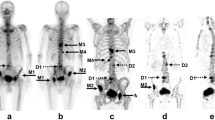
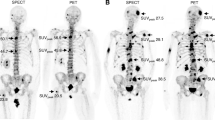
Abstract
The appearance of bone metastasis in patients with breast or prostate cancer makes the skeleton most affected by metastatic cancer. It is estimated that these two cancers lead in 80% of the cases to the appearance of bone metastasis, which is considered the main cause of death. 99mTc-methylene diphosphonate (99mTc-MDP) bone scintigraphy is the most commonly used radionuclide imaging technique for the detection and prognosis of bone carcinoma. With this work, it was intended to develop a new computational solution to extract from 99mTc-MDP bone scintigraphy images quantitative measurements of the affected regions in relation to the non-pathological regions. Hence, the uptake indexes computed from a new imaging exam are compared with the indexes computed from a previous exam of the same patient. Using active shape models, it is possible to segment the regions of the skeleton more prone to be affected by the bone carcinoma. On the other hand, the metastasis is segmented using the region-growing algorithm. Then, the uptake rate is calculated from the relation between the maximum intensity pixel of the metastatic region in relation to the maximum intensity pixel of the skeletal region where the metastasis was located. We evaluated the developed solution using scintigraphic images of 15 patients (7 females and 8 males) with bone carcinoma in two distinct time exams. The bone scans were obtained approximately 3 h after the injection of 740 MBq of 99mTc-MDP. The obtained indexes were compared against the evaluations in the clinical reports of the patients. It was possible to verify that the indexes obtained are according to the clinical evaluations of the 30 exams analyzed. However, there were 2 cases where the clinical evaluation was unclear as to the progression or regression of the disease, and when comparing the indexes, it is suggested the progression of the disease in one case and the regression in the other one. Based on the obtained results, it is possible to conclude that the computed indexes allow a quantitative analysis to evaluate the response to the prescribed therapy. Thus, the developed solution is promising to be used as a tool to help the technicians at the time of clinical evaluation.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
The skeleton is most affected by metastatic cancer, with a higher prevalence for prostate and breast cancer. These two cancers cause in 80% of the cases, the appearance of bone metastases, which is considered the main cause of death. In many cases, the metastatic lesions are multifocal, which means that they are located throughout the skeleton with greater incidence in the axial skeleton [1].
The skeleton is constantly remodeling due to the coordinated activity of osteoclasts and osteoblasts. In normal bone, there is a balanced sequence: first, the osteoclasts absorb the bone and then the osteoblasts form bone in the same place. In cases of metastatic cancer, malignant cells secrete factors that affect this balance leading to osteoblastic stimulation [2].
Premature detection of metastases can prevent complications, control the stage of the disease, and help to determine the treatment to follow, which may result in a higher probability of survival and improvements in quality of life.
Bone scintigraphy with 99mTc methylene diphosphonate (MDP) is currently the most commonly used imaging technique in Nuclear Medicine to determine the extent of these lesions in the skeleton, as it provides a two-dimensional (2D) image of the skeleton showing regions with higher uptake (hotspots) [3]. In addition, it has good sensitivity and has been considered as the first alternative imaging method capable of diagnosing asymptomatic bone metastases, since it is readily available and provides a complete skeletal view at reasonable time and cost [4]. Modern bone scan techniques can detect an increase in bone mineral turnover as small as 10% in regions that are only a few millimeters in size. In contrast, a relatively large volume of bone (1 cm3) must demineralize by about 50% before the change can be detected by radiographs. It is not surprising then, that in regard to prostate cancer, the bone scan is often used to stage patients and monitor the course of bone involvement. However, the interpretation of these exams has significant limitations: The evaluation of the exam is not yet standardized making the interpretations subjective and dependent on the experience of the technician. In numerous situations, these evaluations are described in vague terms as the presence or absence of tumor propagation in the skeleton. Therefore, a quantitative analysis of the images under study is necessary to reduce the variability of the observer in order to determine the extent of the lesions in the bone and to identify posttreatment changes that are clinically relevant.
To improve the monitoring of the treatment of bone lesions, some authors have developed scoring metrics for more objective methods of assessing the extent of bone metastasis, such as counting the number of lesions in the total skeleton, assessing the regional distribution of the metastasis or indexes that measures the tumor burden as a percentage of the total skeletal mass (Bone Scan Index) [5]. Therefore, the aim of the present study was to develop a semiautomatic method for the segmentation, i.e., identification, of regions of interest in 99mTc-MDP scintigraphy images of the skeletal system. The segmented regions allow the posterior assessment of the intensity of the hotspots under study and, therefore, the uptake index calculation.
2 Materials and Methods
2.1 Bone Scintigraphy
The bone scans used to evaluate the developed solution were obtained approximately 3 h after an intravenous (IV) injection of 740 MBq of 99mTc MDP. Whole-body images with anterior and posterior views were acquired according to a matrix size of 256 × 1024 pixels and using a gamma camera equipped with low-energy all-purpose collimators (Discovery NM 360, GE Healthcare). The energy discrimination was provided by a 20% window centered on the 140 keV of 99mTc.
2.2 Training Images Group
A training group of images was randomly chosen to build the Point Distribution Models [6, 7, 8, 9], used in the image segmentation step. The used group consisted of 10 images of patients who had undergone whole-body bone scintigraphy at Lenitudes Medical Center & Research, in Portugal.
2.3 Evaluation Images Group
The evaluation group consists of images acquired from 15 patients (8 males and 7 females), 7 of whom had prostate cancer, the other 7 have breast cancer, and 1 case of lung cancer. All these patients perform whole-body bone scintigraphy at Lenitudes Medical Center & Research every 3 months to evaluate the treatment response.
2.4 Bone Scintigraphy Image Processing
The diagram of the developed solution is depicted in Fig. 1. The proposed solution has three main stages: image preprocessing, image segmentation, and uptake index computation. The image preprocessing stage is adapted to minimize the noisy artifacts and enhance the contrast of the input bone scintigraphy images [10]. Image segmentation is one of the most common steps in image processing and analysis area, which intends to identify features of interested in input images [11, 12, 13]. Therefore, the enhanced images are submitted to the segmentation stage in order to identify the regions of the skeleton; namely, the skull, spine, thorax, clavicle, femur, humerus, pelvis, scapula, and sternum, Fig. 2.
After the segmentation of the regions under analysis, it is necessary to segment the existent metastasis, commonly known as hotspots. Then, each uptake index assessment consists in computing the ratio between the value of the pixel with the highest intensity of the corresponding hotspot and the pixel with the highest intensity of the region where the hotspot is located without considering its region.
In the preprocessing step, an adaptive histogram equalization algorithm [14] is applied to enhance the contrast of the dark regions of the input images. In this step, it is also employed the anisotropic diffusion algorithm, first introduced by [15], which is a process that creates a space-scale system where an image leads to a parameterized family of images increasingly blurred based on a diffusion process [10]. This technique had become a useful tool to smooth image noise, detect image edges, segment images, and highlight them; particularly, anisotropic diffusion can smooth an input image while preserving the boundaries of the regions and the small structures present in the image [16].
As already mentioned, in the segmentation step, the Point Distribution Models (PDMs) proposed by Cootes and Taylor [6, 7, 8, 17] are used. PDMs have been used in statistical modeling of objects to describe, i.e., learning, their shapes from a set of training images. Thus, the built model describes the mean shape of the modeled object together with admissible variations in relation to the same mean shape [9]. PDMs emerged as a way of representing a set of forms of an object through the use of a flexible model of the position of certain landmarks located in image examples. These landmark points should reflect important characteristics of the shape of the object to be modeled, and must be selected in a similar way in all training images. In practice, this selection step is time-consuming and some automatic and semiautomatic methods have been proposed to define the landmark points to use in the PDMs building process [9].
In the process of building a PDM, the shape of the object to be modeled must be defined in a set of training images through a set of landmark points [8]. Once the points are selected, the coordinates of all n points that describe the i shape of the object are concatenated in vector \( x_{i} \):
where i = 1,…, N, with N representing the number of shapes in the set of training images and n the number of landmark points. Then, all the training shapes must be aligned in the same set of coordinates. After the alignment of the training shapes, it is possible to find the mean of the shapes and the variability presented in the training images. The modes of variation characterize the ways according to the landmarks of the built model tend to move, and can be obtained through a principal component analysis (PCA) to the derivations of the mean [9]. Thus, it is possible to rewrite each coordinates vector as
where x represents the number of points of the resultant shape of the modeled object, \( \left( {x_{k} ,y} \right) \) is the position of landmark point k, \( \bar{x} \) is the mean position of the landmark points, \( P_{s} = \left( {p_{s1} p_{s2} \ldots p_{st} } \right) \) is the matrix of the first t modes of variation, \( p_{si} \) correspond to the most significant eigenvectors in a PCA of the position variables, and \( b_{s} = \left( {b_{s1} b_{s2} \ldots b_{st} } \right)^{T} \) is a vector of weights for each variation mode of the shape. Each eigenvector describes how each landmark point moves on the training image set. Equation (2) represents the PDM of an object and can be used to generate new forms of the same.
Considering the existence of a trained model, i.e., a PDM, the corresponding Active Shape Model (ASM) [8] can be used to find, i.e., to segment, the modeled object in a new image. Starting with an approximate position of the object to be segmented, the ASM based segmentation applies an iterative optimization method to move each PDM landmark point to a better position. The decision-making to find the best position is based on finding the best combination of a local model along the normal boundary profile of the object in the image to be segmented [9].
2.5 Uptake Index Determination
In order to segment the metastasis, it was used the region-growing algorithm [12, 13], that allows the user to select a seed point and, from this seed point, i.e., seed pixel, the region to be segmented starts growing by attaching neighbor pixels that have similar properties.
The next step consists in calculating the uptake index based on the following steps (Fig. 3):
-
For each segmented metastasis:
-
a.
Compute the tumor involvement based on the intensity of the image pixels, Fig. 3: in the anatomical region where the tumor is located, identify the pixel with the highest intensity of the region (R) that does not belong to the tumor region (M); identify the highest intensity pixel in the tumor region; calculate the ratio between the two intensities previously found;
-
b.
Add the computed result of the tumor involvement to the exam uptake index.
-
a.
3 Results
3.1 Segmentation
The proposed computational solution was used on bone scintigraphy images, and each segmented region was compared against the corresponding manually segmented region. The Dice coefficient, Hausdorff distance, and centroid distance were used to validate the computational segmentations. Examples of segmentations obtained by the proposed solution along with the corresponding manual segmentations are shown in Fig. 4.
The computational segmentations obtained for the skull, thorax, pelvis, and thigh were then evaluated. The range of the centroid distance obtained for the skull was 4.49 ± 2.48, for the thorax was 4.78 ± 2.71, 3.44 ± 1.62 for the pelvis and 10.91 ± 6.59 for the thigh. On the other hand, for the Dice Coefficient, the obtained range for the skull was 0.89 ± 0.024, 0.89 ± 0.038 for the thorax, 0.91 ± 0.021 for the pelvis and 0.67 ± 0.073 for the thigh. Finally, the range of the Hausdorff distance obtained for the skull was 3.24 ± 0.53, 5.41 ± 0.56 for the thorax, 5.64 ± 0.72 for the pelvis and 3.98 ± 0.48 for the thigh.
3.2 Uptake Index
The uptake indexes were computed based on the approach previously described. For each patient, it was studied the last two exams, and the obtained indexes were compared with evaluations presented in the clinical reports.
As to the patients with metastatic prostate cancer, there was a decrease in the uptake index from the first to the second study in all patients with the exception of patient # 3, where the uptake index increased, as can be seen in Fig. 5. On the other hand, as to the patients of metastatic breast cancer, there was a decrease in the uptake index in the second study in patients # 1, # 2, # 4, and # 7 relatively to the first study. Contrary, in patients # 3, # 5, and # 6 the uptake index increased for the second study, Fig. 6. Finally, the patient with lung cancer in the second study had no metastasis and an uptake index of 1.98 computed in the first study.
4 Discussion
Regarding the distances between the centroids found for the skull, thorax, and pelvis, the mean of this metric ranged from 3 to 4 pixels, with a standard deviation around 2 pixels. Given that the size of the images under study was equal to 256 × 1024 pixels, the values found for this metric indicate high similarity between the computed and manual segmentations. Regarding the Dice coefficient, values close to 1 (one) show that the segmentations under comparison are similar, whereas closer to 0 (zero) means that there is no similarity between the segmentation. The mean of this metric found for the skull, thorax, and pelvis was 0.89 for the first two and 0.91 for the pelvis, which once again indicates that the segmentation under comparison were close. The Hausdorff distance, and the distance between centroids also shows that there is not a high degree of distinction between the segmentations. However, the comparison of the femoral segmentations showed more discrepant results. The distance between centroids was on average of 10 pixels with a standard deviation higher than the ones found in the previous cases, around 6 pixels. In terms of the Dice coefficient, the average of this metric was 0.67, considerably lower in comparison to the Dice coefficient found for the other cases. In fact, the segmentation of the femur obtained using the built Point Distribution Model generated the most distinct results. One way to solve this problem is to increase the number of training images. Another alternative would be increasing the number of landmark points distributed along the femur in the PDM building process; mainly, around the head of the femur, which is the less homogenous region to a segment.
In what concerns to the computed uptake indexes, the patients with prostate cancer had results that are in agreement with their qualitative evaluations. For example, in the cases where the study was described as “lower osteoblastic intensity”, it was possible to verify a decrease in the uptake indexes, which indicates a good response to the prescribed therapy. However, in the case of patient #3, it was possible to verify a rise in the levels of uptake index, and the description of this study was evaluated as a “mixed response”, this was due to the regression of some hotspots in the first examination and the appearance of new ones. However, when comparing the two studies through the uptake indexes, it was possible to verify the progression of the metastatic disease. To note, the case of patient #4 where the qualitative analysis was described as “overlapping hotspots in relation to the last study” and the indexes obtained showed a decrease from 2.65 to 2.1 suggesting improvements in the hypercaptation hotspots, mainly in the left ischium. In the case of metastatic breast cancer patients, all results are in agreement with the clinical evaluations. For the patient with carcinoma in the lung, it went from an index of 1.98 in the first study to an uptake index of 0 (zero), since there were no metastasis in the most recent study.
5 Conclusion
The challenges regarding the development of solutions for the fully automatic segmentation of the skeleton and metastasis in scintigraphy images remains a strong research topic. A semiautomatic solution for the segmentation of the regions of interest and the extraction of the information from these regions in 99mTc-MDP bone scintigraphy images was described. The developed solution proved to be effective in identifying the regions of interested in the input images. Although some difficulties have arisen in segmenting properly in some regions, these difficulties can be overcome by increasing the number of training images.
The developed solution was applied to 30 whole-body bone scans acquired from 15 patients. The computed uptake indexes were compared with the corresponding clinical evaluations, and a very promising matching was found. However, the proposed solution should be tested using more challenging cases in order to further evaluate and interpret critically the computed uptake indexes; mainly, to verify how they indicate properly the progression or regression of bone metastasis from 99mTc-MDP bone scintigraphy images.
References
Coleman RE (2001) Metastatic bone disease: clinical features, pathophysiology and treatment strategies. Cancer Treat Rev 27(3):165–176
Idota A, Sawaki M, Yoshimura A, Hattori M, Inaba Y, Oze I, Kikumori T, Kodera Y, Iwata H (2016) Bone scan index predicts skeletal-related events in patients with metastatic breast cancer. Springerplus 5(1):1095
Anand A, Morris MJ, Larson SM, Minarik D, Josefsson A, Helgstrand JT, Oturai PS, Edenbrandt L, Røder MA, Bjartell A (2016) Automated bone scan index as a quantitative imaging biomarker in metastatic castration-resistant prostate cancer patients being treated with enzalutamide. EJNMMI Res 6:23
Uemura K, Miyoshi Y, Kawahara T, Yoneyama S, Hattori Y, Teranishi J-I, Kondo K, Moriyama M, Takebayashi S, Yokomizo Y, Yao M, Uemura H, Noguchi K (2016) Prognostic value of a computer-aided diagnosis system involving bone scans among men treated with docetaxel for metastatic castration-resistant prostate cancer. BMC Cancer 16:109
Shimada H, Setoguchi T, Nakamura S, Yokouchi M, Ishidou Y, Tominaga H, Kawamura I, Nagano S, Komiya S (2015) Evaluation of prognostic scoring systems for bone metastases using single-center data. Mol Clin Oncol 3(6):1361–1370
Cootes T, Taylor C (2004) Statistical models of appearance for computer vision, University of Manchester
Cootes T, Hill A, Taylor C, Haslam J (1994) Use of active shape models for locating structures in medical images. Image Vis Comput 12(6):355–365
Cootes T, Taylor C, Cooper D, Graham J (1995) Active shape models-their training and application. Comput Vis Image Underst 61(1):38–59
Vasconcelos MJM, Tavares JMRS (2008) Methods to automatically build point distribution models for objects like hand palms and faces represented in images. Comput Model Eng Sci 36(3):213–241
Tsiotsios C, Petrou M (2013) On the choice of the parameters for anisotropic diffusion in image processing. Pattern Recognit 46(5):1369–1381
Jodas DS, Pereira AS, Tavares JMRS (2016) A review of computational methods applied for identification and quantification of atherosclerotic plaques in images. Expert Syst Appl 46:1–14
Ma Z, Tavares JMRS, Jorge RN, Mascarenhas T (2010) A review of algorithms for medical image segmentation and their applications to the female pelvic cavity. Comput Methods Biomech Biomed Eng 13(2):235–246
Oliveira RB, Filho ME, Ma Z, Papa JP, Pereira AS, Tavares JMRS (2016) Computational methods for the image segmentation of pigmented skin lesions: a Review. Comput Methods Programs Biomed 131:127–141
Pizer SM, Amburn EP, Austin JD, Cromartie R, Geselowitz A, Greer T, Romeny BTH, Zimmerman JB (1987) Adaptive histogram equalization and its variations. Comput Vis Graph Image Process 39(3):355–368
Perona P, Malik J (1990) Scale-space and edge detection using anisotropic diffusion. IEEE Trans Pattern Anal Mach Intell 12(7):629–639
Gulo CASJ, de Arruda HF, de Araujo AF, Sementille AC, Tavares JMRS (2016) Efficient parallelization on GPU of an image smoothing method based on a variational model. J R-Time Image Process. https://doi.org/10.1007/s11554-016-0623-x
Lindner C, Thiagarajah S, Wilkinson JM, Wallis GA, Cootes TF (2011) Short-term variability of proximal femur shape in anteroposterior pelvic radiographs. In: Proceedings of medical image understanding and analysis - MIUA 2011, London
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Araújo, V., Faria, D., Tavares, J.M.R.S. (2019). New Computational Solution to Compute the Uptake Index from 99mTc-MDP Bone Scintigraphy Images. In: Tavares, J., Fernandes, P. (eds) New Developments on Computational Methods and Imaging in Biomechanics and Biomedical Engineering. Lecture Notes in Computational Vision and Biomechanics, vol 33. Springer, Cham. https://doi.org/10.1007/978-3-030-23073-9_10
Download citation
DOI: https://doi.org/10.1007/978-3-030-23073-9_10
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-23072-2
Online ISBN: 978-3-030-23073-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)