugates that consist of glycans coupled to T cell-dependent protein antigens, as glycans alone usually elicit T cell-independent responses which are weaker and short-lived. However, a major drawback to this glycan-based vaccine strategy is that current production techniques are technically demanding, costly, and unreliable (Price et al. 2016). As an alternative strategy, E. coli can be engineered to produce and display pathogen-specific polysaccharides on OMVs. For example, Chen et al. introduced the gene cluster required for synthesis of F. tularensis O-Polysaccharide (O-PS), a subunit of LPS, into a hypervesiculating laboratory E. coli strain which is O-PS-deficient but still produces the lipid A core to which O-PS attaches (Chen et al. 2016). In this model, pathogen-specific O-PS is synthesized on the cytoplasmic face of the inner membrane by plasmid-encoded enzymes, to which endogenous E. coli proteins complete the translocation of O-PS to the outer membrane and attach it to the lipid A core. The result of this process is that F. tularensis—specific O-PS is displayed on the E. coli outer membrane and consequently the OMVs as well (Fig. 10.5). In parallel, the E. coli host strain was also engineered to produce the less inflammatory, penta-acylated lipid A to circumvent LPS-based toxicity. Vaccination of mice using the subsequent glycoengineered OMVs significantly delayed time to death after lethal F. tularensis challenge. This is a promising result for the development of a vaccine against F. tularensis, which is a class A bioterrorism agent for which no licensed vaccine currently exists (Oyston et al. 2004). A similar strategy was employed by Price et al., who engineered E. coli to produce OMVs displaying the S. pneumoniae serotype 14 capsular polysaccharide, which elicited an immune response comparable to a commercial pneumococcal vaccine in mice (Price et al. 2016). They also designed glycoengineered OMVs that display the C. jejuni N-glycan, which resulted in an unprecedented level of protection against C. jejuni in chickens (Price et al. 2016). Collectively, these studies highlight the potential of glycoengineering in vaccine development for pathogens that have to date proven incompatible with other methods.
Schematic of the assembly of pathogen-specific antigens on the surface of OMVs. Cellular machinery is responsible for synthesis and transport to the outer membrane where flippase (Wzx) is responsible for membrane translocation and surface expression. Membrane-bound antigen is released in budding vesicles. Reproduced with permission from Chen et al. Outer membrane vesicles displaying engineered glycotopes elicit protection antibodies. PNAS, 2016. 113(26): p. E3609-E3618
10.2.8 OMV Vaccines for Host Glycans
Glycoengineering of OMVs also carries the exciting possibility for use in generating immune responses against clinically important host glycans such as those associated with certain types of cancer. For example, Valentine et al. engineered E. coli OMVs to display two clinically important human glycan structures, namely the tumor-specific carbohydrate antigens polysialic acid (PSA) and Thomsen-Friedenreich antigen (T-antigen) (Valentine et al. 2016). Both of these glycans are highly expressed in several different cancers but not in normal cells and antibodies recognizing these antigens could have clinical benefits, however, on their own, they have low intrinsic immunogenicity (Heimburg-Molinaro et al. 2011). OMVs displaying these antigens could elicit strong IgG antibody titers in mice, indicating that this may be an effective strategy for generating functional antibodies against clinically relevant carbohydrates. These results highlight the advantages of using OMVs to display glycan antigens: their production is less complicated and expensive than traditional glycoconjugate vaccines, the strategy can be easily tailored to various glycan antigens (provided the biosynthetic pathway for the target is known), and they provide the necessary adjuvanticity to activate long-lasting, T cell-dependent immunity.
10.3 OMV-Based Therapeutics
Biologically derived nanoparticles isolated from bacteria or mammalian cells have received substantial attention in recent years for their potential as biodegradable carriers that can specifically deliver cargo to targeted sites (Yoo et al. 2011). While many drugs face limitations in application due to toxicity, poor stability, and inability to cross cell membranes, engineered OMVs/MVs can circumvent many of these drawbacks as they can protect their cargo from degradation, target it to specific cells, and deliver it into those cells efficiently.
The capacity for engineered OMVs to target a specific cell population and deliver cargo was effectively demonstrated by Gujrati et al., who developed E. coli OMVs for use in cancer therapy (Gujrati et al. 2014). They engineered E. coli to express an affibody specific to HER2, a transmembrane receptor overexpressed in many cancers, as a fusion protein with the C-terminus of the native ClyA protein, which targets the resulting protein to the OMV surface (Fig. 10.6). Isolated OMVs were loaded via electroporation with a therapeutic siRNA that targets the kinesin spindle protein (KSP), which is overexpressed in rapidly proliferating cells such as those found in tumor tissue. Silencing of KSP blocks the formation of mitotic spindles, leading to cell-cycle arrest and apoptosis. When injected into mice, the resulting OMVs targeted HER2-overexpressing cells with high affinity and were rapidly internalized, leading to significant inhibition of tumor growth and reduction of tumor size that could be attributed to knockdown of KSP expression. As these OMVs were isolated from an E. coli strain with modified LPS, they have low endotoxicity and did not cause significant side effects, which is a major limiting factor of many drug therapies. This work highlights the potential of vesicle-based nanoparticles as cell-specific delivery vehicles that can overcome many of the drawbacks and limitations that hamper the development and release of new cancer therapies.
OMV as therapeutic agents for cancer. (a) E. coli OMVs were labeled with anti-HER2 affibodies then loaded with cytotoxic siRNA labeled with a fluorophore for visualization. (b) For cell binding and uptake studies, HER2-overexpressing SKOV3 cells and HER2-negative MDA-MB-231 cells were co-incubated with AffiHER2 OMV and stained with an anti-affibody antibody (green). Receptor-specific cell binding and uptake were seen only with HER2-overexpressing SKOV3 cells. Reproduced with permission Gujrati et al. (2014) Bioengineered Bacterial Outer Membrane Vesicles as Cell-Specific Drug-Delivery Vehicles for Cancer Therapy. ACS Nano 8(2): pp. 1525–1537
Various other health benefits have been observed for membrane vesicles, particularly those produced by probiotic bacteria. These include bacterial species that colonize the gastrointestinal tract and confer benefits to the host through either direct communication with host cells or through interaction with other probiotic or pathogenic bacteria (Bron et al. 2011). Often, the beneficial effects result from the interaction of probiotic bacteria (and their associated membrane vesicles) with the host immune system. For example, recent studies have suggested that probiotic bacteria can suppress inflammatory and allergic responses through modulation of immune responses. MVs from Bifidobacterium longum could alleviate a food allergy response in a mouse model by penetrating through intestinal epithelial cells and selectively targeting and inducing apoptosis of mast cells (Kim et al. 2016). The commensal species Bacteroides fragilis delivers the immunomodulatory molecule Polysaccharide A to dendritic cells via OMVs, thereby suppressing immune responses that drive inflammation (Shen et al. 2012). A similar result was also shown for Lactobacillus rhamnosus, for which MVs could recapitulate the previously demonstrated immunoregulatory and neuronal effects of whole bacteria (Al-Nedawi et al. 2015). Finally, MVs of Kefir-derived Lactobacillus strains reduced inflammatory cytokine production and alleviated symptoms in a mouse model of inflammatory bowel disease (Seo et al. 2018).
The potential modulation of the host immune system by probiotic bacteria can also have a protective role against various pathogens. While it has not been definitely shown that these benefits are conveyed solely by membrane vesicles, researchers have performed studies using culture media and other preparations devoid of cells and seen similar results as discussed above. As specific examples of MV-mediated protection, Li et al. showed that MVs derived from L. plantarum provided protection to C. elegans against vancomycin-resistant Enterococcus faecium through upregulation of multiple host defense genes (Li et al. 2017). The authors also performed these experiments with the common human intestinal epithelial cell line Caco-2 and observed similar changes in gene expression. Further exploration into the mechanisms of this and other protective effects induced by probiotic MVs might allow for the development of new treatments for antibiotic-resistant pathogens.
Probiotic bacteria have also been suggested to have cancer prevention properties, particularly against colon cancer (Commane et al. 2005; dos Reis et al. 2017; Paolillo et al. 2009). While the exact mechanisms of this are unknown, it has been reported that many probiotic species exert this effect through the secretion of factors that induce apoptosis, and it is likely that these factors are delivered to host cells via bacterial membrane vesicles (Oelschlaeger 2010). Indeed, it was recently shown that purified MVs from L. rhamnosus have significant cytotoxic effects on hepatic cancer cells (Behzadi et al. 2017). As membrane vesicles derived from the microbiota in the gastrointestinal tract can travel to the liver and other nearby organs through the bloodstream, this provides further evidence for the potent anticancer properties of probiotic bacteria and their membrane vesicles (Salminen et al. 2004). Furthermore, as probiotic bacteria generally do not have deleterious effects on the host, exploitation of these properties may make possible the development of new cancer treatments that do not also cause the damaging side effects of traditional chemotherapeutic agents (Behzadi et al. 2017).
10.3.1 Emerging Therapeutic Applications
In addition to the potential anticancer and other uses discussed above, there are a number of other therapeutics that could benefit from delivery via OMVs/MVs. Compounds such as antimicrobial peptides which would be susceptible to degradation in their free form, or antibiotics that would otherwise not be able to cross the cell membrane, could be packaged into bacterial membrane vesicles for protection and delivery into target cells (Liu et al. 2018b). This may be particularly important for treatment of pathogens that can ordinarily resist antibiotics through mechanisms such as outer membranes with low permeability, or biofilms that delay penetration of the antibiotic or directly inactivate it via secreted enzymes such as β-lactamases (Messiaen et al. 2013). Unlike free molecules, vesicles may be transported through the biofilm and taken up into cells, bypassing these defense mechanisms. For example, packaging of the antibiotic tobramycin into artificial liposomes greatly increased its effectiveness against multiple pathogens including Burkholderia cepacia, Pseudomonas aeruginosa, and S. aureus (Beaulac et al. 1998). Several liposome-based drugs are currently in clinical trials; however OMV/MV-based vesicles may offer additional benefits related to ease of production or the ability to incorporate specific cell targeting motifs through genetic engineering.
Phage therapy has also received significant attention in recent years as an alternative treatment for antibiotic-resistant bacteria (Lin et al. 2017). One challenge to the implementation of phage therapies is that many phages have narrow host ranges, limiting their applicability. As membrane vesicles naturally play a role in broadening phage host ranges through the transfer of phage receptors between bacterial hosts, this capability could be harnessed for clinical use (Tzipilevich et al. 2017). For example, vesicles derived from phage-sensitive bacteria could be given to a patient prior to administration of the phage itself. If those vesicles are taken up by bacteria already infecting the patient, the phage receptor could be transferred and could potentially enhance targeting of the phage to the infectious bacteria (Liu et al. 2018b).
It has also been proposed that vesicles could be used for delivery of genome editing tools, such as the Cas9-guide RNA ribonucleoprotein complex required for CRISPR-based genome editing, to correct genetic disorders or to combat pathogens (Knott and Doudna 2018; Liu et al. 2019).
Membrane vesicles could also be used as vehicles for delivery of nutritional compounds to the host gastrointestinal tract. For example, vitamin K2 (menaquinone) is a cofactor required for the production of blood coagulation factors and osteocalcin (a bone-forming protein) and may also prevent osteoporosis, coronary heart disease, and liver cancer, but recent data indicate that subclinical vitamin K deficiency is not uncommon (DiNicolantonio et al. 2015). Vitamin K cannot be synthesized by humans, but rather is mainly produced by bacteria in the intestine such as B. subtilis and certain strains of lactic acid bacteria (Liu et al. 2018a). In particular, a strain of B. subtilis isolated from the traditional Japanese fermented soybean product natto could be engineered to produce very high amounts of vitamin K2 (Sato et al. 2001). As menaquinones are hydrophobic compounds that accumulate in the bacterial cell membrane, the potential exists for vitamin K-containing MVs isolated from B. subtilis or lactic acid bacteria to be administered as dietary supplements (Liu et al. 2018a). A similar strategy could be used for other important nutritional or medically relevant compounds that are not naturally produced in high amounts by these bacteria, if they could be genetically engineered to produce and package such compounds in vesicles.
10.3.2 Commercial Application of EVs
The protein composition of bacterial membrane vesicles is highly variable dependent upon growth conditions, culture age, and many other factors. Despite the variability, abundant membrane proteins such as porins, membrane channels, and others consistently appear in proteomic analysis of bacterial membrane vesicles and offer potential anchors for their functionalization (Dean et al. 2019; Kroniger et al. 2018; Kwon et al. 2009; Lee et al. 2007; Schwechheimer et al. 2013; Yun et al. 2017). With the targeted loading of bacterial membrane vesicles, researchers can begin to exploit the inherent natural advantages afforded by these vesicles, such as protection from environmental conditions, as well as others of specific design such as enzyme localization and assembly. This engineering of OMVs has the potential for designer probiotics, biological catalysts, and even development for new therapeutic platforms. In the subsequent section, we highlight some of the areas where engineered OMVs and MVs are already making inroads as new tools for a range of commercial, environmental, and health-related applications.
10.3.3 OMVs for Biomass Conversion
The successful engineering of E. coli and other bacteria to express target proteins and display them on the OMV surface makes possible their use not just in biomedical applications, but for other purposes in which nanoparticles are needed. Many biological processes, such as the Krebs TCA cycle in mitochondria or cellulose hydrolysis by cellulosomes on the surface of anaerobic bacteria, are carried out through complex multienzyme cascades that achieve specificity and efficiency through compartmentalization and precise spatial organization of individual proteins. In an effort to mimic this organization for biotechnological applications, several studies have demonstrated synthetic assembly of multiple enzymes onto liposomes or polymersomes; however, the process is complex, costly, and the resulting liposomes are often fragile, making this approach impractical for large-scale applications (Fischer et al. 2002; van Dongen et al. 2009; Vriezema et al. 2007). Conversely, OMVs could provide the ideal backbone for synthetic nanoreactors if some or all of the enzyme production and assembly could be driven by genetic engineering of the E. coli host strain.
This approach was successfully demonstrated by Park et al., who engineered E. coli OMVs to display a functional multienzyme complex similar to a cellulosome (Park et al. 2014). Natural cellulosomes are composed of a structural scaffold containing repeating cohesin domains that are bound individually to cellulases via corresponding dockerin domains (Fontes and Gilbert 2010). To mimic this structure on the surface of OMVs, Park and colleagues engineered E. coli to express a scaffold consisting of three different cohesin domains and a cellulose binding domain, attached to the outer membrane by the ice nucleation protein anchor (Fig. 10.7). Three different cellulases, each possessing a dockerin domain corresponding to one of the cohesins, were produced separately in E. coli and subsequently incubated with the OMVs to allow cohesin–dockerin interactions to assemble the full multienzyme complex. The resulting OMVs showed 23-fold enhancement of cellulose hydrolysis as compared to free enzymes. This result is very promising for the potential of OMVs as nanobioreactors in biotechnology, with the added benefit that the same strategy could be used for various other enzymatic cascades by replacing the dockerin-bound cellulases with other enzymes engineered to contain the dockerin domain.
Multienzyme assembly on engineered OMVs. Left panel: A trivalent scaffold was developed that contained three orthogonal cohesion domains that enabled the assembly of cellulose enzymes tagged with complementary dockerin domains. Right panel: Assembly of the enzyme system on OMV surfaces significantly improved enzyme activity (orange bars) compared to the free enzyme controls (blue bars). Reproduced with permission from Park et al. (2014) Positional Assembly of Enzymes on Bacterial Outer Membrane Vesicles for Cascade Reactions. PLoS ONE 9(5): e97103
10.3.4 OMVS for Bioremediation
Fortunately for mankind, microbial populations are able to exploit ancestral, bi-functional, or newly evolved cellular processes to degrade and consume many of the chemical contaminants we have produced or inadvertently released into the environment. Take for example, the rapid degradation of the oil plume released following the Deep Sea Horizon disaster (Atlas and Hazen 2011; Scoma et al. 2016) or the more recent identification of marine bacteria capable of degrading some of the plastics released into the ocean (Dash et al. 2013; Urbanek et al. 2018). Observations such as these and others stimulate the continued efforts of researchers and government agencies to develop biological tools for environmental remediation. Natural bioremediation, where an indigenous organism degrades the environmental contaminant is, of course, the ideal scenario. Unfortunately, this is often not achievable as spills and targeted release of toxic compounds can occur in locations where such microbes are not native. As an alternative, the enzymatic systems from these organisms can be isolated, produced recombinantly, and deployed at the point of concern to facilitate decontamination. Enzymes, however, have their own limitations such as stability and cost of manufacture.
Similar to their function in the natural world, engineered bacterial membrane vesicles afford protection to encapsulated biomolecules from harsh environmental conditions. The controlled loading of enzymes into bacterial OMVs/MVs provides for a method of producing reagents that can easily be isolated from bacterial cultures, lyophilized for storage and distribution, then rehydrated to facilitate environmental cleanup. In a series of publications, Alves et al. showed that a protein–protein ligation system could be used to direct the packaging of a phosphotriesterase (PTE) enzyme capable of degrading an organophosphate compound into E. coli OMVs (Alves et al. 2015a, 2016). The authors employed a recombinant version of a native porin protein (ompA) presenting a small peptide (SpyTag) and a PTE fusion with its counterpart (SpyCatcher) (Fig. 10.8). As described by Zakeri and colleagues, the SpyCatcher/SpyTag system allows for the spontaneous formation of an isopeptide bond between the two components which in this system, facilitated the anchoring of PTE to the outer membrane and subsequent packaging of the enzyme into the OMVs (Zakeri et al. 2012). The authors demonstrated that not only did the enzyme maintain activity but also survived a number of storage and environmental conditions better than the free enzyme. In subsequent studies, Alves et al. also showed that these materials had relevance outside the laboratory by testing their materials in environmental water samples spiked with substrate and on a number of materials chosen to mimic military vehicle paint and surfaces (Alves et al. 2018). In these studies, the authors also examined enzyme activity under nonideal conditions such as variable pH, high salinity, and in environmental water samples with varying microbial populations and debris composition. Given the aversion of many societies to employ genetically modified organisms or release nonindigenous species into the wild, OMV-based reagents offer an alternative approach to the development of Green reagents for environmental remediation.
Directed packaging of the OMV lumen. A protein–protein ligation system (SpyCatcher/SpyTag) was used to anchor a recombinant phosphotriesterase (PTE) enzyme to the bacterial outer membrane enabling the directed loading of nascent OMVs. Reproduced with permission from Alves, N.J., et al., Bacterial Nanobioreactors—Directing Enzyme Packaging int Bacterial Outer Membrane Vesicles. ACS Appl Mater Interfaces, 2015. 7(44): p. 24963–72
10.3.5 OMVs for Imaging and Biosensing
Today, synthetically manufactured liposomes are routinely used for the delivery of therapeutics and in many diagnostic assays (Akbarzadeh et al. 2013; Alavi et al. 2017; Xing et al. 2016). Despite their many successes, liposomes can be difficult to manufacture, load, and store for prolonged periods of time (Alves et al. 2015b). In contrast, engineered OMVs offer a potential path to a simple manufacture platform that allows for controlled packaging and a final product that exhibits biophysical properties making them ideal reagents for drug delivery and the building of biological sensors. As an example, Chen et al. demonstrated how the ability to modify both the exterior proteins and interior cargo could be used in combination to develop reagents that have both assay and imaging capabilities (Fig. 10.9) (Chen et al. 2017). Here the authors expanded upon a previous construct for OMV modification that presents a cohesion–dockerin scaffold on the OMV exterior, adding a terminal domain (Z-domain) that allows for attachment of an antibody (Chen et al. 2010; Park et al. 2014). Interior packaging is accomplished using a modified bacterial lipoprotein (SlyB) fusion which is known to localize to the outer membrane of E. coli (Tokuda and Matsuyama 2004). This system allows for versatility in assay and imaging development as any number of antibodies could be added to the exterior and used in combination with a wide array of reporter proteins, from fluorescent proteins to luminescent proteins.
OMV-based reagents for imaging and biosensing. OMVs from Gram-negative bacteria offer numerous opportunities for modification by targeting both the exterior and interior domains. (a) Researchers have shown that exterior domains of OMVs can be modified to add targeting moieties while the interior cavity can be loaded with a range of reporter molecules using membrane anchors, fusions, and various other methods. (b) Multifunctional OMVs produced in this manner can be used as a one-pot material for bioassays such as the traditional ELISA or used in cell targeting and imaging
10.3.6 Future Commercial Applications of Bacterial Membrane Vesicles
While therapeutic applications of OMVs/MVs are at the forefront of research efforts, the encapsulation of biomolecules within bacterial membrane vesicles has great potential for commercial applications. The ease of production and the protection afforded to encapsulated proteins make this a highly versatile system that could easily be adapted to benefit many applications that are not currently explored.
Recombinant production of proteins, enzymes, and other biomolecules is a critical component of both the commercial and medical industries (Adrio and Demain 2010; Demain and Vaishnav 2009; Ferrer-Miralles and Villaverde 2013; Sanchez-Garcia et al. 2016). While successes such as the E. coli-produced insulin serve as banners for this technology, there are numerous other biomolecules that are never able to come to market due to complications associated with biomanufacturing. Often, large-scale production of biomolecules is hindered by low yields that can be associated with toxicity, insolubility, or any number of other issues. Membrane vesicles could potentially serve as a method of “off-loading” recombinant products before they could accumulate in the engineered organism leading to toxicity. Enclosed within OMVs, these biomolecules could be isolated directly from batch cultures through engineered epitopes on the OMV surface as shown by Alves et al. (2017).
Researchers have shown that OMVs function in many microbial community interactions including interspecies communication and regulation of microbial populations. Additionally, antimicrobial peptides and other cytotoxic compounds are readily packaged within vesicles and purified OMVs alone can display bactericidal activity (Dean et al. 2019; Park 2018; Schulz et al. 2018). While this has obvious therapeutic benefits, these properties of OMVs and MVs could also be harnessed for nonmedical purposes. Microbial communities and the biofilms they form also plague many industrial, military, and environmental systems. In addition to obvious examples such as food production facilities (Glass et al. 1983; Marchand et al. 2012), damaging biofilms can also be found in fuel storage containers (Bücker et al. 2014), oil and gas pipelines (Tingyue Gu 2015), on the hulls of ships (Schultz et al. 2011), and countless other locations. The use of biological reagents that are easily resorbed into the environment to eliminate or reduce biofilms could prove beneficial in developing technologies that can readily be implemented without concern for additional harmful environmental effects.
10.4 Conclusion
Scientists continue to gain a greater understanding of the microbial world and the valuable tools it provides to society. We have long exploited them for food processing, as a pipeline for drug discovery, and as miniature factories for the production of target biomolecules; however, as shown here, bacteria can begin to aid the development of new vaccines or even as reagents for environmental cleanup. With the rapid emergence of antimicrobial resistance, OMV/MV derived vaccines may serve as mankind’s next line of defense in the battle against pathogenic microbes, particularly those pathogens that can be weaponized such as B. anthracis or intracellular pathogens such as F. tularensis that are difficult to treat with conventional methods. Beyond vaccines, the observations and theories that support the presence of OMVs in the bloodstream and passing through the blood-brain barrier suggest that with advances in synthetic biology and a greater understanding of the human microbiome, scientists may be able to harness microbial communities to effect changes and treat disorders. Well beyond traditional probiotics, advances in these areas of research will dramatically alter the way we think about the foods we eat as well as how medical therapies are administered. Finally, by mimicking the bacteria themselves and loading enzyme systems into OMVs, we may be able to develop cell-free catalytic reagents that can be used to remediate environmental disasters, treat waste water, or even synthesize novel materials. Both natural and engineered OMVs and MVs have great potential in many areas of human society and will be an area of scientific investigation for years to come.
References
Adrio J-L, Demain AL (2010) Recombinant organisms for production of industrial products. BIoeng Bugs 1:116–131
Akbarzadeh A et al (2013) Liposome: classification, preparation, and applications. Nanoscale Res Lett 8:102
Alaniz RC, Deatherage BL, Lara JC, Cookson BT (2007) Membrane vesicles are immunogenic facsimiles of Salmonella typhimurium that potently activate dendritic cells, prime B and T cell responses, and stimulate protective immunity in vivo. J Immunol 179:7692–7701
Alavi M, Karimi N, Safaei M (2017) Application of various types of liposomes in drug delivery systems. Adv Pharm Bull 7:3–9
Al-Nedawi K et al (2015) Gut commensal microvesicles reproduce parent bacterial signals to host immune and enteric nervous systems. FASEB J 29:684–695
Alves NJ, Turner KB, Daniele MA, Oh E, Medintz IL, Walper SA (2015a) Bacterial nanobioreactors—directing enzyme packaging into bacterial outer membrane vesicles. ACS Appl Mater Interfaces 7:24963–24972
Alves NJ, Turner KB, Medintz IL, Walper SA (2015b) Emerging therapeutic delivery capabilities and challenges utilizing enzyme/protein packaged bacterial vesicles. Ther Delv 6:873–887
Alves NJ, Turner KB, Medintz IL, Walper SA (2016) Protecting enzymatic function through directed packaging into bacterial outer membrane vesicles. Sci Rep 6:24866
Alves NJ, Turner KB, DiVito KA, Daniele MA, Walper SA (2017) Affinity purification of bacterial outer membrane vesicles (OMVs) utilizing a His-tag mutant. Res Microbiol 168:139–146
Alves NJ, Moore M, Johnson BJ, Dean SN, Turner KB, Medintz IL, Walper SA (2018) Environmental decontamination of a chemical warfare simulant utilizing a membrane vesicle-encapsulated phosphotriesterase. ACS Appl Mater Interfaces 10:15712–15719
Arigita C, Jiskoot W, Westdijk J, van Ingen C, Hennink WE, Crommelin DJA, Kersten GFA (2004) Stability of mono- and trivalent meningococcal outer membrane vesicle vaccines. Vaccine 22:629–642
Asensio CJA et al (2011) Outer membrane vesicles obtained from Bordetella pertussis Tohama expressing the lipid a deacylase PagL as a novel acellular vaccine candidate. Vaccine 29:1649–1656
Atlas RM, Hazen TC (2011) Oil biodegradation and bioremediation: a tale of the two worst spills in U.S. history. Environ Sci Technol 45:6709–6715
Avila-Calderón ED et al (2012) Characterization of outer membrane vesicles from Brucella melitensis and protection induced in mice. Clin Dev Immunol 2012:352493–352493
Barak I, Muchova K (2013) The role of lipid domains in bacterial cell processes. Int J Mol Sci 14:4050–4065
Bartolini E et al (2013) Recombinant outer membrane vesicles carrying Chlamydia muridarum HtrA induce antibodies that neutralize chlamydial infection in vitro. J Extracell Vesicles 2:20181
Beaulac C, Sachetelli S, Lagace J (1998) In-vitro bactericidal efficacy of sub-MIC concentrations of liposome-encapsulated antibiotic against gram-negative and gram-positive bacteria. J Antimicrob Chemother 41:35–41
Behzadi E, Mahmoodzadeh Hosseini H, Imani Fooladi AA (2017) The inhibitory impacts of Lactobacillus rhamnosus GG-derived extracellular vesicles on the growth of hepatic cancer cells. Microb Pathog 110:1–6
Berleman JE et al (2014) The lethal cargo of Myxococcus xanthus outer membrane vesicles. Front Microbiol 5:474
Bishop AL, Camilli A (2011) Vibrio cholerae: lessons for mucosal vaccine design. Expert Rev Vaccines 10:79–94
Bolan GA, Sparling PF, Wasserheit JN (2012) The emerging threat of untreatable gonococcal infection. New Engl J Med 366:485–487
Bottero D et al (2013) Outer membrane vesicles derived from Bordetella parapertussis as an acellular vaccine against Bordetella parapertussis and Bordetella pertussis infection. Vaccine 31:5262–5268
Bron PA, van Baarlen P, Kleerebezem M (2011) Emerging molecular insights into the interaction between probiotics and the host intestinal mucosa. Nat Rev Microbiol 10:66
Brown L, Wolf JM, Prados-Rosales R, Casadevall A (2015) Through the wall: extracellular vesicles in gram-positive bacteria, mycobacteria and fungi. Nat Rev Microbiol 13:620
Bücker F et al (2014) Fuel biodegradation and molecular characterization of microbial biofilms in stored diesel/biodiesel blend B10 and the effect of biocide. Int Biodeterior Biodegradation 95:346–355
Chatterjee D, Chaudhuri K (2011) Association of cholera toxin with Vibrio cholerae outer membrane vesicles which are internalized by human intestinal epithelial cells. FEBS Lett 585:1357–1362
Chen DJ, Osterrieder N, Metzger SM, Buckles E, Doody AM, DeLisa MP, Putnam D (2010) Delivery of foreign antigens by engineered outer membrane vesicle vaccines. Proc Natl Acad Sci U S A 107:3099–3104
Chen L et al (2016) Outer membrane vesicles displaying engineered glycotopes elicit protective antibodies. Proc Natl Acad Sci U S A 113:E3609–E3618
Chen Q, Rozovsky S, Chen W (2017) Engineering multi-functional bacterial outer membrane vesicles as modular nanodevices for biosensing and bioimaging. Chem Commun (Camb) 53:7569–7572
Choi SJ et al (2015) Active immunization with extracellular vesicles derived from Staphylococcus aureus effectively protects against staphylococcal lung infections, mainly via Th1 cell-mediated immunity. PLoS One 10:e0136021–e0136021. https://doi.org/10.1371/journal.pone.0136021
Claassen I et al (1996) Production, characterization and control of a Neisseria meningitidis hexavalent class 1 outer membrane protein containing vesicle vaccine. Vaccine 14:1001–1008. d
Clemens JD et al (1990) Breast feeding and the risk of severe cholera in rural Bangladeshi children. Am J Epidemiol 131:400–411
Coffman RL, Sher A, Seder RA (2010) Vaccine adjuvants: putting innate immunity to work. Immunity 33:492–503
Comanducci M et al (2002) NadA, a novel vaccine candidate of Neisseria meningitidis. J Exp Med 195:1445
Commane D, Hughes R, Shortt C, Rowland I (2005) The potential mechanisms involved in the anti-carcinogenic action of probiotics. Mutat Res 591:276–289
Copeland S, Warren HS, Lowry SF, Calvano SE, Remick D (2005) Acute inflammatory response to endotoxin in mice and humans. Clin Diagn Lab Immunol 12:60–67. https://doi.org/10.1128/cdli.12.1.60-67.2005
Daleke-Schermerhorn MH et al (2014) Decoration of outer membrane vesicles with multiple antigens by using an autotransporter approach. Appl Environ Microbiol 80:5854–5865
Dash HR, Mangwani N, Chakraborty J, Kumari S, Das S (2013) Marine bacteria: potential candidates for enhanced bioremediation. Appl Microbiol Biotechnol 97:561–571
de Kleijn E et al (2001) Serum bactericidal activity and isotype distribution of antibodies in toddlers and schoolchildren after vaccination with RIVM hexavalent PorA vesicle vaccine. Vaccine 20:352–358
Dean SN, Leary DH, Sullivan CJ, Oh E, Walper SA (2019) Isolation and characterization of lactobacillus-derived membrane vesicles. Sci Rep 9:877. https://doi.org/10.1038/s41598-018-37120-6
Deatherage BL, Lara JC, Bergsbaken T, Rassoulian Barrett SL, Lara S, Cookson BT (2009) Biogenesis of bacterial membrane vesicles. Mol Microbiol 72:1395–1407
Del Campo J et al (2010) Intranasal immunization with a proteoliposome-derived cochleate containing recombinant gD protein confers protective immunity against genital herpes in mice. Vaccine 28:1193–1200
Demain AL, Vaishnav P (2009) Production of recombinant proteins by microbes and higher organisms. Biotechnol Adv 27:297–306
DiNicolantonio JJ, Bhutani J, O’Keefe JH (2015) The health benefits of vitamin K. Open Heart 2:e000300–e000300. https://doi.org/10.1136/openhrt-2015-000300
dos Reis SA, da Conceição LL, Siqueira NP, Rosa DD, da Silva LL, Peluzio MCG (2017) Review of the mechanisms of probiotic actions in the prevention of colorectal cancer. Nutr Res 37:1–19
Edwards JL, Jennings MP, Apicella MA, Seib KL (2016) Is gonococcal disease preventable? The importance of understanding immunity and pathogenesis in vaccine development. Crit Rev Microbiol 42:928–941
Elhenawy W, Bording-Jorgensen M, Valguarnera E, Haurat MF, Wine E, Feldman MF (2016) LPS remodeling triggers formation of outer membrane vesicles in Salmonella. MBio 7:e00940–e00916. https://doi.org/10.1128/mBio.00940-16
Ellis TN, Kuehn MJ (2010) Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol Mol Biol Rev 74:81–94 d
Fantappiè L et al (2014) Antibody-mediated immunity induced by engineered Escherichia coli OMVs carrying heterologous antigens in their lumen. J Extracell Vesicles 3:24015. https://doi.org/10.3402/jev.v3.24015
Ferrer-Miralles N, Villaverde A (2013) Bacterial cell factories for recombinant protein prodcution; expanding the catalogue. Microb Cell Factories 12:113
Finne J, Leinonen M, Mäkelä PH (1983) Antigenic similarities between brain components and bacteria causing meningitis: implications for vaccine development and pathogenesis. Lancet 322:355–357
Fischer A, Frnaco A, Oberholzer T (2002) Giant vesicles as microreactors for enzymatic mRNA synthesis. Chembiochem 3:409–417
Fisseha M, Chen P, Brandt B, Kijek T, Moran E, Zollinger W (2005) Characterization of native outer membrane vesicles from mutant strains of Neisseria meningitidis for use in parenteral vaccination. Infect Immun 73:4070–4080
Fontes CM, Gilbert HJ (2010) Cellulosomes: highly efficient nanomachines designed to deconstruct plant cell wall omplex carbohydrates. Annu Rev Biochem 79:655–681
Gala RP, Zaman RU, D’Souza MJ, Zughaier SM (2018) Novel whole-cell inactivated Neisseria gonorrhoeae microparticles as vaccine formulation in microneedle-based transdermal immunization. Vaccine 6:60
Gerritzen MJH, Martens DE, Wijffels RH, van der Pol L, Stork M (2017) Bioengineering bacterial outer membrane vesicles as vaccine platform. Biotechnol Adv 35:565–574
Geurtsen J et al (2006) Expression of the lipopolysaccharide-modifying enzymes pagP and pagL modulates the endotoxic activity of Bordetella pertussis. Infect Immun 74:5574–5585
Giuliani MM et al (2006) A universal vaccine for serogroup B meningococcus. Proc Natl Acad Sci U S A 103:10834
Glass RI, Svennerholm A-M, Stoll BJ, Khan MR, Hossain KMB, Hug MI, Holmgren J (1983) Protection against Cholera in breast-fed children by antibodies in breast milk. New Engl J Med 308:1389–1392
Gnopo YMD, Watkins HC, Stevenson TC, DeLisa MP, Putnam D (2017) Designer outer membrane vesicles as immunomodulatory systems – reprogramming bacteria for vaccine delivery. Adv Drug Del Rev 114:132–142
Gorringe AR, Pajón R (2012) Bexsero. Hum Vaccin Immunother 8:174–183
Grande R et al (2017) Detection and physicochemical characterization of membrane vesicles (MVs) of Lactobacillus reuteri DSM 17938. Front Microbiol 8:1040. https://doi.org/10.3389/fmicb.2017.01040
Gujrati V, Kim S, Kin S-H, Min JJ, Choy HE, Kim SC, Jon S (2014) Bioengineered bacterial outer membrane vesicles as cell-specific drug-delivery vehicles for cancer therapy. ACS Nano 8:1525–1537
Hadad R, Jacobsson S, Pizza M, Rappuoli R, Fredlund H, Olcén P, Unemo M (2012) Novel meningococcal 4CMenB vaccine antigens – prevalence and polymorphisms of the encoding genes in Neisseria gonorrhoeae. APMIS 120:750–760
Hanson LA, Hofvander Y, Lindquist B, Zetterstrom R (1985) Breast-feeding as a protection against gastroenteritis and other infections. Acta Paediatr Scand 74:641–642
Heimburg-Molinaro J, Lum M, Vijay G, Jain M, Almogren A, Rittenhouse-Olson K (2011) Cancer vaccines and carbohydrate epitopes. Vaccine 29:8802–8826
Holst J, Martin D, Arnold R, Huergo CC, Oster P, O’Hallahan J, Rosenqvist E (2009) Properties and clinical performance of vaccines containing outer membrane vesicles from Neisseria meningitidis. Vaccine 27:B3–B12
Holst J et al (2013) Vaccines against meningococcal serogroup B disease containing outer membrane vesicles (OMV): lessons from past programs and implications for the future. Hum Vaccin Immunother 9:1241–1253. d
Huang W et al (2016) Employing Escherichia coli-derived outer membrane vesicles as an antigen delivery platform elicits protective immunity against Acinetobacter baumannii infection. Sci Rep 6:37242
Jan AT (2017) Outer membrane vesicles (OMVs) of Gram-negative bacteria: a perspective update. Front Microbiol 8:1053. https://doi.org/10.3389/fmicb.2017.01053
Jiang Y, Kong Q, Roland KL, Curtiss R (2014) Membrane vesicles of Clostridium perfringens type A strains induce innate and adaptive immunity. Int J Med Microbiol 304:431–443
Kaparakis-Liaskos M, Ferrero RL (2015) Immune modulation by bacterial outer membrane vesicles. Nat Rev Immunol 15:375
Keenan JI, Allardyce RA, Bagshaw PF (1998) Lack of protection following immunisation with H. pylori outer membrane vesicles highlights antigenic differences between H. felis and H. pylori. FEMS Microbiol Lett 161:21–27
Kesavalu L, Ebersole JL, Machen RL, Holt SC (1992) Porphyromonas gingivalis virulence in mice: induction of immunity to bacterial components. Infect Immun 60:1455–1464
Kim J-H et al (2016) Extracellular vesicle–derived protein from Bifidobacterium longum alleviates food allergy through mast cell suppression. J Allegy Clin Immunol 137:507–516.e508
Klugman KP, Gotschlich EC, Blake MS (1989) Sequence of the structural gene (rmpM) for the class 4 outer membrane protein of Neisseria meningitidis, homology of the protein to gonococcal protein III and Escherichia coli OmpA, and construction of meningococcal strains that lack class 4 protein. Infect Immun 57:2066–2071
Knott GJ, Doudna JA (2018) CRISPR-Cas guides the future of genetic engineering. Science 361:866–869
Knox KW, Cullen J, Work E (1967) An extracellular lipopolysaccharide-phospholipid-protein complex produced by Escherichia coli grown under lysine-limiting conditions. Biochem J 103:192–200
Koeberling O, Seubert A, Granoff DM (2008) Bactericidal antibody responses elicited by a meningococcal outer membrane vesicle vaccine with overexpressed factor H-binding protein and genetically attenuated endotoxin. J Infect Dis 198:262–270
Kroniger T, Otto A, Becher D (2018) Proteomic analysis of bacterial (outer) membrane vesicles: progress and clinical potential. Expert Rev Proteomics 15:623–626
Kuipers K et al (2015) Salmonella outer membrane vesicles displaying high densities of pneumococcal antigen at the surface offer protection against colonization. Vaccine 33:2022–2029
Kulp A, Kuehn MJ (2010) Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol 64:163–184
Kwon SO, Gho YS, Lee JC, Kim SI (2009) Proteome analysis of outer membrane vesicles from a clinical Acinetobacter baumannii isolate. FEMS Microbiol Lett 297:150–156
Lamb RA, Zebedee SL, Richardson CD (1985) Influenza virus M2 protein is an integral membrane protein expressed on the infected-cell surface. Cell 40:627–633
Lee EY et al (2007) Global proteomic profiling of native outer membrane vesicles derived from Escherichia coli. Proteomics 7:3143–3153
Lee EY, Choi DS, Kim KP, Gho YS (2008) Proteomics in gram-negative bacterial outer membrane vesicles. Mass Spectrom Rev 27:535–555
Lee E-Y et al (2009) Gram-positive bacteria produce membrane vesicles: proteomics-based characterization of Staphylococcus aureus-derived membrane vesicles. Proteomics 9:5425–5436
Lee JC et al (2012) Klebsiella pneumoniae secretes outer membrane vesicles that induce the innate immune response. FEMS Microbiol Lett 331:17–24
Leitner DR, Feichter S, Schild-Prüfert K, Rechberger GN, Reidl J, Schild S (2013) Lipopolysaccharide modifications of a cholera vaccine candidate based on outer membrane vesicles reduce endotoxicity and reveal the major protective antigen. Infect Immun 81:2379–2393
Leitner D et al (2015) A combined vaccine approach against Vibrio cholerae and ETEC based on outer membrane vesicles. Front Microbiol 6:823. https://doi.org/10.3389/fmicb.2015.00823
Li M, Lee K, Hsu M, Nau G, Mylonakis E, Ramratnam B (2017) Lactobacillus-derived extracellular vesicles enhance host immune responses against vancomycin-resistant enterococci. BMC Microbiol 17:66–66
Lin DM, Koskella B, Lin HC (2017) Phage therapy: an alternative to antibiotics in the age of multi-drug resistance. World J Gastrointest Pharmacol Ther 8:162–173
Liu Y, Alexeeva S, Defourny KA, Smid EJ, Abee T (2018a) Tiny but mighty: bacterial membrane vesicles in food biotechnological applications. Curr Opin Biotechnol 49:179–184
Liu Y, Defourny KAY, Smid EJ, Abee T (2018b) Gram-positive bacterial extracellular vesicles and their impact on health and disease. Front Microbiol 9:1502–1502 d
Liu Y, Smid EJ, Abee T, Notebaart RA (2019) Delivery of genome editing tools by bacterial extracellular vesicles. Microb Biotechnol 12:71–73
Madico G et al (2006) The meningococcal vaccine candidate GNA1870 binds the complement regulatory protein factor H and enhances serum resistance. J Immunol 177:501–510
Manolova V, Flace A, Bauer M, Schwarz K, Saudan P, Bachmann MF (2008) Nanoparticles target distinct dendritic cell populations according to their size. Eur J Immunol 38:1404–1413. d
Marchand S, De Block J, De Jonghe V, Coorevits A, Heyndrickx M, Herman L (2012) Biofilm formation in milk production and processing environments; influence on milk quality and safety. Compr Rev Food Sci Food Saf 11:133–147
Masignani V et al (2003) Vaccination against Neisseria meningitidis using three variants of the lipoprotein GNA1870. J Exp Med 197:789–799
McConnell MJ, Rumbo C, Bou G, Pachón J (2011) Outer membrane vesicles as an acellular vaccine against Acinetobacter baumannii. Vaccine 29:5705–5710
Messiaen A-S, Forier K, Nelis H, Braeckmans K, Coenye T (2013) Transport of nanoparticles and tobramycin-loaded liposomes in Burkholderia cepacia complex biofilms. PLoS One 8:e79220–e79220. https://doi.org/10.1371/journal.pone.0079220
Mitra S, Chakrabarti MK, Koley H (2013) Multi-serotype outer membrane vesicles of Shigellae confer passive protection to the neonatal mice against shigellosis. Vaccine 31:3163–3173
Muralinath M, Kuehn MJ, Roland KL, Curtiss R 3rd (2011) Immunization with Salmonella enterica serovar Typhimurium-derived outer membrane vesicles delivering the pneumococcal protein PspA confers protection against challenge with Streptococcus pneumoniae. Infect Immun 79:887–894
Newman L et al (2015) Global estimates of the prevalence and incidence of four curable sexually transmitted infections in 2012 based on systematic review and global reporting. PLoS One 10:e0143304. https://doi.org/10.1371/journal.pone.0143304
Nieves W et al (2014) A Burkholderia pseudomallei outer membrane vesicle vaccine provides protection against lethal sepsis. Clin Vaccine Immunol 21:747–754
Oelschlaeger TA (2010) Mechanisms of probiotic actions – a review. Int J Med Microbiol 300:57–62
Olaya-Abril A et al (2014) Characterization of protective extracellular membrane-derived vesicles produced by Streptococcus pneumoniae. J Proteome 106:46–60
Osterholm MT, Kelley NS, Sommer A, Belongia EA (2012) Efficacy and effectiveness of influenza vaccines: a systematic review and meta-analysis. Lancet Infect Dis 12:36–44
Oyston PCF, Sjöstedt A, Titball RW (2004) Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat Rev Microbiol 2:967
Paolillo R, Romano Carratelli C, Sorrentino S, Mazzola N, Rizzo A (2009) Immunomodulatory effects of lactobacillus plantarum on human colon cancer cells. Int Immunopharmacol 9:1265–1271
Park K (2018) Inherent antimicrobial activity by bacteria-derived vesicles. J Control Release 290:180
Park M, Sun Q, Liu F, DeLisa MP, Chen W (2014) Positional assembly of enzymes on bacterial outer membrane vesicles for cascade reactions. PLoS One 9:e97103. https://doi.org/10.1371/journal.pone.0097103
Pérez O et al (2009) Mucosal approaches in Neisseria vaccinology. Vaccimonitor 18:55–57
Petousis-Harris H, Paynter J, Morgan J, Saxton P, McArdle B, Goodyear-Smith F, Black S (2017) Effectiveness of a group B outer membrane vesicle meningococcal vaccine against gonorrhoea in New Zealand: a retrospective case-control study. Lancet 390:1603–1610
Pierson T, Matrakas D, Taylor YU, Manyam G, Morozov VN, Zhou W, van Hoek ML (2011) Proteomic characterization and functional analysis of outer membrane vesicles of Francisella novicida suggests possible role in virulence and use as a vaccine. J Proteome Res 10:954–967
Pizza M et al (2000) Identification of vaccine candidates against Serogroup B Meningococcus by whole-genome sequencing. Science 287:1816
Prados-Rosales R et al (2014) Mycobacterial membrane vesicles administered systemically in mice induce a protective immune response to surface compartments of Mycobacterium tuberculosis. MBio 5:e01921
Price NL, Goyette-Desjardins G, Nothaft H, Valguarnera E, Szymanski CM, Segura M, Feldman MF (2016) Glycoengineered outer membrane vesicles: a novel platform for bacterial vaccines. Sci Rep 6:24931. https://doi.org/10.1038/srep24931
Pritsch M et al (2016) Comparison of intranasal outer membrane vesicles with cholera toxin and injected MF59C.1 as adjuvants for malaria transmission blocking antigens AnAPN1 and Pfs48/45. J Immunol Res 2016:11
Pupo E, Hamstra H-J, Meiring H, van der Ley P (2014) Lipopolysaccharide engineering in Neisseria meningitidis: Structural analysis of different pentaacyl lipid A mutants and comparison of their modified agonist properties. J Biol Chem 289:8668–8680
Qureshi K et al (2006) Breast milk reduces the risk of illness in children of mothers with cholera: observations from an epidemic of cholera in Guinea-Bissau. Pediatr Infect Dis J 25:1163–1166
Raetz CRH, Whitfield C (2002) Lipopolysaccharide endotoxins. Annu Rev Biochem 71:635–700
Raposo G, Stoorvogel W (2013) Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 200:373–383
Rappazzo CG et al (2016) Recombinant M2e outer membrane vesicle vaccines protect against lethal influenza A challenge in BALB/c mice. Vaccine 34:1252–1258
Rappuoli R (2000) Reverse vaccinology. Curr Opin Microbiol 3:445–450
Rivera J, Cordero RJB, Nakouzi AS, Frases S, Nicola A, Casadevall A (2010) Bacillus anthracis produces membrane-derived vesicles containing biologically active toxins. Proc Natl Acad Sci U S A 107:19002–19007
Roberts R et al (2008) Outer membrane vesicles as acellular vaccine against pertussis. Vaccine 26:4639–4646
Roier S et al (2016) A novel mechanism for the biogenesis of outer membrane vesicles in Gram-negative bacteria. Nat Commun 7:10515. https://doi.org/10.1038/ncomms10515
Rosenstein NE, Perkins BA, Stephens DS, Popovic T, Hughes JM (2001) Meningococcal disease. N Engl J Med 344:1378–1388. d
Salminen MK, Rautelin H, Tynkkynen S, Poussa T, Saxelin M, Valtonen V, Järvinen A (2004) Lactobacillus bacteremia, clinical significance, and patient outcome, with special focus on probiotic L. Rhamnosus GG. Clin Infect Dis 38:62–69
Salverda MLM et al (2016) Surface display of a borrelial lipoprotein on meningococcal outer membrane vesicles. Vaccine 34:1025–1033
Sanchez-Garcia L, Martin L, Mangues R, Ferrer-Miralles N, Vazquez E, Villaverde A (2016) Recombinant pharmaceuticals from microbial cells: a 2015 update. Microb Cell Factories 15:33. https://doi.org/10.1186/s12934-016-0437-3
Sato T, Yamada Y, Ohtani Y, Mitsui N, Murasawa H, Araki S (2001) Efficient production of menaquinone (vitamin K2) by a menadione-resistant mutant of Bacillus subtilis. J Ind Microbiol Biotechnol 26:115–120
Schild S, Nelson EJ, Camilli A (2008) Immunization with Vibrio cholerae outer membrane vesicles induces protective immunity in mice. Infect Immun 76:4554–4563
Schild S, Nelson EJ, Bishop AL, Camilli A (2009) Characterization of Vibrio cholerae outer membrane vesicles as a candidate vaccine for cholera. Infect Immun 77:472–484
Schroeder J, Aebischer T (2009) Recombinant outer membrane vesicles to augment antigen-specific live vaccine responses. Vaccine 27:6748–6754
Schultz MP, Bendick JA, Holm ER, Hertel WM (2011) Economic impact of biofouling on a naval surface ship. Biofouling 27:87–98
Schulz E et al (2018) Biocompatible bacteria-derived vesicles show inherent antimicrobial activity. J Control Release 290:46–55
Schwechheimer C, Sullivan CJ, Kuehn MJ (2013) Envelope control of outer membrane vesicle production in gram-negative bacteria. Biochemistry 52:3031–3040
Scoma A, Yakimov MM, Boon N (2016) Challenging oil bioremediation at deep-sea hydrostatic pressure. Front Microbiol 7:1203
Semchenko EA, Day CJ, Seib KL (2017) MetQ of Neisseria gonorrhoeae is a surface-expressed antigen that elicits bactericidal and functional blocking antibodies. Infect Immun 85:e00898–e00816
Semchenko EA, Tan A, Borrow R, Seib KL (2018) The serogroup B meningococcal vaccine Bexsero elicits antibodies to Neisseria gonorrhoeae. Clin Infect Dis 69:1101–1111. https://doi.org/10.1093/cid/ciy1061
Seo MK, Park EJ, Ko SY, Choi EW, Kim S (2018) Therapeutic effects of kefir grain lactobacillus-derived extracellular vesicles in mice with 2,4,6-trinitrobenzene sulfonic acid-induced inflammatory bowel disease. J Dairy Sci 101:8662–8671
Serruto D et al (2010) Neisseria meningitidis GNA2132, a heparin-binding protein that induces protective immunity in humans. Proc Natl Acad Sci 107:3770–3775. https://doi.org/10.1073/pnas.0915162107
Shen Y, Giardino Torchia ML, Lawson GW, Karp CL, Ashwell JD, Mazmanian SK (2012) Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe 12:509–520
Steeghs L et al (2008) Differential activation of human and mouse toll-like receptor 4 by the adjuvant candidate LpxL1 of Neisseria meningitidis. Infect Immun 76:3801–3807. https://doi.org/10.1128/iai.00005-08
Tan K, Li R, Huang X, Liu Q (2018) Outer membrane vesicles: current status and future direction of these novel vaccine adjuvants. Front Microbiol 9:783–783
Tingyue Gu DX (2015) The war against problematic biofilms in the oil and gas industry. J Microb Biochem Technol 07:1000e124. https://doi.org/10.4172/1948-5948.1000e124
Tinsley CR, Nassif X (1996) Analysis of genetic difference between Neisseria meningitidis and Neisseria gonorrhoeae: two closely related bacteria expressing two different pathogenicities. Proc Natl Acad Sci U S A 93:11109–11114
Tokuda H, Matsuyama S (2004) Sorting of lipoproteins to the outer membrane in E. coli. Biochim Biophys Acta 1693:5–13. https://doi.org/10.1016/j.bbamcr.2004.02.005
Treanor JJ (2015) Prospects for broadly protective influenza vaccines. Vaccine 33:D39–D45
Tricarico C, Clancy J, D’Souza-Schorey C (2017) Biology and biogenesis of shed microvesicles. Small GTPases 8:220–232
Tzipilevich E, Habusha M, Ben-Yehuda S (2017) Acquisition of phage sensitivity by bacteria through exchange of phage receptors. Cell 168:186–199.e112
Urbanek AK, Rymowicz W, Mironczuk AM (2018) Degradation of plastics and plastic-degrading bacteria in cold marine habitats. Appl Microbiol Biotechnol 102:7669–7678
Valentine Jenny L et al (2016) Immunization with outer membrane vesicles displaying designer glycotopes yields class-switched, glycan-specific antibodies. Cell Chem Biol 23:655–665
van de Waterbeemd B et al (2010) Improved OMV vaccine against Neisseria meningitidis using genetically engineered strains and a detergent-free purification process. Vaccine 28:4810–4816
van de Waterbeemd B, Mommen GPM, Pennings JLA, Eppink MH, Wijffels RH, van der Pol LA, de Jong APJM (2013) Quantitative proteomics reveals distinct differences in the protein content of outer membrane vesicle vaccines. J Proteome Res 12:1898–1908
van den Dobbelsteen GPJM, van Dijken HH, Pillai S, van Alphen L (2007) Immunogenicity of a combination vaccine containing pneumococcal conjugates and meningococcal PorA OMVs. Vaccine 25:2491–2496
van der Ley P, van der Biezen J, Poolman JT (1995) Construction of Neisseria meningitidis strains carrying multiple chromosomal copies of the porA gene for use in the production of a multivalent outer membrane vesicle vaccine. Vaccine 13:401–407. d
van der Ley P, Steeghs L, Hamstra HJ, ten Hove J, Zomer B, van Alphen L (2001) Modification of lipid a biosynthesis in Neisseria meningitidis lpxL mutants: influence on lipopolysaccharide structure, toxicity, and adjuvant activity. Infect Immun 69:5981–5990. https://doi.org/10.1128/iai.69.10.5981-5990.2001
van der Pol L, Stork M, van der Ley P (2015) Outer membrane vesicles as platform vaccine technology. Biotechnol J 10:1689–1706
van Dongen SFM, Nallani M, Cornelissen JJLM, Nolte RJM, van Hest JCM (2009) A three-enzyme cascade reaction through positional assembly of enzymes in a Polymersome Nanoreactor. Chem Eur J 15:1107–1114
Vriezema DM et al (2007) Positional assembly of enzymes in polymersome nanoreactors for cascade reactions. Angew Chem 119:7522–7526
Watkins HC et al (2017) Safe recombinant outer membrane vesicles that display M2e elicit heterologous influenza protection. Mol Ther 25:989–1002
Whelan J, Kløvstad H, Haugen IL, Holle MR-DRB, Storsaeter J (2016) Ecologic study of meningococcal B vaccine and Neisseria gonorrhoeae infection, Norway. Emerg Infect Dis 22:1137–1139. https://doi.org/10.3201/eid2206.151093
Xiang SD, Scholzen A, Minigo G, David C, Apostolopoulos V, Mottram PL, Plebanski M (2006) Pathogen recognition and development of particulate vaccines: does size matter? Methods 40:1–9
Xing H, Hwang K, Lu Y (2016) Recent developments of liposomes as nanocarriers for theranostic applications. Theranostics 6:1336–1352
Yoo J-W, Irvine DJ, Discher DE, Mitragotri S (2011) Bio-inspired, bioengineered and biomimetic drug delivery carriers. Nat Rev Drug Discov 10:521
Yun SH et al (2017) Proteomic characterization of the outer membrane vesicle of the halophilic marine bacterium Novosphingobium pentaromativorans US6-1. J Microbiol 55:56–62
Zakeri B, Fierer JO, Celik E, Chittock EC, Schwarz-Linek U, Moy VT, Howarth M (2012) Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc Natl Acad Sci U S A 109:E690–E697
Zariri A et al (2016a) Meningococcal outer membrane vesicle composition-dependent activation of the innate immune response. Infect Immun 84:3024–3033
Zariri A, Pupo E, van Riet E, van Putten JPM, van der Ley P (2016b) Modulating endotoxin activity by combinatorial bioengineering of meningococcal lipopolysaccharide. Sci Rep 6:36575. https://doi.org/10.1038/srep36575
Zepp F (2010) Principles of vaccine design—lessons from nature. Vaccine 28:C14–C24
Zomer A, Vendrig T, Hopmans ES, van Eijndhoven M, Middeldorp JM, Pegtel DM (2010) Exosomes: Fit to deliver small RNA. Commun Integr Biol 3:447–450
Acknowledgments
Funding for this work was provided by Core funds of the Naval Research Laboratory and the Office of the Secretary of Defense through the Applied Research for the Advancement of S&T Priorities Synthetic Biology for Military-Relevant Environments program. The authors would also like to thank and acknowledge Dr. Claretta Sullivan (Air Force Research Laboratory) for previous efforts in imaging OMV/MV formation in Lactobacillus acidophilus which contributed to Fig. 10.1.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Caruana, J.C., Walper, S.A. (2020). Bacterial Membrane Vesicles and Their Applications as Vaccines and in Biotechnology. In: Kaparakis-Liaskos, M., Kufer, T. (eds) Bacterial Membrane Vesicles. Springer, Cham. https://doi.org/10.1007/978-3-030-36331-4_10
Download citation
DOI: https://doi.org/10.1007/978-3-030-36331-4_10
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-36330-7
Online ISBN: 978-3-030-36331-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)