Abstract
Emerged as a highly versatile and applicable vertebrate animal model to study embryonic development, zebrafish over the last two decades became a valuable human disease model for cardiovascular research. The unprecedented in vivo imaging capabilities allowing live analysis of organ formation and organ function combined with the ease of precise genetic interrogation established zebrafish as a recognized cardiovascular disease model. This chapter provides an overview of zebrafish’s history in biomedical and particularly cardiovascular research, delivers an outline of available methodologies and resources when using zebrafish, and gives examples of the versatility of zebrafish in cardiovascular research and how it progressed our understanding of a given disease.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Emerged as a highly versatile and applicable vertebrate animal model to study embryonic development, zebrafish over the last two decades became a valuable human disease model for cardiovascular research. The unprecedented in vivo imaging capabilities allowing live analysis of organ formation and organ function combined with the ease of precise genetic interrogation established zebrafish as a recognized cardiovascular disease model. This chapter provides an overview of zebrafish’s history in biomedical and particularly cardiovascular research, delivers an outline of available methodologies and resources when using zebrafish, and gives examples of the versatility of zebrafish in cardiovascular research and how it progressed our understanding of a given disease.
9.1 From a Developmental Model to a Disease Model
9.1.1 The Beginnings and the Rise in Popularity
In the 1980s, Dr. George Streisinger and his colleagues at the University of Oregon laid the groundwork for using the zebrafish as a genetic model. He focused initially his pioneering work on developmental genetics based on several practical advantages and genetic amenities. The idea of having a vertebrate model with which genome-wide forward genetic screens are feasible as performed before in worms, flies, and plants to uncover novel gene functions ultimately materialized in 1989 was carried out by a colleague of Streisinger at the University of Oregon [1]. Inspired by the success and the potential of this first forward genetic screen, scientists from two groups in Boston and Tübingen conducted the first large-scale forward genetic screen in a vertebrate model. This screen recovered over 800 mutants with mutations in 372 genes uniquely involved in the development of the central nervous system, intestines, blood, bone, vasculature, and the heart to only name a few [2, 3]. Even over 20 years after conducting this screen, zebrafish mutants from it are still being used and analyzed in labs worldwide, illustrating the huge impact and the value this screen had and still has in the community.
Zebrafish are approximately 3–5-cm-long fish belonging to the teleost family with inexpensive housing and modest space requirements. When mated, a single female can produce between 100 and 300 eggs per week. Sexual maturity is reached with the age of 3 months. Particularly tempting as a developmental model is its extrauterine development and its fast ontogeny. Its small larval size of 1–2 mm allows analysis throughout the entire development, while macroscopic cell experiments like cell transplantations remain feasible and efficient. Gastrulation begins at around 5 h postfertilization (hpf) and segmentation, and with that somitogenesis, by 10 hpf (Table 9.1) [4]. Embryonic vasculogenesis is completed by 18 hpf, and angiogenesis can be observed before 20 hpf, and the first primitive heart tube initiates first contractions within the first day of development. Already 3 days postfertilization (dpf), the cardiovascular system of the zebrafish is comparable to that of a newborn mammal and reaches full maturity by 5 dpf, in a by-then organism exhibiting most of the internal organs found in humans [5]. In contrast to mammalian models, zebrafish does not rely on a functioning cardiovascular system during the first 5 days of life, since oxygen and nutrient supply is achieved exclusively through passive diffusion. Even substantial cardiovascular deficiencies can thus be tolerated and fish, even without any observable heart activity or blood flow, develop overly normal without systemic compromising impact due to secondary adverse effects within the first 5 dpf [6].
9.1.2 The Uprising Disease Model
Not only because of these obvious advantages but also with the 1996 mutagenesis screens and the recovery of mutant alleles modeling known human diseases, the zebrafish began its journey toward challenging the till-then dominating mouse model in recapitulating human disease conditions. As a nonmammalian model though, the zebrafish comes with apparent limitation, the most obvious being that zebrafish lack some of the organs found in humans, like lungs and mammary gland. The first zebrafish mutant reported modeling a human pathology and contributed to a better understanding of the underlying pathomechanism was yquem (ype) [7]. Zebrafish and humans share remarkable conservation in hematopoiesis, and ype presented a photosensitive porphyria syndrome as seen in patients. Although porphyria in humans was already associated to a gene encoding uroporphyrinogen decarboxylase (UROD), the study of ype now explained the exact enzymatic defect leading to the disease also in humans [7].
Subsequently, publications on zebrafish modeling various human pathologies dramatically increased. While modeling blood disorders in zebrafish dominated initially, zebrafish models for human diseases ranging from ocular disorders and neurodegenerative diseases to muscular dystrophies, cancer, addiction, behavioral disorders, and cardiovascular diseases have been described [8,9,10]. In particular, concerning congenital heart malformations, like valve deficiencies, to aneurisms, cardiomyopathies, and arrhythmias, zebrafish is now a well-established cardiovascular disease model. Although having a two-chambered heart instead of a four-chambered heart as in mammals, the zebrafish was proved to be particularly valuable for cardiac electropathophysiology (see section below) as compared to mice, since zebrafish action potential shape, heart rate, and the electrocardiogram closely resemble that of humans [11].
9.2 Genetics and Genetic Interrogation in Zebrafish
The ease of performing even large-scale genetic studies in zebrafish, which in the past also contributed significantly to a more detailed understanding of genetics in human disease, makes a high-quality genome sequence and complete annotation essential. The zebrafish is now further frequently used as an independent functional validation tool for findings from human genetics studies [12]. A recent study, initiated in 2001 by the Wellcome Sanger Trust Institute, identified and annotated 26,000 individual genes encoded on the zebrafish’s 25 chromosomes compared to the roughly 20,500 genes in the human genome [13]. Sequencing additionally revealed that teleosts underwent a whole-genome duplication, with 25–30% of the duplicated genes still retained in zebrafish [13, 14]. Thus, frequently duplicated genes are found when screening for human disease gene orthologues, adding potentially to a functional validation experimentation since duplicated genes often exhibit subfunctionalization properties. Seventy percent of the human genes have a direct homologue in the zebrafish. More astounding is that 84% of the known human disease genes have a direct homologue in fish [13].
Along with this solid foundation, the zebrafish comes with a highly versatile tool set to interrogate gene functions and identify genes with so far unappreciated functions. As traditionally introduced and envisioned by its pioneers, the zebrafish was and is a powerful forward genetics model.
9.2.1 Forward Genetics
Classical chemical mutagenesis screens, in particular deploying N-ethyl-N-nitrosourea (ENU), are easy and unprecedented in zebrafish compared to any other vertebrate model and enable production of thousands of mutants in a relatively short time (reviewed here [15]). While being easy and producing mutations very efficiently with little genomic bias, chemical mutagenesis screens have the disadvantage that subsequent identification of the gene mutation causative for the observed phenotype is time- and resource-consuming. To overcome this limitation, insertional mutagenesis techniques using replication-deficient pseudotyped retroviruses or transposons were introduced by several groups [16,17,18]. In addition, and initially adapted from mouse, the Sleeping Beauty DNA transposon somatic mutagenesis system was successfully introduced in zebrafish and led to the identification of conserved and novel human cancer genes in zebrafish [19]. Insertional mutagenesis, due to its predesigned insertional cassettes, including reporters, enhancers, silencers, and amplification tags, allows rapid identification of the targeted gene. However, this comes with the price that retroviruses and transposons typically achieve a very much lower mutagenic efficiency than ENU, requiring considerably larger libraries of mutagenized fish.
Using sensitized genetic backgrounds to perform modifier screens is a powerful way to isolate genetic interactions of genes. Since screening of several genomes and large libraries of mutagenized animals is necessary, this was longtime reserved for invertebrate models, including Drosophila and C. elegans. Kramer et al. performed the first enhancer screen for modifiers affecting dorsoventral patterning by using heterozygous zebrafish mutants for the chordin gene and identified initially seven enhancer mutations [20]. Bai et al. screened for genetic suppressors of a mutation in the moonshine (mon) gene, which encodes for the transcriptional intermediary factor 1 gamma (tf1γ) [21]. mon mutants exhibit anemia and larval lethality. They utilized a complex haploid screening strategy and identified the sunrise mutation that fully normalized globin content in mon mutants. Interestingly, sunrise encodes for cdc73, a component of the Pol II transcriptional elongation complex, for the first time linking tf1γ function and transcriptional elongation to erythroid development.
9.2.2 Reverse Genetics
The so far numerous forward screens performed in zebrafish to date, despite the significant time, staff, and resource efforts, have led to the identification of plentiful new genes and conserved pathways involved in human disease, including cardiovascular disease [22,23,24]. However, not only because zebrafish became a valuable model to functionally validate candidate genes but also because of advances in sequencing and other large-scale gene identification technologies, including high-throughput automated gene expression analysis, i.e., in situ hybridization screenings, reverse genetic tools to interrogate the function of distinct genes have been established [25, 26]. Development of available technologies adapted to zebrafish research accelerated dramatically during recent years (Table 9.2).
First reverse genetics in zebrafish was performed based on technologies that were well established and combined with advanced sequencing leading to targeting induced local lesions in genomes, or TILLING for short [28]. In TILLING, libraries of ENU-mutagenized F1 families are sequenced and sperm cryopreserved. Sperm can be revitalized by in vitro fertilization when a locus of interest is affected by an induced mutation. Hundreds and thousands of cryopreserved sperm from TILLING efforts are stored in huge facilities and catalogued. They are available to the research community through the Zebrafish International Research Center (ZIRC, www.zebrafish.org) at the University of Oregon, USA, and the European Zebrafish Research Center (EZRC, http://www.ezrc.kit.edu) at the Karlsruhe Institute for Technology (KIT), Germany [29].
With the introduction of morpholino-modified antisense oligonucleotides (MO) adapted from Xenopus, an easy, fast, efficient, and specific gene silencing tool when well controlled [30] to analyze the function of a distinct gene became available [31]. MOs do not target the genomic locus of a gene but rather bind to the transcript to cause aberrant translation or splicing. They do so either by targeting the translational start site to block translation completely or by targeting splice-site junctions in the pre-mRNA causing either exon skipping or intron integration resulting in frameshift and premature stop codon generation [31, 32]. While translation blocking MOs require an antibody specific for the targeted protein to assess efficacy, the efficiency of disruption of splicing by the splice-site MOs can be easily evaluated by RT-PCR. The development of photoactivatable MOs (caged MOs) helped to overcome the limitation that MOs act globally and constitutively [33]. Caged MO activity can be temporarily and even spatially controlled by applying cellular resolution light, i.e., using two-photon microscopes [34]. MOs proved to be a versatile tool not only to inhibit gene function but also to confirm genetic interactions of genes when titrated and co-injected, for gene dosage experiments, to suppress miRNAs by targeting miRNA maturation or for miRNA target validation by interfering with miRNA target 3′-UTR binding [35,36,37]. Despite being extremely valuable, MOs come with the disadvantage of having transient effects because they do not cause genetic lesions, allowing gene knockdown only for about 3 days [31]. Further, MO experiments require well-trained and experienced staff to control for potential off-target effects and toxicity [30, 38]. Recently, discrepancies between the observed phenotypes induced by MOs and stable mutants were reported, which can be in part explained by genetic compensation mechanisms occurring in mutants but not when the gene is transiently inactivated as in morphants, possibly thereby obscuring phenotypes otherwise visible after MO use [27, 39].
The latest development of advanced genome-editing technologies complements the existing reverse genetics tools in zebrafish. First, zinc finger nucleases (ZFN), then TALE nucleases, and ultimately CRISPR/Cas9 have been shown to be highly effective in zebrafish, and particularly, CRISPR enables cheap and relatively effortless generation of mutants in virtually every lab [40,41,42,43,44,45]. Narayanan et al. recently demonstrated the feasibility of simultaneous in vivo mutagenesis of entire gene and miRNA gene families by multiplex CRISPR/Cas9, highlighting the versatility of this system [46]. Multiplexed CRISPR/Cas9 was subsequently employed to demonstrate that miRNAs provide phenotypic robustness and protect the vascular systems from changing environmental conditions [47].
Besides inducing simple lesions, CRISPR/Cas9 enables targeted genome editing allowing introduction and correction of distinct mutations, integration of exogenous sequences like LoxP, and even introduction of reporter genes, including GFP [48]. However, besides the very well and efficiently performing NHEJ (non-homologues end-joining), HDR (homology-directed repair) seems to work rather inefficient in zebrafish [48, 49].
As in other animal models, transgenic strategies are another valuable reverse genetics tool to elucidate gene function. Either wild-type forms or constitutive active or dominant-negative isoforms of the gene of interest can be transiently, by simple RNA injections, or stably, by stably integrated transgenesis, expressed. Generation of a stable transgenic line takes between 6 and 12 months. Here, also a diverse tool set, including GAL4/UAS- and Cre/LoxP-based systems, and a diverse set of promoters, including cell-type-specific promoter elements or tamoxifen and heat shock-responsive elements to drive spatially and temporally controlled expression of the transgene, are available [50].
9.2.3 Genetic Diagnostic Testing Using Zebrafish
Current large-scale genomic sequencing studies of human patients have identified many disease-associated genetic variants at an accelerating pace and at ever-decreasing costs. One of the major challenges that we are facing with all these genomic data is to determine the pathogenicity of newly identified candidate disease genes and genetic variants. This is particularly true for the ones that have not been previously evaluated in an experimental model system. Owing to the abovementioned advantages as a vertebrate model system, zebrafish has emerged as an attractive model to determine the cause-effect relationship between the genetic variants and the human defects and the underlying molecular disease mechanisms.
Giving the versatile genetic tool set in zebrafish, the ability of genetic diagnostic testing ranges from simple gene causality and single variant testing to the identification and characterization of more complex disease genetics. We will focus here on how the zebrafish can be used to test for a variety of genetic testing scenarios; distinct examples can be found in the disease entity sections below.
(I) Using transient MO-mediated knockdown or stable CRISPR/Cas9 knockouts as a tool to identify disease causality of a gene from single genes or from a set of candidates isolated through, i.e., GWAS, enables simple and fast causality screening. Gene candidates mapped for disorders segregating under a recessive mode of inheritance can be easily and rapidly tested. Phenocopy of large parts of the phenotypic spectrum found in the respective patient cohort allows for identification and functional verification of identified null variants with predicted nonsense, frameshift, or splice-site mutations. (II) The power of zebrafish is the ability to transiently overexpress genes or gene variants by injection of in vitro generated RNA carrying the desired mutation. This allows very specific in vivo variant testing. In vivo complementation experiments by combining mRNA injection of transcripts carrying the to-be tested mutation with MO-mediated knockdown or stable knockout result in more specific mutation-focused testing. The loss-of-function effect of the variant is proven by the inability to rescue the induced gene loss. This method is very helpful to identify null variants in a scenario of recessively inherited disorder. Injection of variants into wild-type animals enables identification of dominant-negative acting gene mutations. If small amounts of the mutant variant cause a disease phenotype, while injection of equal amounts of the wild-type variant has no effect, this indicates toxicity of the mutant allele and hence dominant-negative mode of action [51]. (III) The ease to generate stable mutants in single or multiple genes at once and the availability of a huge variety of mutant alleles through international resource centers as mentioned above enable even the unraveling of more complex genetic traits in vivo. Combining mutant alleles by multiplex targeting through CRISPR or compound mutation generation by crossing of different mutant lines allows demonstration of genetic interactors and modifiers of phenotypic traits [47, 52]. Convenient is the use of MOs, since here the amount of gene reduction can be titrated. Combining carefully titrated MOs targeting different specific genes can thus be used to prove genetic interaction and disease modifiers in an in vivo setting with little time effort [35].
The applicability of zebrafish in the diverse genetic scenarios is summarized in Table 9.3.
9.3 Cardiovascular Disease Models
9.3.1 Congenital Heart Disease Modeling in Zebrafish
Congenital heart diseases (CHDs) are the leading cause of human birth defects, and many of these diseases originate from genetic defects that impact cardiac development and morphogenesis [58, 59]. During development, the heart is the first organ to form and function. After its initial formation at the ventral midline, the vertebrate heart undergoes a series of complex morphogenetic processes that transform the linear heart tube into a functional pumping organ (Fig. 9.1a–g) [5, 63, 64]. Although the zebrafish heart appears to be structural simpler than its counterpart in higher vertebrates, much of the molecular and cellular mechanisms governing cardiac formation and further maturation are conserved between zebrafish and higher vertebrates [5, 23, 60, 64,65,66].
The embryonic and adult zebrafish hearts. (a) The zebrafish linear heart tube at 24 hpf. (b) The looping zebrafish linear heart tube at 36 hpf. (c) The two-chambered zebrafish heart at 48 hpf. (d) The zebrafish heart grows in size by 5 dpf. (e) Cross-sectional view of 5 dpf zebrafish heart showing ventricular protruding trabeculae, cardiac valves, and the outmost epicardial layer. Modified from Brown et al. JCC 2016 [60]. (f, g) Confocal projections of 3 and 5 dpf zebrafish hearts, respectively. From Samsa et al., Development 2015 [61]. (h) Schematics of juvenile and adult zebrafish hearts at the top and an isolated adult heart at the bottom. (i–l) Control hearts (i) and hearts with left-right patterning defects (j–l) stained by in situ hybridization labeling the cardiac-specific myosin light chain 2. From Lai et al. Development 2012 [62]
CHDs in humans feature a wide variety of structural abnormalities that affect nearly every structure of the developing heart including the myocardium, the valves, and the great vessels [59]. If left uncorrected, in severe cases, the structural defects will lead to cardiac dysfunction and thus negatively impact life quality of the affected individual. In zebrafish, myocardial wall abnormalities manifest as either hypoplastic ventricle or hypotrabeculation, both of which cause progressive reduction in cardiac contractility [67]. Cardiac valve malformations often arise from defective endocardial cell migration and/or remodeling, leading to improper endocardial cushion and/or valve leaflet formation [23, 68, 69]. These defects can result in blood regurgitation and altered intracardiac fluid dynamics [68, 69]. Other cardiovascular defects in zebrafish include heterotaxy whereby the relative position of the zebrafish cardiac chambers is altered [70, 71], and malformations of the pharyngeal arch arteries (PAAs), the transient embryonic blood vessels that eventually give rise to the carotid arteries and great vessels of the heart [72, 73].
Hypoplastic left heart syndrome (HLHS) is a rare but complex CHD characterized by severe hypoplastic left ventricle and extremely underdeveloped mitral and aortic valves [74, 75]. These severe structural defects render the left side of the heart less efficient in pumping blood to the body, and surgical corrections are needed to improve the survival of HLHS infants [76]. HLHS usually occurs as isolated disease and the associated genetic loci include, among others, those encoding the transcriptional factors NKX2.5 and HAND1 and the gap junction protein GJA1 [77,78,79]. Interestingly, recent studies in zebrafish revealed an important role for Nkx2.5 and its homologue Nkx2.7 in chamber identity maintenance [80, 81]. Although the numbers of differentiated ventricular and atrial cardiomyocytes (CMs) are not affected by the loss of nkx2.5 function at the linear heart tube stage, zebrafish nkx2.5 mutant demonstrates a nearly 50% decrease in the number of ventricular CMs with a corresponding increase in the number of atrial CMs starting from cardiac looping stage. These nkx2.5 mutant phenotypes are further enhanced by a loss of nkx2.7 function. These observations suggest that these two Nkx factors play critical role in maintaining ventricular and atrial cell identity during the time of cardiac loop and chamber emergence [80, 81]. Although no studies have reported an enlargement of atria in HLHS human patients, studies using zebrafish might still provide mechanistic clue as to why HLHS patients develop miniature left ventricles. Combing mouse forward genetics and further validation by CRISPR/Cas9 gene editing in mouse and zebrafish, a recent study showed that Sap130, a histone deacetylase complex subunit, mediates left ventricular hypoplasia, further demonstrating the value of zebrafish as a powerful genetic system to model HLHS [82].
Cardiac valves are critical components in the heart that function to ensure unidirectional blood flow (Fig. 9.2e). Human patients with congenital valve defects would present either obstructed or backward blood flow due to valvular stenosis and valvular insufficiency, respectively [85,86,87]. In zebrafish, the atrioventricular (AV) valve that forms at the AV junction is the most well-studied valvular structure [23, 68, 69, 88, 89]. At 48 hours postfertilization (hpf), as the AV junction constricts and demarcates the two cardiac chambers, the endocardial cells at the junction begin to differentiate and form a specialized ringlike structure, the AV canal, to help reduce retrograde blood flow [68, 69]. Around 72 hpf, the AV endocardial cells further invaginate and generate primitive valve leaflets allowing for complete prevention of regurgitation [69]. Due to the conserved molecular mechanisms underlying valve formation between zebrafish and higher vertebrates, zebrafish has been utilized to evaluate the potential in vivo effect of genetic variants associated with human congenital valve defects. By sequencing 32 candidate genes implicated in atrioventricular septum (AVS) development, a recent study identified 11 detrimental genetic variants associated with AVS defects [57]. Further functional analysis demonstrated that expression of a patient-specific ALK2 L343P variant in zebrafish embryos led to aberrant expression of endocardial cushion marker genes and disrupted AV canal formation. More recently, Durst et al. identified DCHS1 missense mutations as an inherited risk factor for mitral valve prolapse (MVP) [53]. DCHS1 encodes a member of the cadherin superfamily involved in cell polarity. MO-mediated knockdown of the zebrafish homologue dchs1b compromised AV canal formation and resulted in retrograde blood flow [53]. Intriguingly, the wild-type human DCHS1, but not the two DCHS1 variants (P197L, R2513H), rescued the AV canal defects caused by dchs1b knockdown in zebrafish embryos, further supporting the link between the familiar mutations with MVP [53].
Zebrafish as a vascular model. (a) Vasculature of a transgenic Tg(kdrl:EGFP) embryo at 5dpf. (b, c) Cardiovascular defects in zebrafish ccm2 mutants, characterized by endocardial overproliferation (b′) and increased sprout- and branch point formation (c′) at 48hpf. From Renz et al. [83] (d) Zebrafish as a model for atherosclerosis. Lipid deposits labeled by fluorescent lipids in the vascular wall of Tg(kdrl:EGFP) transgenic animal [84]
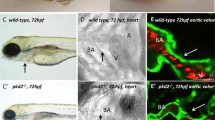
Heterotaxy is a rare birth defect featuring randomization of visceral organ situs [90]. Although situs inversus leads to little or no medical problems, heterotaxy often causes structural abnormalities in the heart and/or other major visceral organs. The heart is the first organ that responds to laterality signals and thus exhibits left-right asymmetry (Fig. 9.1h–k). In addition to the positioning defect, other cardiac defects seen in heterotaxy patients are transposition of the great arteries and septal defect [91,92,93]. Because of an essential role of the motile cilia in left-right patterning, genetic mutations that affect motile cilia formation and/or function and the downstream nodal signaling that controls left-right patterning result in heterotaxy and cardiac structural abnormalities [94]. To date, targeted candidate exome sequencing has identified many human mutations in ciliary dynein components, such as dynein intermediate chain 1 (DNAI1) [95, 96], dynein heavy chain 5 (DANH5) [95], dynein heavy chain 6 (DNAH6) [97], and left-right dynein (DNAH11) [96], in heterotaxy patients. Selection of the candidate genes was based on genetic data obtained from model organisms, in particular Chlamydomonas mutants with the defective structure and/or function of flagella [98]. Intriguingly, recent human genetic studies have identified novel heterotaxy-causing genes, some of which have unanticipated roles in cilia structure and function or potentially affect the downstream signaling controlling left-right patterning [99].
9.3.2 Cardiomyopathy Modeling in Zebrafish
In 2002, with the zebrafish mutants silent heart (sih) and pickwick (pik) harboring mutations in cardiac troponin and titin, respectively, rendering the hearts a contractile and recapitulating human cardiomyopathy, the usefulness of zebrafish as a cardiomyopathy model became obvious [100, 101]. Both genes are associated with human hypertrophic (HCM) and dilated cardiomyopathy (DCM) [102, 103]. In subsequent years, many more cardiomyopathy-causing genes were evaluated using the zebrafish. Roughly 96% of all known human cardiomyopathy-causing genes have a direct zebrafish orthologue, supporting the usefulness of zebrafish [104]. The zebrafish even helped to identify novel DCM genes, i.e., Nexilin, which was first identified as a novel component of the sarcomeric z-disk in zebrafish and then confirmed to be a molecular base of human DCM by a genetic screening of patients with idiopathic DCM [51]. Strikingly, even the ultrastructural characteristics of Nexilin loss described in zebrafish were mimicked in human Nexilin mutation carriers. Subsequently, Nexilin was also linked to human HCM [105]. Vogel et al. systematically evaluated the usefulness of zebrafish as a disease animal model for human DCM [106]. They knocked down a diverse set of known human DCM genes and evaluated the resulting phenotypes. Inactivation of all genes not only reliably caused DCM, but zebrafish morphants even recapitulated gene-specific disease characteristics, highlighting the value of zebrafish as a diagnostic model to evaluate human cardiomyopathy genetics. Genes evaluated in zebrafish quickly expanded from merely sarcomeric proteins to signaling molecules, transcription factors, and cytoskeletal and other regulatory proteins [106,107,108,109,110,111]. Zebrafish models contributed dramatically to novel mechanistic insights of human cardiomyopathy, as exemplified by a study from Zou et al. The authors used CRISPR technology to generate distinct titin mutants, leading to the identification of a human conserved titin internal promoter that can rescue favorably mutations residing in the N-terminus of the protein. This renders N-terminal mutations less severe than mutations affecting the C-terminus and might serve as an explanation for the observed variability in expressivity and severity in human titin mutation carriers [112].
Besides larval zebrafish, adults recently are used to model cardiomyopathy. Human cardiomyopathy usually affects adult individuals with disease mutations predominantly being recessively inherited. While previously homozygous zebrafish mutants with manifestation of disease during larval developmental stages were predominantly analyzed, adult zebrafish might represent a model closer modeling the human pathology. The adult zebrafish heart consists of all cell types found in the human heart, including cardiomyocytes, fibroblasts and endothelial cells, while the larval heart mainly contains cardiomyocytes. Common pathways involved in disease progression in human heart disease, including the β-adrenergic system, were shown to function more similarly in the adult than in the larval heart [113]. A few adult cardiomyopathy models exist that contributed significantly to a deeper understanding of disease progression and potential therapeutic opportunities [114,115,116,117]. A recent study aimed at understanding the basis for the heterogeneity of observed phenotypes in essential myosin light chain (MYL3) mutation carriers, which can be observed even in patients carrying the same mutation [118]. The authors used adult, heterozygous mutant fish, mimicking human dominant mutation carriers. Combining functional analyses with molecular and biochemical assays, the study provides evidence that MYL3 is essential to adapt heart function to physical stress. Heterozygous mutant fish developed cardiomyopathy and increased lethality only after exposure to forced swimming while being unremarkable under normal conditions. Variability in expressivity and severity of MYL3-associated cardiomyopathy in humans hence depends on the degree of the stress the heart experiences during lifetime, and controlling cardiovascular stress might represent a possibility to reduce disease symptoms [118].
While being valuable to more closely resemble human cardiomyopathy, using adult zebrafish comes with disadvantages. The biggest disadvantage is time, since generating stable mutants is significantly more time-consuming than MO-based larval studies. Further, analysis of heart function in larvae is very easy and can be performed by using a simple light microscope. Because adult zebrafish lack transparency, more advanced imaging modalities have to be used, demanding higher costs and highly trained personnel. In accordance to mammalian studies, echocardiography was adapted to analyze heart function in adult zebrafish, now allowing the assessment of conventional echocardiographic parameters to advanced analysis of detailed myocardial mechanics deploying modern speckle tracking [118,119,120]. And last but not least, the ability of using larval zebrafish to perform high-throughput screens (see section below) is unprecedented and is impossible to match with adult zebrafish. Nevertheless, adult zebrafish cardiomyopathy models are rising and will more and more complement larval studies in the future.
9.3.3 Arrhythmias
While murine hearts beat with up to 900 beats per minute (bpm), zebrafish hearts contract with a frequency of 60–100 bpm, thus much more closely resembling heart rates of humans. This resemblance is furthermore evident in the very similar form and kinetic of a zebrafish ventricular action potential and the echocardiogram [11]. Drugs targeting late repolarization channels induce confound arrhythmias in zebrafish while being only marginally functional in mice. Although being very small, the larval zebrafish heart is able to present a variety of different arrhythmic flavors, including tachycardia, bradycardia, atrioventricular block (AV block), atrial fibrillation, and sinus exit block. First evidence for the applicability of zebrafish as an arrhythmia model came from the island beat (isl) mutant in 2001, harboring a mutation in the cardiac L-type calcium channel [121]. isl larvae display a noncontractile ventricle and an asynchronously contracting atrium, resembling atrial fibrillation. Breakdance mutant zebrafish (bre), carrying loss-of-function mutations in the zebrafish orthologue of the human HERG channel, model long-QT syndrome 2 (LQT2) [122, 123]. bre hearts develop a second-degree AV block, with the ventricle skipping every other beat of the atrium (2,1 rhythm). Using a morpholino targeting kcnh6 or high dosages of HERG-blocking drugs, such as terfenadine or E-4031, enables to induce a 3:1 and 4:1 and even up to a third-degree AV block. bre fish are very useful to screen for modifiers of LQT2 in vivo [124, 125]. Besides LQT, with reggae (reg) mutant zebrafish, one of the first animal models for the human short-QT syndrome 1 (SQT1) was introduced [126]. reg mutant fish carry a gain-of-function mutation in the zebrafish HERG channel and display a whole range of phenotypes, including complete cessation of contractility over up to hours, to atrial fibrillation and sinus exit block. A sodium-calcium exchanger (NCX1) deficiency was reported for the tremblor (tre) mutant that displays chaotic and dyssynchronized cardiac contractions and atrial fibrillation due to abnormal calcium transients [127]. A second tre mutant, besides being important for rhythmicity, indicated a role for NCX1 for normal cardiac development and sarcomere formation, since this mutant displayed malformed hearts and severely disrupted sarcomeres [128].
A direct implication for human genetics came from a zebrafish transgenic line that expressed a mutation in the SCN5A sodium channel frequently associated with conduction disease, sinus node dysfunction, atrial and ventricular arrhythmias, and dilated cardiomyopathy in patients [129]. Mutant SCN5A expressing zebrafish developed bradycardia, conduction system abnormalities, and premature death, suggesting conserved functions in the heart and demonstrating the usefulness of zebrafish as an in vivo screening model to distinguish benign from functional genetic variants found in humans with arrhythmias [129].
9.3.4 Vascular Disease
Zebrafish offers the ability to choose from a diverse range of transgenic reporter lines labeling vascular endothelial cells, lymphatic endothelial cells, or vascular smooth muscle cells and pericytes [68, 130,131,132,133,134]. Combined with its exceptional imaging capabilities, this facilitates unprecedented visualization of organ formation and malformation as well as vascular function and malfunction in vivo (Fig. 9.2a). Studies in zebrafish have contributed greatly to advance our understanding of vascular biology and disease. The prominent role of Notch signaling for arterial-venous specification and its function as a determining factor to select tip and stalk cell fates during angiogenesis was first described in zebrafish [135, 136]. Further, our views of early formation of the first embryonic artery and vein were reinvented with observations in zebrafish that the first embryonic vein forms by selective sprouting of progenitor cells from a common arterial and venous precursor vessel subsequently undergoing fate segregation regulated through the ligand EphrinB2 and its receptor EphB4 [137].
Defects in vascular integrity and resulting hemorrhage were among the first pathologies described in zebrafish, advancing our understanding of factors essential for vessel integrity and of genes involved in human disease conditions involving hemorrhage formation [24, 138,139,140,141,142,143,144,145]. The cerebral cavernous malformation (CCM) protein complex plays a crucial role for normal blood vessel development and vascular integrity [146]. CCMs are vascular lesions characterized by enlarged thin-walled blood vessels and lack of supporting subendothelial cells such as smooth muscle or astrocytic foot processes. In patients, CCMs are primarily found within the neurovasculature of the central nervous system and often cause headaches, seizure, or often lethal cerebral hemorrhages due to a loss of function of at least one of the three genes, KRIT1/CCM1, CCM2/OSM, or CCM3/PDCD10. In zebrafish, ccm mutants exhibit proliferation and sprouting defects in endocardial and endothelial cells (Fig. 9.2b, c). Mechanistically, the loss of CCM proteins results in a ß1 integrin-dependent overexpression of the zinc finger transcription factors Klf2a and Klf2b, which in turn causes an upregulation of endothelial-specific factor egfl7, thereby promoting excessive angiogenesis. Pharmacological inhibition of VEGF signaling or klf2a/b knockdown by antisense oligonucleotide morpholino injection rescued ccm mutant cardiovascular defects [83]. Thus, zebrafish can help to further unravel so far unrecognized pathomechanistic insights of human diseases and enables identification of potential therapeutic targets.
Worldwide, atherosclerosis is the leading cause of death. It is a pathological process of inflammation and progressive deposition of cholesterol, cellular debris, and calcium in the artery walls. Zebrafish, with its unique visualization abilities by combining transgenic reporter lines with biological indicator dyes, can help to identify novel mechanisms and candidate genes and their role in the pathogenesis of the disease to ultimately screen for new druggable targets [147, 148]. Recent studies have shown that zebrafish fed with high-cholesterol diet (HCD) mimic lipid deposition within arterial vessels in humans (Fig. 9.2d) [84]. Combined with transgenic lines marking leukocytes or macrophages, zebrafish becomes a powerful tool to analyze inflammatory modulating drugs on the progression of atherosclerosis in vivo [149].
9.4 Zebrafish in High-Throughput Drug Screens
Over the last 100 years or so, classical drug development has been employing two broad types of small molecule screening strategies, the phenotypic screening and the target-based screening, to identify small molecules that can be used as lead compound for novel pharmaceutical drugs [150]. Historically, new drugs discovery has been mostly relied on phenotype-based screening that utilizes cellular or animal models to search for compounds that induce desirable phenotypic change(s). Since 1980s, owing to the advances in molecular biology and genomics, target-based screening, which aims to identify small molecules against defined molecular targets implicated in human diseases, has immediately gained popularity. Nevertheless, there are a few advantages for phenotypic screening over the target-based screening as a tool of choice for lead discovery [151, 152]. For instance, phenotypic screening doesn’t require prior knowledge about the molecular target to identify drugs that produce therapeutic effects. This strategy also has the potential to identify compound that alleviates a diseased phenotype through targeting multiple biological targets. As such, the last two decades has witnessed the emergency of zebrafish as an animal model to the forefront of phenotype-based small molecule screening [152].
In 2000, Peterson et al. published the very first whole-organism-based small molecule screen and identified compounds that affected various aspects of early zebrafish development [153]. The same group also performed the first chemical suppressor screen aiming to reverse the coarctation phenotype observed in zebrafish gridlock mutants [154,155,156]. gridlock harbors a hypomorphic mutation in the hey2 gene that encodes a bHLH transcriptional repressor [155]. After screening 5000 small molecules, two structurally related compounds were identified to suppress the gridlock coarctation phenotype likely through upregulating VEGF expression [154]. Given the resemblance of zebrafish cardiac electrophysiology to that of human hearts [11], zebrafish has been served as a particularly valuable model for identifying therapeutics that can rescue arrhythmia defects. By screening 1200 commercially available small molecules, Peal et al. identified two compounds that reproducibly rescued breakdance mutant long-QT defect [125]. In recent years, phenotype-based screening using zebrafish has benefited significantly from the latest developed motorized robotics systems. These high-throughput tools and technologies facilitate embryo dispensation, compound delivery, and incubation, as well as image acquisition and analysis of a variety of parameters to grasp the complexity of cardiac function, and will undoubtedly accelerate the identification of cardiovascular disease therapeutics in whole-organism-based systems.
9.5 Conclusions
-
A variety of human cardiovascular diseases can be modeled in zebrafish.
-
Availability of a highly versatile genetic tool set allows easy and fast interrogation of gene function and evaluation of disease causality of gene candidates and even provides insights into functional genetic networks.
-
The unprecedented visualization capabilities of zebrafish enable phenotypic characterization from whole organ level down to cell and subcellular characterization.
-
As a future prediction, zebrafish will urge into high-throughput drug discovery and will accelerate the identification of disease, including cardiovascular disease, therapeutics in whole-organism-based systems in the future.
References
Kimmel CB. Genetics and early development of zebrafish. Trends Genet. 1989;5:283–8.
Driever W, Solnica-Krezel L, Schier AF, Neuhauss SC, Malicki J, Stemple DL, Stainier DY, Zwartkruis F, Abdelilah S, Rangini Z, Belak J, Boggs C. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37–46.
Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, van Eeden FJ, Jiang YJ, Heisenberg CP, Kelsh RN, Furutani-Seiki M, Vogelsang E, Beuchle D, Schach U, Fabian C, Nusslein-Volhard C. The identification of genes with unique and essential functions in the development of the zebrafish, danio rerio. Development. 1996;123:1–36.
Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310.
Stainier DY, Lee RK, Fishman MC. Cardiovascular development in the zebrafish. I. Myocardial fate map and heart tube formation. Development. 1993;119:31–40.
Just S, Meder B, Berger IM, Etard C, Trano N, Patzel E, Hassel D, Marquart S, Dahme T, Vogel B, Fishman MC, Katus HA, Strahle U, Rottbauer W. The myosin-interacting protein smyd1 is essential for sarcomere organization. J Cell Sci. 2011;124:3127–36.
Wang H, Long Q, Marty SD, Sassa S, Lin S. A zebrafish model for hepatoerythropoietic porphyria. Nat Genet. 1998;20:239–43.
Phillips JB, Westerfield M. Zebrafish models in translational research: tipping the scales toward advancements in human health. Dis Model Mech. 2014;7:739–43.
Ablain J, Zon LI. Of fish and men: using zebrafish to fight human diseases. Trends Cell Biol. 2013;23:584–6.
Santoriello C, Zon LI. Hooked! Modeling human disease in zebrafish. J Clin Invest. 2012;122:2337–43.
Verkerk AO, Remme CA. Zebrafish: A novel research tool for cardiac (patho)electrophysiology and ion channel disorders. Front Physiol. 2012;3:255.
Davis EE, Frangakis S, Katsanis N. Interpreting human genetic variation with in vivo zebrafish assays. Biochim Biophys Acta. 1842;2014:1960–70.
Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, Collins JE, Humphray S, McLaren K, Matthews L, McLaren S, Sealy I, Caccamo M, Churcher C, Scott C, Barrett JC, Koch R, Rauch GJ, White S, Chow W, Kilian B, Quintais LT, Guerra-Assuncao JA, Zhou Y, Gu Y, Yen J, Vogel JH, Eyre T, Redmond S, Banerjee R, Chi J, Fu B, Langley E, Maguire SF, Laird GK, Lloyd D, Kenyon E, Donaldson S, Sehra H, Almeida-King J, Loveland J, Trevanion S, Jones M, Quail M, Willey D, Hunt A, Burton J, Sims S, McLay K, Plumb B, Davis J, Clee C, Oliver K, Clark R, Riddle C, Elliot D, Threadgold G, Harden G, Ware D, Begum S, Mortimore B, Kerry G, Heath P, Phillimore B, Tracey A, Corby N, Dunn M, Johnson C, Wood J, Clark S, Pelan S, Griffiths G, Smith M, Glithero R, Howden P, Barker N, Lloyd C, Stevens C, Harley J, Holt K, Panagiotidis G, Lovell J, Beasley H, Henderson C, Gordon D, Auger K, Wright D, Collins J, Raisen C, Dyer L, Leung K, Robertson L, Ambridge K, Leongamornlert D, McGuire S, Gilderthorp R, Griffiths C, Manthravadi D, Nichol S, Barker G, Whitehead S, Kay M, Brown J, Murnane C, Gray E, Humphries M, Sycamore N, Barker D, Saunders D, Wallis J, Babbage A, Hammond S, Mashreghi-Mohammadi M, Barr L, Martin S, Wray P, Ellington A, Matthews N, Ellwood M, Woodmansey R, Clark G, Cooper J, Tromans A, Grafham D, Skuce C, Pandian R, Andrews R, Harrison E, Kimberley A, Garnett J, Fosker N, Hall R, Garner P, Kelly D, Bird C, Palmer S, Gehring I, Berger A, Dooley CM, Ersan-Urun Z, Eser C, Geiger H, Geisler M, Karotki L, Kirn A, Konantz J, Konantz M, Oberlander M, Rudolph-Geiger S, Teucke M, Lanz C, Raddatz G, Osoegawa K, Zhu B, Rapp A, Widaa S, Langford C, Yang F, Schuster SC, Carter NP, Harrow J, Ning Z, Herrero J, Searle SM, Enright A, Geisler R, Plasterk RH, Lee C, Westerfield M, de Jong PJ, Zon LI, Postlethwait JH, Nusslein-Volhard C, Hubbard TJ, Roest Crollius H, Rogers J, Stemple DL. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498–503.
Postlethwait JH, Woods IG, Ngo-Hazelett P, Yan YL, Kelly PD, Chu F, Huang H, Hill-Force A, Talbot WS. Zebrafish comparative genomics and the origins of vertebrate chromosomes. Genome Res. 2000;10:1890–902.
Amsterdam A, Hopkins N. Mutagenesis strategies in zebrafish for identifying genes involved in development and disease. Trends Genet. 2006;22:473–8.
Nagayoshi S, Hayashi E, Abe G, Osato N, Asakawa K, Urasaki A, Horikawa K, Ikeo K, Takeda H, Kawakami K. Insertional mutagenesis by the tol2 transposon-mediated enhancer trap approach generated mutations in two developmental genes: Tcf7 and synembryn-like. Development. 2008;135:159–69.
Sivasubbu S, Balciunas D, Davidson AE, Pickart MA, Hermanson SB, Wangensteen KJ, Wolbrink DC, Ekker SC. Gene-breaking transposon mutagenesis reveals an essential role for histone h2afza in zebrafish larval development. Mech Dev. 2006;123:513–29.
Gaiano N, Amsterdam A, Kawakami K, Allende M, Becker T, Hopkins N. Insertional mutagenesis and rapid cloning of essential genes in zebrafish. Nature. 1996;383:829–32.
McGrail M, Hatler JM, Kuang X, Liao HK, Nannapaneni K, Watt KE, Uhl JD, Largaespada DA, Vollbrecht E, Scheetz TE, Dupuy AJ, Hostetter JM, Essner JJ. Somatic mutagenesis with a sleeping beauty transposon system leads to solid tumor formation in zebrafish. PLoS One. 2011;6:e18826.
Kramer C, Mayr T, Nowak M, Schumacher J, Runke G, Bauer H, Wagner DS, Schmid B, Imai Y, Talbot WS, Mullins MC, Hammerschmidt M. Maternally supplied smad5 is required for ventral specification in zebrafish embryos prior to zygotic bmp signaling. Dev Biol. 2002;250:263–79.
Bai X, Kim J, Yang Z, Jurynec MJ, Akie TE, Lee J, LeBlanc J, Sessa A, Jiang H, DiBiase A, Zhou Y, Grunwald DJ, Lin S, Cantor AB, Orkin SH, Zon LI. Tif1gamma controls erythroid cell fate by regulating transcription elongation. Cell. 2010;142:133–43.
Asnani A, Peterson RT. The zebrafish as a tool to identify novel therapies for human cardiovascular disease. Dis Model Mech. 2014;7:763–7.
Bakkers J. Zebrafish as a model to study cardiac development and human cardiac disease. Cardiovasc Res. 2011;91:279–88.
Stainier DY, Fouquet B, Chen JN, Warren KS, Weinstein BM, Meiler SE, Mohideen MA, Neuhauss SC, Solnica-Krezel L, Schier AF, Zwartkruis F, Stemple DL, Malicki J, Driever W, Fishman MC. Mutations affecting the formation and function of the cardiovascular system in the zebrafish embryo. Development. 1996;123:285–92.
Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3:59–69.
Vogel G. Genomics. Sanger will sequence zebrafish genome. Science. 2000;290:1671.
Rossi A, Kontarakis Z, Gerri C, Nolte H, Holper S, Kruger M, Stainier DY. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015;524:230–3.
Wienholds E, van Eeden F, Kosters M, Mudde J, Plasterk RH, Cuppen E. Efficient target-selected mutagenesis in zebrafish. Genome Res. 2003;13:2700–7.
Sprague J, Doerry E, Douglas S, Westerfield M. The zebrafish information network (zfin): a resource for genetic, genomic and developmental research. Nucleic Acids Res. 2001;29:87–90.
Eisen JS, Smith JC. Controlling morpholino experiments: don’t stop making antisense. Development. 2008;135:1735–43.
Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet. 2000;26:216–20.
Draper BW, Morcos PA, Kimmel CB. Inhibition of zebrafish fgf8 pre-mrna splicing with morpholino oligos: a quantifiable method for gene knockdown. Genesis. 2001;30:154–6.
Shestopalov IA, Sinha S, Chen JK. Light-controlled gene silencing in zebrafish embryos. Nat Chem Biol. 2007;3:650–1.
Ouyang X, Shestopalov IA, Sinha S, Zheng G, Pitt CL, Li WH, Olson AJ, Chen JK. Versatile synthesis and rational design of caged morpholinos. J Am Chem Soc. 2009;131:13255–69.
Montalbano A, Juergensen L, Roeth R, Weiss B, Fukami M, Fricke-Otto S, Binder G, Ogata T, Decker E, Nuernberg G, Hassel D, Rappold GA. Retinoic acid catabolizing enzyme cyp26c1 is a genetic modifier in shox deficiency. EMBO Mol Med. 2016;8:1455–69.
Hassel D, Cheng P, White MP, Ivey KN, Kroll J, Augustin HG, Katus HA, Stainier DY, Srivastava D. Microrna-10 regulates the angiogenic behavior of zebrafish and human endothelial cells by promoting vascular endothelial growth factor signaling. Circ Res. 2012;111:1421–33.
Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of nodal agonist and antagonist by mir-430. Science. 2007;318:271–4.
Blum M, De Robertis EM, Wallingford JB, Niehrs C. Morpholinos: antisense and sensibility. Dev Cell. 2015;35:145–9.
Kok FO, Shin M, Ni CW, Gupta A, Grosse AS, van Impel A, Kirchmaier BC, Peterson-Maduro J, Kourkoulis G, Male I, DeSantis DF, Sheppard-Tindell S, Ebarasi L, Betsholtz C, Schulte-Merker S, Wolfe SA, Lawson ND. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev Cell. 2015;32:97–108.
Hwang WY, Fu Y, Reyon D, Maeder ML, Kaini P, Sander JD, Joung JK, Peterson RT, Yeh JR. Heritable and precise zebrafish genome editing using a crispr-cas system. PLoS One. 2013;8:e68708.
Chang N, Sun C, Gao L, Zhu D, Xu X, Zhu X, Xiong JW, Xi JJ. Genome editing with rna-guided cas9 nuclease in zebrafish embryos. Cell Res. 2013;23:465–72.
Sander JD, Cade L, Khayter C, Reyon D, Peterson RT, Joung JK, Yeh JR. Targeted gene disruption in somatic zebrafish cells using engineered talens. Nat Biotechnol. 2011;29:697–8.
Huang P, Xiao A, Zhou M, Zhu Z, Lin S, Zhang B. Heritable gene targeting in zebrafish using customized talens. Nat Biotechnol. 2011;29:699–700.
Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Amacher SL. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26:702–8.
Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to fok i cleavage domain. Proc Natl Acad Sci U S A. 1996;93:1156–60.
Narayanan A, Hill-Teran G, Moro A, Ristori E, Kasper DM, Roden CA, Lu J, Nicoli S. In vivo mutagenesis of mirna gene families using a scalable multiplexed crispr/cas9 nuclease system. Sci Rep. 2016;6:32386.
Kasper DM, Moro A, Ristori E, Narayanan A, Hill-Teran G, Fleming E, Moreno-Mateos M, Vejnar CE, Zhang J, Lee D, Gu M, Gerstein M, Giraldez A, Nicoli S. Micrornas establish uniform traits during the architecture of vertebrate embryos. Dev Cell. 2017;40:552–565 e555.
Albadri S, Del Bene F, Revenu C. Genome editing using crispr/cas9-based knock-in approaches in zebrafish. Methods. 2017;121–122:77–85.
Won M, Dawid IB. Pcr artifact in testing for homologous recombination in genomic editing in zebrafish. PLoS One. 2017;12:e0172802.
Kawakami K. Tol2: a versatile gene transfer vector in vertebrates. Genome Biol. 2007;8(Suppl 1):S7.
Hassel D, Dahme T, Erdmann J, Meder B, Huge A, Stoll M, Just S, Hess A, Ehlermann P, Weichenhan D, Grimmler M, Liptau H, Hetzer R, Regitz-Zagrosek V, Fischer C, Nurnberg P, Schunkert H, Katus HA, Rottbauer W. Nexilin mutations destabilize cardiac z-disks and lead to dilated cardiomyopathy. Nat Med. 2009;15:1281–8.
Wang X, Yu Q, Wu Q, Bu Y, Chang NN, Yan S, Zhou XH, Zhu X, Xiong JW. Genetic interaction between pku300 and fbn2b controls endocardial cell proliferation and valve development in zebrafish. J Cell Sci. 2013;126:1381–91.
Durst R, Sauls K, Peal DS, deVlaming A, Toomer K, Leyne M, Salani M, Talkowski ME, Brand H, Perrocheau M, Simpson C, Jett C, Stone MR, Charles F, Chiang C, Lynch SN, Bouatia-Naji N, Delling FN, Freed LA, Tribouilloy C, LeTourneau T, LeMarec H, Fernandez-Friera L, Solis J, Trujillano D, Ossowski S, Estivill X, Dina C, Bruneval P, Chester A, Schott JJ, Irvine KD, Mao Y, Wessels A, Motiwala T, Puceat M, Tsukasaki Y, Menick DR, Kasiganesan H, Nie X, Broome AM, Williams K, Johnson A, Markwald RR, Jeunemaitre X, Hagege A, Levine RA, Milan DJ, Norris RA, Slaugenhaupt SA. Mutations in dchs1 cause mitral valve prolapse. Nature. 2015;525:109–13.
Dina C, Bouatia-Naji N, Tucker N, Delling FN, Toomer K, Durst R, Perrocheau M, Fernandez-Friera L, Solis J, Investigators P, Le Tourneau T, Chen MH, Probst V, Bosse Y, Pibarot P, Zelenika D, Lathrop M, Hercberg S, Roussel R, Benjamin EJ, Bonnet F, Lo SH, Dolmatova E, Simonet F, Lecointe S, Kyndt F, Redon R, LeMarec H, Froguel P, Ellinor PT, Vasan RS, Bruneval P, Markwald RR, Norris RA, Milan DJ, Slaugenhaupt SA, Levine RA, Schott JJ, Hagege AA, MVP F, Jeunemaitre X, Leducq Transatlantic MN. Genetic association analyses highlight biological pathways underlying mitral valve prolapse. Nat Genet. 2015;47:1206–11.
Ding Y, Sun X, Huang W, Hoage T, Redfield M, Kushwaha S, Sivasubbu S, Lin X, Ekker S, Xu X. Haploinsufficiency of target of rapamycin attenuates cardiomyopathies in adult zebrafish. Circ Res. 2011;109:658–69.
Group CCHW. Meta-analysis of rare and common exome chip variants identifies s1pr4 and other loci influencing blood cell traits. Nat Genet. 2016;48:867–76.
Smith KA, Joziasse IC, Chocron S, van Dinther M, Guryev V, Verhoeven MC, Rehmann H, van der Smagt JJ, Doevendans PA, Cuppen E, Mulder BJ, Ten Dijke P, Bakkers J. Dominant-negative alk2 allele associates with congenital heart defects. Circulation. 2009;119:3062–9.
Hoffman JI, Kaplan S. The incidence of congenital heart disease. J Am Coll Cardiol. 2002;39:1890–900.
Bruneau BG. The developmental genetics of congenital heart disease. Nature. 2008;451:943–8.
Brown DR, Samsa LA, Qian L, Liu J. Advances in the study of heart development and disease using zebrafish. J Cardiovasc Dev Dis. 2016;3:13.
Samsa LA, Givens C, Tzima E, Stainier DY, Qian L, Liu J. Cardiac contraction activates endocardial notch signaling to modulate chamber maturation in zebrafish. Development. 2015;142:4080–91.
Lai SL, Yao WL, Tsao KC, Houben AJ, Albers HM, Ovaa H, Moolenaar WH, Lee SJ. Autotaxin/lpar3 signaling regulates kupffer’s vesicle formation and left-right asymmetry in zebrafish. Development. 2012;139:4439–48.
Srivastava D. Making or breaking the heart: from lineage determination to morphogenesis. Cell. 2006;126:1037–48.
Stainier DY. Zebrafish genetics and vertebrate heart formation. Nat Rev Genet. 2001;2:39–48.
Liu J, Stainier DY. Zebrafish in the study of early cardiac development. Circ Res. 2012;110:870–4.
Glickman NS, Yelon D. Cardiac development in zebrafish: coordination of form and function. Semin Cell Dev Biol. 2002;13:507–13.
Samsa LA, Yang B, Liu J. Embryonic cardiac chamber maturation: trabeculation, conduction, and cardiomyocyte proliferation. Am J Med Genet C Semin Med Genet. 2013;163C:157–68.
Beis D, Bartman T, Jin SW, Scott IC, D’Amico LA, Ober EA, Verkade H, Frantsve J, Field HA, Wehman A, Baier H, Tallafuss A, Bally-Cuif L, Chen JN, Stainier DY, Jungblut B. Genetic and cellular analyses of zebrafish atrioventricular cushion and valve development. Development. 2005;132:4193–204.
Scherz PJ, Huisken J, Sahai-Hernandez P, Stainier DY. High-speed imaging of developing heart valves reveals interplay of morphogenesis and function. Development. 2008;135:1179–87.
Bisgrove BW, Snarr BS, Emrazian A, Yost HJ. Polaris and polycystin-2 in dorsal forerunner cells and kupffer’s vesicle are required for specification of the zebrafish left-right axis. Dev Biol. 2005;287:274–88.
Vetrini F, D'Alessandro LC, Akdemir ZC, Braxton A, Azamian MS, Eldomery MK, Miller K, Kois C, Sack V, Shur N, Rijhsinghani A, Chandarana J, Ding Y, Holtzman J, Jhangiani SN, Muzny DM, Gibbs RA, Eng CM, Hanchard NA, Harel T, Rosenfeld JA, Belmont JW, Lupski JR, Yang Y. Bi-allelic mutations in pkd1l1 are associated with laterality defects in humans. Am J Hum Genet. 2016;99:886–93.
Paffett-Lugassy N, Singh R, Nevis KR, Guner-Ataman B, O'Loughlin E, Jahangiri L, Harvey RP, Burns CG, Burns CE. Heart field origin of great vessel precursors relies on nkx2.5-mediated vasculogenesis. Nat Cell Biol. 2013;15:1362–9.
Nagelberg D, Wang J, Su R, Torres-Vazquez J, Targoff KL, Poss KD, Knaut H. Origin, specification, and plasticity of the great vessels of the heart. Curr Biol. 2015;25:2099–110.
Barron DJ, Kilby MD, Davies B, Wright JG, Jones TJ, Brawn WJ. Hypoplastic left heart syndrome. Lancet. 2009;374:551–64.
Noonan JA, Nadas AS. The hypoplastic left heart syndrome; an analysis of 101 cases. Pediatr Clin N Am. 1958;5:1029–56.
Norwood WI, Lang P, Hansen DD. Physiologic repair of aortic atresia-hypoplastic left heart syndrome. N Engl J Med. 1983;308:23–6.
Hinton RB, Martin LJ, Rame-Gowda S, Tabangin ME, Cripe LH, Benson DW. Hypoplastic left heart syndrome links to chromosomes 10q and 6q and is genetically related to bicuspid aortic valve. J Am Coll Cardiol. 2009;53:1065–71.
Dasgupta C, Martinez AM, Zuppan CW, Shah MM, Bailey LL, Fletcher WH. Identification of connexin43 (alpha1) gap junction gene mutations in patients with hypoplastic left heart syndrome by denaturing gradient gel electrophoresis (dgge). Mutat Res. 2001;479:173–86.
Iascone M, Ciccone R, Galletti L, Marchetti D, Seddio F, Lincesso AR, Pezzoli L, Vetro A, Barachetti D, Boni L, Federici D, Soto AM, Comas JV, Ferrazzi P, Zuffardi O. Identification of de novo mutations and rare variants in hypoplastic left heart syndrome. Clin Genet. 2012;81:542–54.
Targoff KL, Colombo S, George V, Schell T, Kim SH, Solnica-Krezel L, Yelon D. Nkx genes are essential for maintenance of ventricular identity. Development. 2013;140:4203–13.
Targoff KL, Schell T, Yelon D. Nkx genes regulate heart tube extension and exert differential effects on ventricular and atrial cell number. Dev Biol. 2008;322:314–21.
Liu X, Yagi H, Saeed S, Bais AS, Gabriel GC, Chen Z, Peterson KA, Li Y, Schwartz MC, Reynolds WT, Saydmohammed M, Gibbs B, Wu Y, Devine W, Chatterjee B, Klena NT, Kostka D, de Mesy Bentley KL, Ganapathiraju MK, Dexheimer P, Leatherbury L, Khalifa O, Bhagat A, Zahid M, Pu W, Watkins S, Grossfeld P, Murray SA, Porter GA Jr, Tsang M, Martin LJ, Benson DW, Aronow BJ, Lo CW. The complex genetics of hypoplastic left heart syndrome. Nat Genet. 2017;49:1152–9.
Renz M, Otten C, Faurobert E, Rudolph F, Zhu Y, Boulday G, Duchene J, Mickoleit M, Dietrich AC, Ramspacher C, Steed E, Manet-Dupe S, Benz A, Hassel D, Vermot J, Huisken J, Tournier-Lasserve E, Felbor U, Sure U, Albiges-Rizo C, Abdelilah-Seyfried S. Regulation of beta1 integrin-klf2-mediated angiogenesis by ccm proteins. Dev Cell. 2015;32:181–90.
Fang L, Green SR, Baek JS, Lee SH, Ellett F, Deer E, Lieschke GJ, Witztum JL, Tsimikas S, Miller YI. In vivo visualization and attenuation of oxidized lipid accumulation in hypercholesterolemic zebrafish. J Clin Invest. 2011;121:4861–9.
Markwald RR, Norris RA, Moreno-Rodriguez R, Levine RA. Developmental basis of adult cardiovascular diseases: valvular heart diseases. Ann N Y Acad Sci. 2010;1188:177–83.
Nishimura RA, Otto CM, Bonow RO, Carabello BA, Erwin JP III, Guyton RA, O’Gara PT, Ruiz CE, Skubas NJ, Sorajja P, Sundt TM III, Thomas JD, Anderson JL, Halperin JL, Albert NM, Bozkurt B, Brindis RG, Creager MA, Curtis LH, DeMets D, Guyton RA, Hochman JS, Kovacs RJ, Ohman EM, Pressler SJ, Sellke FW, Shen WK, Stevenson WG, Yancy CW, American College of C, American College of Cardiology/American Heart A, American Heart A. 2014 AHA/ACC guideline for the management of patients with valvular heart disease: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. J thorac Cardiovasc Surg. 2014;148:e1–e132.
Nishimura RA, Otto CM, Bonow RO, Carabello BA, Erwin JP III, Fleisher LA, Jneid H, Mack MJ, McLeod CJ, O’Gara PT, Rigolin VH, Sundt TM III, Thompson A. 2017 AHA/ACC focused update of the 2014 aha/acc guideline for the management of patients with valvular heart disease: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. J Am Coll Cardiol. 2017;70:252–89.
Staudt D, Stainier D. Uncovering the molecular and cellular mechanisms of heart development using the zebrafish. Annu Rev Genet. 2012;46:397–418.
Pestel J, Ramadass R, Gauvrit S, Helker C, Herzog W, Stainier DY. Real-time 3d visualization of cellular rearrangements during cardiac valve formation. Development. 2016;143:2217–27.
Zhu L, Belmont JW, Ware SM. Genetics of human heterotaxias. Eur J Hum Genet. 2006;14:17–25.
Amula V, Ellsworth GL, Bratton SL, Arrington CB, Witte MK. Heterotaxy syndrome: impact of ventricular morphology on resource utilization. Pediatr Cardiol. 2014;35:38–46.
Unolt M, Putotto C, Silvestri LM, Marino D, Scarabotti A, Valerio M, Caiaro A, Versacci P, Marino B. Transposition of great arteries: new insights into the pathogenesis. Front Pediatr. 2013;1:11.
Lin AE, Krikov S, Riehle-Colarusso T, Frias JL, Belmont J, Anderka M, Geva T, Getz KD, Botto LD. National Birth Defects Prevention S. Laterality defects in the national birth defects prevention study (1998–2007): birth prevalence and descriptive epidemiology. Am J Med Genet A. 2014;164A:2581–91.
Pennekamp P, Menchen T, Dworniczak B, Hamada H. Situs inversus and ciliary abnormalities: 20 years later, what is the connection? Cilia. 2015;4:1.
Kennedy MP, Omran H, Leigh MW, Dell S, Morgan L, Molina PL, Robinson BV, Minnix SL, Olbrich H, Severin T, Ahrens P, Lange L, Morillas HN, Noone PG, Zariwala MA, Knowles MR. Congenital heart disease and other heterotaxic defects in a large cohort of patients with primary ciliary dyskinesia. Circulation. 2007;115:2814–21.
Nakhleh N, Francis R, Giese RA, Tian X, Li Y, Zariwala MA, Yagi H, Khalifa O, Kureshi S, Chatterjee B, Sabol SL, Swisher M, Connelly PS, Daniels MP, Srinivasan A, Kuehl K, Kravitz N, Burns K, Sami I, Omran H, Barmada M, Olivier K, Chawla KK, Leigh M, Jonas R, Knowles M, Leatherbury L, Lo CW. High prevalence of respiratory ciliary dysfunction in congenital heart disease patients with heterotaxy. Circulation. 2012;125:2232–42.
Li Y, Yagi H, Onuoha EO, Damerla RR, Francis R, Furutani Y, Tariq M, King SM, Hendricks G, Cui C, Saydmohammed M, Lee DM, Zahid M, Sami I, Leatherbury L, Pazour GJ, Ware SM, Nakanishi T, Goldmuntz E, Tsang M, Lo CW. Dnah6 and its interactions with pcd genes in heterotaxy and primary ciliary dyskinesia. PLoS Genet. 2016;12:e1005821.
Ostrowski LE, Dutcher SK, Lo CW. Cilia and models for studying structure and function. Proc Am Thorac Soc. 2011;8:423–9.
Guimier A, Gabriel GC, Bajolle F, Tsang M, Liu H, Noll A, Schwartz M, El Malti R, Smith LD, Klena NT, Jimenez G, Miller NA, Oufadem M, Moreau de Bellaing A, Yagi H, Saunders CJ, Baker CN, Di Filippo S, Peterson KA, Thiffault I, Bole-Feysot C, Cooley LD, Farrow EG, Masson C, Schoen P, Deleuze JF, Nitschke P, Lyonnet S, de Pontual L, Murray SA, Bonnet D, Kingsmore SF, Amiel J, Bouvagnet P, Lo CW, Gordon CT. Mmp21 is mutated in human heterotaxy and is required for normal left-right asymmetry in vertebrates. Nat Genet. 2015;47:1260–3.
Xu X, Meiler SE, Zhong TP, Mohideen M, Crossley DA, Burggren WW, Fishman MC. Cardiomyopathy in zebrafish due to mutation in an alternatively spliced exon of titin. Nat Genet. 2002;30:205–9.
Sehnert AJ, Huq A, Weinstein BM, Walker C, Fishman M, Stainier DY. Cardiac troponin t is essential in sarcomere assembly and cardiac contractility. Nat Genet. 2002;31:106–10.
Siu BL, Niimura H, Osborne JA, Fatkin D, MacRae C, Solomon S, Benson DW, Seidman JG, Seidman CE. Familial dilated cardiomyopathy locus maps to chromosome 2q31. Circulation. 1999;99:1022–6.
Thierfelder L, Watkins H, MacRae C, Lamas R, McKenna W, Vosberg HP, Seidman JG, Seidman CE. Alpha-tropomyosin and cardiac troponin t mutations cause familial hypertrophic cardiomyopathy: a disease of the sarcomere. Cell. 1994;77:701–12.
Shih YH, Zhang Y, Ding Y, Ross CA, Li H, Olson TM, Xu X. Cardiac transcriptome and dilated cardiomyopathy genes in zebrafish. Circ Cardiovasc Genet. 2015;8:261–9.
Wang H, Li Z, Wang J, Sun K, Cui Q, Song L, Zou Y, Wang X, Liu X, Hui R, Fan Y. Mutations in nexn, a z-disc gene, are associated with hypertrophic cardiomyopathy. Am J Hum Genet. 2010;87:687–93.
Vogel B, Meder B, Just S, Laufer C, Berger I, Weber S, Katus HA, Rottbauer W. In-vivo characterization of human dilated cardiomyopathy genes in zebrafish. Biochem Biophys Res Commun. 2009;390:516–22.
Ramspacher C, Steed E, Boselli F, Ferreira R, Faggianelli N, Roth S, Spiegelhalter C, Messaddeq N, Trinh L, Liebling M, Chacko N, Tessadori F, Bakkers J, Laporte J, Hnia K, Vermot J. Developmental alterations in heart biomechanics and skeletal muscle function in desmin mutants suggest an early pathological root for desminopathies. Cell Rep. 2015;11:1564–76.
Liu J, Bressan M, Hassel D, Huisken J, Staudt D, Kikuchi K, Poss KD, Mikawa T, Stainier DY. A dual role for erbb2 signaling in cardiac trabeculation. Development. 2010;137:3867–75.
Meder B, Laufer C, Hassel D, Just S, Marquart S, Vogel B, Hess A, Fishman MC, Katus HA, Rottbauer W. A single serine in the carboxyl terminus of cardiac essential myosin light chain-1 controls cardiomyocyte contractility in vivo. Circ Res. 2009;104:650–9.
Bendig G, Grimmler M, Huttner IG, Wessels G, Dahme T, Just S, Trano N, Katus HA, Fishman MC, Rottbauer W. Integrin-linked kinase, a novel component of the cardiac mechanical stretch sensor, controls contractility in the zebrafish heart. Genes Dev. 2006;20:2361–72.
Rottbauer W, Just S, Wessels G, Trano N, Most P, Katus HA, Fishman MC. Vegf-plcgamma1 pathway controls cardiac contractility in the embryonic heart. Genes Dev. 2005;19:1624–34.
Zou J, Tran D, Baalbaki M, Tang LF, Poon A, Pelonero A, Titus EW, Yuan C, Shi C, Patchava S, Halper E, Garg J, Movsesyan I, Yin C, Wu R, Wilsbacher LD, Liu J, Hager RL, Coughlin SR, Jinek M, Pullinger CR, Kane JP, Hart DO, Kwok PY, Deo RC. An internal promoter underlies the difference in disease severity between n- and c-terminal truncation mutations of titin in zebrafish. elife. 2015;4:e09406.
Kossack M, Hein S, Juergensen L, Siragusa M, Benz A, Katus HA, Most P, Hassel D. Induction of cardiac dysfunction in developing and adult zebrafish by chronic isoproterenol stimulation. J Mol Cell Cardiol. 2017;108:95–105.
Kalogirou S, Malissovas N, Moro E, Argenton F, Stainier DY, Beis D. Intracardiac flow dynamics regulate atrioventricular valve morphogenesis. Cardiovasc Res. 2014;104:49–60.
Asimaki A, Kapoor S, Plovie E, Karin Arndt A, Adams E, Liu Z, James CA, Judge DP, Calkins H, Churko J, Wu JC, MacRae CA, Kleber AG, Saffitz JE. Identification of a new modulator of the intercalated disc in a zebrafish model of arrhythmogenic cardiomyopathy. Sci Transl Med. 2014;6:240ra274.
Ding Y, Sun X, Xu X. Tor-autophagy signaling in adult zebrafish models of cardiomyopathy. Autophagy. 2012;8:142–3.
Sun X, Hoage T, Bai P, Ding Y, Chen Z, Zhang R, Huang W, Jahangir A, Paw B, Li YG, Xu X. Cardiac hypertrophy involves both myocyte hypertrophy and hyperplasia in anemic zebrafish. PLoS One. 2009;4:e6596.
Scheid LM, Mosqueira M, Hein S, Kossack M, Juergensen L, Mueller M, Meder B, Fink RH, Katus HA, Hassel D. Essential light chain s195 phosphorylation is required for cardiac adaptation under physical stress. Cardiovasc Res. 2016;111:44–55.
Wang LW, Huttner IG, Santiago CF, Kesteven SH, Yu ZY, Feneley MP, Fatkin D. Standardized echocardiographic assessment of cardiac function in normal adult zebrafish and heart disease models. Dis Model Mech. 2017;10:63–76.
Hein SJ, Lehmann LH, Kossack M, Juergensen L, Fuchs D, Katus HA, Hassel D. Advanced echocardiography in adult zebrafish reveals delayed recovery of heart function after myocardial cryoinjury. PLoS One. 2015;10:e0122665.
Rottbauer W, Baker K, Wo ZG, Mohideen MA, Cantiello HF, Fishman MC. Growth and function of the embryonic heart depend upon the cardiac-specific l-type calcium channel alpha1 subunit. Dev Cell. 2001;1:265–75.
Arnaout R, Ferrer T, Huisken J, Spitzer K, Stainier DY, Tristani-Firouzi M, Chi NC. Zebrafish model for human long qt syndrome. Proc Natl Acad Sci U S A. 2007;104:11316–21.
Langheinrich U, Vacun G, Wagner T. Zebrafish embryos express an orthologue of herg and are sensitive toward a range of qt-prolonging drugs inducing severe arrhythmia. Toxicol Appl Pharmacol. 2003;193:370–82.
Milan DJ, Kim AM, Winterfield JR, Jones IL, Pfeufer A, Sanna S, Arking DE, Amsterdam AH, Sabeh KM, Mably JD, Rosenbaum DS, Peterson RT, Chakravarti A, Kaab S, Roden DM, MacRae CA. Drug-sensitized zebrafish screen identifies multiple genes, including gins3, as regulators of myocardial repolarization. Circulation. 2009;120:553–9.
Peal DS, Mills RW, Lynch SN, Mosley JM, Lim E, Ellinor PT, January CT, Peterson RT, Milan DJ. Novel chemical suppressors of long qt syndrome identified by an in vivo functional screen. Circulation. 2011;123:23–30.
Hassel D, Scholz EP, Trano N, Friedrich O, Just S, Meder B, Weiss DL, Zitron E, Marquart S, Vogel B, Karle CA, Seemann G, Fishman MC, Katus HA, Rottbauer W. Deficient zebrafish ether-a-go-go-related gene channel gating causes short-qt syndrome in zebrafish reggae mutants. Circulation. 2008;117:866–75.
Langenbacher AD, Dong Y, Shu X, Choi J, Nicoll DA, Goldhaber JI, Philipson KD, Chen JN. Mutation in sodium-calcium exchanger 1 (ncx1) causes cardiac fibrillation in zebrafish. Proc Natl Acad Sci U S A. 2005;102:17699–704.
Ebert AM, Hume GL, Warren KS, Cook NP, Burns CG, Mohideen MA, Siegal G, Yelon D, Fishman MC, Garrity DM. Calcium extrusion is critical for cardiac morphogenesis and rhythm in embryonic zebrafish hearts. Proc Natl Acad Sci U S A. 2005;102:17705–10.
Huttner IG, Trivedi G, Jacoby A, Mann SA, Vandenberg JI, Fatkin D. A transgenic zebrafish model of a human cardiac sodium channel mutation exhibits bradycardia, conduction-system abnormalities and early death. J Mol Cell Cardiol. 2013;61:123–32.
Chen X, Gays D, Milia C, Santoro MM. Cilia control vascular mural cell recruitment in vertebrates. Cell Rep. 2017;18:1033–47.
Ando K, Fukuhara S, Izumi N, Nakajima H, Fukui H, Kelsh RN, Mochizuki N. Clarification of mural cell coverage of vascular endothelial cells by live imaging of zebrafish. Development. 2016;143:1328–39.
Karpanen T, Schulte-Merker S. Zebrafish provides a novel model for lymphatic vascular research. Methods Cell Biol. 2011;105:223–38.
Santoro MM, Pesce G, Stainier DY. Characterization of vascular mural cells during zebrafish development. Mech Dev. 2009;126:638–49.
Jin SW, Beis D, Mitchell T, Chen JN, Stainier DY. Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development. 2005;132:5199–209.
Siekmann AF, Lawson ND. Notch signalling limits angiogenic cell behaviour in developing zebrafish arteries. Nature. 2007;445:781–4.
Hellstrom M, Phng LK, Hofmann JJ, Wallgard E, Coultas L, Lindblom P, Alva J, Nilsson AK, Karlsson L, Gaiano N, Yoon K, Rossant J, Iruela-Arispe ML, Kalen M, Gerhardt H, Betsholtz C. Dll4 signalling through notch1 regulates formation of tip cells during angiogenesis. Nature. 2007;445:776–80.
Herbert SP, Huisken J, Kim TN, Feldman ME, Houseman BT, Wang RA, Shokat KM, Stainier DY. Arterial-venous segregation by selective cell sprouting: an alternative mode of blood vessel formation. Science. 2009;326:294–8.
Xu B, Zhang Y, Du XF, Li J, Zi HX, Bu JW, Yan Y, Han H, Du JL. Neurons secrete mir-132-containing exosomes to regulate brain vascular integrity. Cell Res. 2017;27:882–97.
Tobia C, Chiodelli P, Nicoli S, Dell'era P, Buraschi S, Mitola S, Foglia E, van Loenen PB, Alewijnse AE, Presta M. Sphingosine-1-phosphate receptor-1 controls venous endothelial barrier integrity in zebrafish. Arterioscler Thromb Vasc Biol. 2012;32:e104–16.
Butler MG, Gore AV, Weinstein BM. Zebrafish as a model for hemorrhagic stroke. Methods Cell Biol. 2011;105:137–61.
Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY, Srivastava D. Mir-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272–84.
Buchner DA, Su F, Yamaoka JS, Kamei M, Shavit JA, Barthel LK, McGee B, Amigo JD, Kim S, Hanosh AW, Jagadeeswaran P, Goldman D, Lawson ND, Raymond PA, Weinstein BM, Ginsburg D, Lyons SE. Pak2a mutations cause cerebral hemorrhage in redhead zebrafish. Proc Natl Acad Sci U S A. 2007;104:13996–4001.
Gu Y, Jin P, Zhang L, Zhao X, Gao X, Ning Y, Meng A, Chen YG. Functional analysis of mutations in the kinase domain of the tgf-beta receptor alk1 reveals different mechanisms for induction of hereditary hemorrhagic telangiectasia. Blood. 2006;107:1951–4.
Hall CJ, Flores MV, Davidson AJ, Crosier KE, Crosier PS. Radar is required for the establishment of vascular integrity in the zebrafish. Dev Biol. 2002;251:105–17.
Liu J, Fraser SD, Faloon PW, Rollins EL, Vom Berg J, Starovic-Subota O, Laliberte AL, Chen JN, Serluca FC, Childs SJ. A betapix pak2a signaling pathway regulates cerebral vascular stability in zebrafish. Proc Natl Acad Sci U S A. 2007;104:13990–5.
Kleaveland B, Zheng X, Liu JJ, Blum Y, Tung JJ, Zou Z, Sweeney SM, Chen M, Guo L, Lu MM, Zhou D, Kitajewski J, Affolter M, Ginsberg MH, Kahn ML. Regulation of cardiovascular development and integrity by the heart of glass-cerebral cavernous malformation protein pathway. Nat Med. 2009;15:169–76.
Gut P, Baeza-Raja B, Andersson O, Hasenkamp L, Hsiao J, Hesselson D, Akassoglou K, Verdin E, Hirschey MD, Stainier DY. Whole-organism screening for gluconeogenesis identifies activators of fasting metabolism. Nat Chem Biol. 2013;9:97–104.
Weger BD, Weger M, Nusser M, Brenner-Weiss G, Dickmeis T. A chemical screening system for glucocorticoid stress hormone signaling in an intact vertebrate. ACS Chem Biol. 2012;7:1178–83.
Stoletov K, Fang L, Choi SH, Hartvigsen K, Hansen LF, Hall C, Pattison J, Juliano J, Miller ER, Almazan F, Crosier P, Witztum JL, Klemke RL, Miller YI. Vascular lipid accumulation, lipoprotein oxidation, and macrophage lipid uptake in hypercholesterolemic zebrafish. Circ Res. 2009;104:952–60.
Swinney DC, Anthony J. How were new medicines discovered? Nat Rev Drug Discov. 2011;10:507–19.
Eggert US. The why and how of phenotypic small-molecule screens. Nat Chem Biol. 2013;9:206–9.
MacRae CA, Peterson RT. Zebrafish as tools for drug discovery. Nat Rev Drug Discov. 2015;14:721–31.
Peterson RT, Link BA, Dowling JE, Schreiber SL. Small molecule developmental screens reveal the logic and timing of vertebrate development. Proc Natl Acad Sci U S A. 2000;97:12965–9.
Peterson RT, Shaw SY, Peterson TA, Milan DJ, Zhong TP, Schreiber SL, MacRae CA, Fishman MC. Chemical suppression of a genetic mutation in a zebrafish model of aortic coarctation. Nat Biotechnol. 2004;22:595–9.
Zhong TP, Rosenberg M, Mohideen MA, Weinstein B, Fishman MC. Gridlock, an hlh gene required for assembly of the aorta in zebrafish. Science. 2000;287:1820–4.
Weinstein BM, Stemple DL, Driever W, Fishman MC. Gridlock, a localized heritable vascular patterning defect in the zebrafish. Nat Med. 1995;1:1143–7.
Acknowledgments
We would like to thank the members of the Hassel and Liu lab for the vivid discussions and valuable input for the book chapter. We would further like to deeply thank all authors whose work we cited and at the same time apologize to all authors whose papers we could not reference due to space limitations. This work was supported by R56HL133081 to J.L.
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Liu, J., Renz, M., Hassel, D. (2019). Interrogating Cardiovascular Genetics in Zebrafish. In: Erdmann, J., Moretti, A. (eds) Genetic Causes of Cardiac Disease. Cardiac and Vascular Biology, vol 7. Springer, Cham. https://doi.org/10.1007/978-3-030-27371-2_9
Download citation
DOI: https://doi.org/10.1007/978-3-030-27371-2_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-27370-5
Online ISBN: 978-3-030-27371-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)