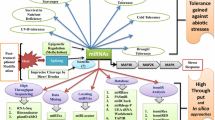
Abstract
MicroRNAs (miRNAs) are small endogenous non-coding RNAs with 20–22 nucleotides acts as the regulatory RNA. Since their discovery in model plant Arabidopsis, curiosity in understanding the function of plant miRNAs increased and regulatory role of miRNA are now being understood by researchers. The ability of miRNA to repress or induce the expression of several genes based on homology of few nucleotides has intrigued the scientific community. The detailed study has elucidated the step-wise biogenesis of miRNA across the species. Plants has evolved to respond to various external stimuli just by tinkering the expression of master regulators like transcription factors, miRNAs, etc. These master regulators further regulate the expression of several hundreds of downstream genes. There are several miRNAs has been identified as regulators of expression of various abiotic stresses viz., drought, cold, salt and high temperature and biotic stresses viz., viruses, bacteria, fungi, nematodes and insect pests. Additionally, paradigm shift in terms of sequencing technology and computational approaches led to identification of differentially expressed miRNAs for various stresses in plants. This led to the identification of thousands of miRNAs across the species which in turn helps to understand the molecular mechanisms involved in providing the stress tolerance. The present review discusses the mechanism of miRNA biogenesis, and role of identified miRNAs in regulating stress response in plants. The understanding of molecular mechanism of tolerance mediated through miRNA may help in improving the crop yields during various stresses.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Plants being sessile in nature have developed the ability to cope up with different growth habitats and fluctuating climatic conditions by improvising myriad regulatory mechanisms. There are many classes of small endogenous RNA molecules , such as small transfer RNA (tRNA), ribosomal RNA (rRNA), small nucleolar RNA (snoRNA), small interfering RNA (siRNA) and microRNA (miRNA). miRNA and siRNA are biochemically and functionally indistinguishable. Both are 19–20 nucleotides (nt) in length with 5′-phosphate and 3′-hydroxyl ends, and assemble into RISC to silence specific gene expression. MicroRNAs (miRNAs) are small non-coding RNAs with 20–22 nucleotides discovered as the regulatory RNA in C. elegans. First plant miRNAs were discovered in Arabidopsis in 2002 and over the past three decades they have been reported in about 120 plant species. A transcribed miRNA acts by different mechanisms like feedback and feedforward loop regulations and has the ability to control its own transcription as well as other genes. A single miRNA may regulate hundreds of mRNAs and in turn may effect a network of interactions. The length of miRNA genes varies from miRNAs to miRNs and from species to species. For example, miRNA genes in plant species are usually longer than in animals. The initial tools like genetic screening for miRNA identification were often time consuming, expensive, and cumbersome. There was a tectonic shift in the technology of the sequencing and computational methods. These advances aided in completion of draft genome sequences with less cost. By using bioinformatic software and tools with the combination of the next-generation deep sequencing, miRNA identification and expression studies in plants have increased dramatically. Computational approaches have estimated that organisms probably contain about 1–5% miRNA genes of the total protein-coding genes (Lai 2003; Lim et al. 2003; Lewis et al. 2005). Notably, various miRNAs are now known to play a role in biotic and abiotic stress, which has led researchers to consider them as a promising tool to develop stress-resistant crops.
According to the United Nations reports present world population is at 7.8 billion and expected to reach 9.8 billion by 2050 (The World Population Prospects: The 2017 Revision). Besides population growth, there is an increase in the prosperity across the world. If the present trend continues, because of the richer diets we should double the amount of crops we grow by 2050. Consequently, to feed the ever-increasing world population it is highly important to find solutions to increase the global food production by developing stress tolerant crop plants. Biotic stresses account for up to 30% crop loss worldwide (Bebber and Gurr 2015). To deal with these devastating pathogens and pests, plants have specialized defense mechanisms, which get induced when there is a stress (rewrite). Plants have developed various physiological and molecular mechanisms to deal with abiotic stresses such as drought, salinity, heat, cold and dehydration by minimizing water loss and photosynthesis. The role of the genes were elucidated either through overexpression or through silencing.
In this chapter, we aimed to describe briefly biogenesis of plant miRNAs and different online tools available for discovering and expression profiling of miRNAs based on computational methods and also understanding their role in tolerance mechanism against abiotic and biotic stress.
2 Biogenesis of miRNAs
There are four steps in biogenesis of miRNAs: (1) MIR genes transcription, (2) miRNA precursor processing, (3) miRNA stabilization and (4) RISC formation.
-
1.
MIR Genes Transcription
The genes which are coding for miRNAs are known as MIR genes . The promoters of MIR genes contain typical TATA box motifs and transcription factor binding motifs indicate that transcription MIR genes is regulated by general and specific transcription factors (Xie et al. 2005; Megraw et al. 2006).The first step in biogenesis of miRNAs starts in the nucleus with primary miRNA (pri-miRNA) transcribed from MIR genes by RNA polymerase II (Xie et al. 2005). Pol II activity in MIR transcription is probably subject to phosphoregulation (Hajheidari et al. 2013). pri-miRNAs can be more than 1 kb in length, they can undergo canonical splicing, polyadenylation, and capping. Just like mRNA, nascent pri-miRNAs are capped at the 5′ end and polyadenylated at the 3′ end, and intron-containing pri-miRNAs are spliced or alternatively spliced (Stepien et al. 2017). The pri-miRNA is processed within the nucleus by a multiprotein complex consisting of DCL1/HYL1/SE called the Microprocessor .
-
2.
miRNA Precursor Processing
The second step involves cleavage of the pri-miRNA into the pre-miRNA, the hairpin structure in the pri-miRNA (Lee et al. 2003). DAWDLE (DDL) is a fork head-associated protein, required for pri-RNA accumulation and recruitment of RNase III family enzyme DICER-like protein 1 (DCL1) to pri-miRNA for downstream processing (Yu et al. 2008). DICER-LIKE1 (DCL1) makes a cut from 15 to 17 nt away from the base of the stem or a bulge or unstructured region within the loop-distal stem. HYPONASTIC LEAVES1 (HYL1) is one of the family member of DOUBLE STRANDED RNA BINDING PROTEINS (DRBs). HYL1 interacts with DCL1 to facilitate efficient and precise miRNA precursor processing (Yang et al. 2014).The resulting precursor-miRNA (pre-miRNA) is further cleaved by DCL1 to produce a 21-nt miRNA/miRNA∗ duplex (Zhu et al. 2013). Alternative processing modes include loop-to-base processing (Bologna et al. 2009). Homodimerization of HYL1 is essential for its functions in miRNA precursor processing (Yang et al. 2014). HYL1 also affects the splicing of some pri-miRNAs and strand selection from miRNA/miRNA∗ duplexes in AGO1 loading (Ben Chaabane et al. 2012). The DCL1 together with HYL1 (HYPONASTIC LEAVES 1) and the zinc-finger protein SE (SERRATE) were required for processing of pre-miRNA into miRNA duplex .
-
3.
miRNA Stabilization
The miRNA/miRNA∗ duplex is stabilized through 3′-terminal 2′-O-methylation by HEN1. The export of miRNAs from the nucleus to the cytoplasm is fundamental for miRNA activity (Köhler and Hurt 2007; Rogers and Chen 2013).The 2-nt 3′ overhang, characteristic of RNase III-mediated cleavage gets methylated by HEN1 (HUA ENHANCER 1), that is recognized by exportin 5, HASTY (HST), is proposed to export the miRNA/miRNA∗ duplex to the cytoplasm based on the assumption that the duplex is produced by DCL1 in the nucleus (Bollman et al. 2003).
-
4.
RISC Formation
In the cytoplasm, miRNAs are unwound into single strand mature miRNAs by helicase. The miRNA strand with relatively lower stability of base-pairing at its 5′ end act as guide molecule to reach the target mRNA and is incorporated into a ribonucleoprotein complex RISC, whereas the other miRNA strand is typically degraded (Du and Zamore 2005). Once incorporated into RISC, the miRNA directs AGO1 (or AGO10) containing RISCs to its target mRNA for cleavage or translational repression on the basis of sequence complementarity. In cases of perfect or near-perfect complementarity to the miRNA, target mRNA scan be cleaved (sliced) and degraded; otherwise, their translation is repressed (Martinez and Tuschl 2004; Treiber et al. 2012). Therefore, miRNAs control gene expression by regulating mRNA stability and translation (Eulalio et al. 2008).
3 List of Bioinformatics Tools for miRNAs Prediction, Identification and Characterization
4 Role of miRNAs in Plant Abiotic Stress Tolerance
Plants have evolved highly sophisticated molecular machinery to cope up and adapt to the challenging environmental conditions. In addition to various mechanisms, miRNAs-mediated rapid response plays crucial role in plant adaption. Various studies have shown that several miRNAs were downregulated in order to increase their target stress-responsive genes during stress conditions in a range of plant species (López and Pérez-Quintero 2012).
4.1 Drought
miRNAs play significant role in sensing the drought stress and imparting tolerance in plants (Ferdous et al. 2015). Drought-responsive miRNAs and their mechanism in drought tolerance is well established in crop plants including Arabidopsis (Clauw et al. 2016), tomato (Liu et al. 2017), rice (Zhou et al. 2010), maize (Aravind et al. 2017), sorghum (Katiyar et al. 2015) and grasses (Zhou et al. 2013). Thirteen miRNAs expression were up-regulated and six miRNAs were downregulated, under drought stress in Arabidopsis. All these differentially expressed miRNAs also play significant role in key developmental process, suggesting that the existence of tight regulation of plant growth and development and drought tolerance (Ferdous et al. 2015; Muthusamy et al. 2017). Under drought stress conditions, miR166, miR167, miR169, miR383 and miR398 family members displayed differential expression pattern in drought tolerant and drought susceptible genotypes (Katiyar et al. 2015). Balyan et al. (2017) studied the drought tolerance mechanism in the set of rice cultivars comprising drought tolerance and susceptible genotypes and showed the role of Cultivar-specific drought responsive (CSDR)-miRNAs networks involving seven family members (osa-miR159f, osa-miR1871, osa-miR398b, osa-miR408-3p, osa-miR2878-5p, osa-miR528-5p and osa-miR397a) by modulating the Cu and ROS homeostasis. This finding shed a novel insight on Cultivar-specific drought responsiveness network which can potentially be targeted in breeding programs in regulating drought responsive genes for the development of new drought tolerant genotypes (Lenka et al. 2018).
4.2 Cold
In Arabidopsis, 11 miRNAs (miR156/157, miR159/319, miR164, miR165/166, miR169, miR172, miR393, miR394, miR396, miR397 and miR398) were induced under cold stress (Sunkar and Zhu 2004; Zhou et al. 2008; Liu et al. 2008; Chinnusamy et al. 2010). Song et al. (2017) identified 34 conserved and 5 novel miRNAs family members that showed a differential expression pattern between the cold-stressed and control spikelet samples of wheat. These miRNAs were known to target the floral organ pattern homeotic transcription factors members including ARF, SPB, MYB and MADS-box. Melatonin induced downregulation of miR159, miR858 and miR8029 increases the cold tolerance ability of Citrullus lanatus L. (Li et al. 2016). Melatonin-mediated miRNA downregulation increases the transcript levels of the target cold tolerance genes involved in signaling, protection and detoxification. In tomato, four miRNAs (miR167, miR169, miR172 and miR393) expression were increased immediately under cold stress (Koc et al. 2015). Cold stress-responsive miRNAs target wide range of proteins with diverse cellular function, indicating an intricate regulation molecular network in responses to cold stress (Chinnusamy and Zhu 2009; Chinnusamy et al. 2010; Megha et al. 2018). Cis-regulatory analysis in the promoters of cold-responsive miRNAs showed the presence of conserved regulatory elements including ABRE, LTRs, MYB binding sites, and HSE (Liu et al. 2008; Zhou et al. 2008).
4.3 Salt
Salt stress inhibits the plant growth and development. High concentration of salts in the plant cells modulates the ABA synthesis which in turn results in closure of stomata, reduction of photosynthesis activity and increase in ROS (Chinnusamy et al. 2006; Mangrauthia et al. 2013). Several salt-stress responsive genes (transcription factors, transporters, ROS enzymes, etc.) were targeted by the miRNAs (Chinnusamy and Zhu 2003; Mondal and Ganie 2014). The expression pattern of the salt stress responsive genes NADP-dependent malic enzyme, cytochrome oxidase and sulfurylase were modulated by miRNAs (Ding et al. 2009; Mangrauthia et al. 2013). The role of miRNAs in imparting tolerance to salt stress were documented in plants (Ferdous et al. 2015). Ten miRNAs (miR156, miR165, miR319, miR393, miR396, miR167, miR168, miR171, miR152 and miR394) were reported to play a pivotal role in salt tolerance in Arabidopsis and chickpea (Liu et al. 2008; Kohli et al. 2014). In Populus, 15 miRNAs targeting the key developmental salt-stress responsive genes regulating auxin signaling, light or circadian rhythms and tissue morphogenesis were differentially expressed under salt-stress condition (Li et al. 2013). A total of 259 miRNAs were differentially expressed in chickpea under salinity and moisture stress conditions (Khandal et al. 2017). Seventy one miRNAs were differentially expressed under salinity in radish (Sun et al. 2015).
4.4 High Temperature
Heat shock responsive transcription factors HSFA1b and HSFA7b induce the expression of high temperature responsive miR398 in Arabidopsis (Guan et al. 2013). In rice, miRNA genes belonging to 162 miRNA families were differentially expressed under high temperature stress, 33 families displayed shoot-specific expression, 12 displayed root-specific expressions, and 117 displayed expression in both shoot and root tissues. Seventy-nine miRNAs were differentially expressed under heat stress conditions in wheat. These results suggest the presence of wider role of miRNA mediated regulation in imparting stress tolerance under heat stress conditions (Mangrauthia et al. 2017). Several heat stress responsive genes including ClpATPase (Muthusamy et al. 2016), HSF (Guan et al. 2013) and HSP (Muthusamy et al. 2017) expression were under regulation of miRNA. MiR396b-3p expression were increased under both heat and drought conditions, suggesting a wider scope for utilization in crop improvement programs in developing climate resilient crop plants (Barciszewska-Pacak et al. 2015).
5 Role of miRNAs in Plant Biotic Stress Tolerance
Plants are faced with innumerable biotic stresses caused by pests, parasites and pathogens. Fungi, bacteria, nematodes and viruses are the pathogens primarily accountable for plant diseases and major concern is of their continuous and fast evolution. Plants have different lines of defense to all these biotic stresses and they respond through several morphological, biochemical, and molecular mechanisms and interactions among their respective signaling pathways (Nejat et al. 2017). One of the lines of plants defense in response to biotic stresses through miRNAs by expressing or regulating stress responsive genes and transcription factors strive to mitigate the stress.
In genomics era, the whole genome, transcriptome, proteome and interactome sequencing and analysis has become a baseline for different areas of research. The small RNA sequencing of organisms identified putative and novel RNAs which might be involved in regulatory pathways. Deep sequencing of stress treated and untreated plant samples showed regulation of small RNAs which could be studied further to improve the conventional approaches for development of stress resistant crops. High throughput sequencing of tomato microRNAs in 2011 identified conserved and novel miRNAs expressed in tomato (Zuo et al. 2011), which regulates the expression of genes involved in biotic stresses. miRNAs are explicitly employed by plants in response to pathogenic attacks.
5.1 Viruses
Viruses contain DNA or RNA as a genetic material in either double-stranded or single-stranded form. The viruses affect host transcriptome levels (Reyes et al. 2016) by transferring genomic DNA/RNA into the host genome. The virus utilizes the host machinery to amplify the genomic content and synthesize proteins by using RNA-dependent RNA polymerase and reverse transcriptases (retroviruses). The viruses also affect the miRNA levels which in-turn affects the fate of the target genes (Pradhan et al. 2015).
In soyabean, 12 potential miRNAs were identified and through 5′-RNA-ligase-mediated rapid amplification of cDNA ends (5′-RLM-RACE) analyses showed 9 miRNAs (miR395, miR530, miR1510, miR1514, miR1515, miR1535, miR2109, miR3522 and miR2118-3p) responded to SMV soybean mosaic virus infection (Yin et al. 2013). In tomato, 40 novel miRNAs were identified in response to cucumber mosaic virus (CMV) and functional analysis revealed miRNA related to defense response and photosynthesis (Feng et al. 2014). In watermelon, by using small RNA sequencing RNA technology, 246 novel miRNAs were identified as differentially expressed in response to cucumber green mottle mosaic virus (CGMMV) infection. Further analysis of these miRNAs revealed, these miRNAs influenced wide array of biological functions like cell-wall enhancing, changes in levels of phytohormones, intracellular transport and modulation of different R genes (Sun et al. 2017). miR168 is ubiquitously up-regulated in most of plant-virus combinations. For example in Malus hupehensis resistance against Botryosphaeria dothidea is conferred by miR168 targeting AGO1 (Yu et al. 2017). Similarly, in rice AGO18 sequesterartion by miR168 confers resistance against viruses (Wu et al. 2015). In response to Mungbean Yellow Mosaic India virus (MYMIV) infection gma-miR5787 maintains AGO homeostasis and targets viral genome in soyabean (Ramesh et al. 2017). In Vigna mungo 14 novel and 53 known miRNAs were identified V. mungo Mungbean Yellow Mosaic India virus (MYMIV). Among the 53 known miRNAs, induction of miR396 suppresses JA signaling there by activating the SA-mediated pathway (Kundu et al. 2017). Besides SA, auxin also regulates plant-pathogen interactions through two candidates miR160 and miR393. PVX-potyvirus synergistic infections alters miRNAs (miR156, miR171, miR398 and miR168) and targeted mRNA levels in Nicotiana benthamiana (Pacheco et al. 2012). Comprehensive genome-wide analyses of miRNA revealed that plants modulate the expression of known, constitutively expressed miRNAs in a spatiotemporal specific manner during viral infection .
5.2 Bacteria
Apart from positive interaction like nitrogen fixation, bacteria also causes diseases through negative interactions. Plant activates pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI), when exposed to flagella or elicitors released from bacterium and further express disease resistance genes in response to Effector triggered susceptibility ETS (Schwessinger and Zipfel 2008).
In Arabidopsis miRNAs were globally profiled in response to infection of Pseudomonas syringae pv. tomato (Pst) and identified several miRNAs that regulate plant hormone signaling and biosynthesis (Zhang et al. 2011). The involvement of these hormone pathways in against bacterial defense has been well established (Berens et al. 2017). For example, SA signaling pathways regulates the anti-biotrophic pathogen defense in plants while positive regulation of JA triggers and regulates the anti-necrotrophs (Tamaoki et al. 2013) defense Pseudomonas syringae and Xanthomonas axonopodis induces miRNAs, for example miR160, miR167 and miR390, miR393 all regulate genes involved in the auxin signaling pathway, including different ARFs and F-boxauxin receptors TIR1, AFB2, and AFB3 mRNAs (Zhang et al. 2011; Snyman et al. 2017; Jodder et al. 2017). Evidently, Auxin response factors are the major targets of most of the upregulated miRNAs whilst downregulated miRNAs targets disease resistance genes. In-fact, miR393 involvement in the regulation of auxin signaling pathway was first discovered in anti-bacterial response of Arabidopsis thaliana through active contribution in PTI (Zhang et al. 2006). Besides auxin, some miRNAs were identified regulated other hormonal pathways, like miR159 was involved in abscisic acid (ABA) signaling pathway and miR319 was involved in jasmonic acid (JA) signaling cascade (Li et al. 2010; Fahlgren et al. 2007). In Arabidopsis, mi393 and SA pathway act synergistically to provide tolerance to bacterial infections (Chen et al. 2014). Further experiments revealed that miR393 down regulate MEMB12 (SNARE) gene that encodes protein involved in membrane fusion.
5.3 Fungal
In Arabidopsis , miR773 was functionally characterized and found that concomitant upregulation of miR773 target gene METHYLTRANSFERASE 2 (MET2) considerably increased resistance to Plectosphaerrella cucumerina, Fusarium oxysporum and Colletototrichum higginianum infection (Salvador-Guirao et al. 2017). In Rice, a total of 33 potential miRNAs were identified in providing immunity against the Blast Fungus Magnaporthe oryzae. Among them miR160a and miR398b were functionally characterized in providing suppression against fungal infection (Li et al. 2014). In cotton, 65 miRNAs were identified as differentially expressed in response to the Verticillium. Among them, Ptc-miR482, Ptc-miR1444 and Ptc-miR1448 were found to specific to cotton cultivars which cleaves the PPO (Polyphenol oxidase) gene in providing resistance (Chi et al. 2014; Tran et al. 2012). In Populus, 74 conserved miRNAs along with 27 novel miRNAs from 37 different miRNA families were identified in response to Dothiorella gregaria. Further analysis revealed miR472, miR1447 and miR1448 were targeting the disease resistance gene (Chen et al. 2012).
The change in hormonal pathways is common to all the biotic stresses. In wheat, enhanced auxin-mediated response was observed against powdery mildew infection by miR393 targeting transport inhibitor response 1 (TIR1), i.e., a negative regulator of auxin signaling (Nowara et al. 2010). In case of infection with Puccinia graminis three independent responses (lignin biosynthesis, hormone signaling, and protein biosynthesis) were regulated through eight miRNAs namely miR159, miR164, miR167, miR171, miR408, miR444, miR1129 and miR1138 (Liu and Chen 2009).
In Brassica species, 62 novel miRNAs were differently expressed under Verticillium longisporeum infection. Among them it was found that miR168 negatively regulates the expression of argonaute1 (AGO1). In most of the fungal infections, pathogens change the expression of DCL1 and AGO1 by overtaking the host machinery and cellular homeostasis. But over the period, plants have acclimatized to the situation and started overexpressing miR162 and miR168 in response to fungal elicitors to maintain the homeotic balance of DCL1 and AGO1 as host derived PTI (Baldrich et al. 2014).
5.4 Nematodes
Over the year, Nematodes have been proved as menace for crop growth, development, yield and productivity. Incidentally, it was in the nematode, Caenorhabditis elegans that MicroRNAs (miRNAs) were first discovered (Lee et al. 1993) and subsequently several miRNAs were discovered in response to nematode infection. For example, in Arabidopsis upon the infection of Heterodera schachtii, miR161, miR164, miR167a, miR172c, miR396c, miR396a,b, and miR398a were downregulated (Kammerhofer et al. 2015) whereas over expression miR827 silences NLA (Nitrogen Limitation Adaptation), which encodes for ubiquitin E3 ligase enzyme leading to susceptibility to Heterodera schachtii (Hewezi et al. 2016). In soyabean, 537 known and 70 putative novel miRNAs were in response to Soybean cyst nematode (SCN) infection of which 60 miRNAs belonging to 25 families were shown to be significantly differentially expressed. After in-depth analysis of these differentially expressed, it was revealed that miR159 and miR399 likely targeting different genes in root during SCN Infection (Tian et al. 2017). In Arabidopsis, miR390/TAS3 discovered as regulatory module for proper gall formation through auxin-responsive factors during infection of Meloidogyne javanica (Cabrera et al. 2016). In a recent study more number of gene regulatory modules were identified, i.e., miRNA172/TOE1, miRNA159/MYB33, miRNA390/TAS3-derived-tasiRNAs/ARFs, miRNA319/TCP4 or miRNA396/GRFs during the gall formation (Cabrera et al. 2018).
5.5 Insect Pests
In Chrysanthemum, a total of 303 conserved miRNAs belonging to 276 miRNAs families and 234 potential novel miRNAs were identified. Among them miR159a, miR160a and miR393a (abundant miRNAs) were found to be responsive to the Chrysanthemum morifolium and aphid interaction. (Xia et al. 2015). In tea plant, 512 novel miRNAs were identified in response to Ectropis oblique feeding. A hypothetical model for miRNA regulatory pathways and their target genes was constructed using the data obtained. This will help to uncover the molecular mechanism involved in stress (Jeyaraj et al. 2017). In most of the cases, pathways were studies in the insect biology and information used for RNAi-based insect control (Xu et al. 2013; Burand and Hunter 2013).
6 Conclusion
Next generation sequencing technologies have enabled to generate voluminous data regarding miRNAs. In combination with the cutting edge computational technologies, researchers able to decipher the role of miRNAs in conferring tolerance to different biotic and abiotic stresses. These findings help to map the detailed molecular mechanism involved in providing the resistance. After considerable meta-analysis, researchers will be enabled to identify conserved pathways and specific pathway. With artificial miRNA (amiRNA) technology emerging as potential tool for gene silencing. The information obtained through different high-throughput sequencing technologies can be useful to construct amiRNAs. With the proper application of genome editing and gene silencing, better varieties could be develop to thrive in adverse conditions and provide good yield.
References
Aravind J, Rinku S, Pooja B, Shikha M, Kaliyugam S, Mallikarjuna MG, Kumar A, Rao AR, Nepolean T (2017) Identification, characterization, and functional validation of drought-responsive microRNAs in subtropical maize inbreds. Front Plant Sci 8:941
Baldrich P, Kakar K, Siré C, Moreno AB, Berger A, García-Chapa M, López-Moya JJ, Riechmann JL, San Segundo B (2014) Small RNA profiling reveals regulation of Arabidopsis miR168 and heterochromatic siRNA415 in response to fungal elicitors. BMC Genomics 15(1):1083
Balyan S, Kumar M, Mutum RD, Raghuvanshi U, Agarwal P, Mathur S, Raghuvanshi S (2017) Identification of miRNA-mediated drought responsive multi-tiered regulatory network in drought tolerant rice, Nagina 22. Sci Rep 7(1):15446
Barciszewska-Pacak M, Milanowska K, Knop K, Bielewicz D, Nuc P, Plewka P, Pacak AM, Vazquez F, Karlowski W, Jarmolowski A, Szweykowska-Kulinska Z (2015) Arabidopsis microRNA expression regulation in a wide range of abiotic stress responses. Front Plant Sci 6:410
Bebber DP, Gurr SJ (2015) Crop-destroying fungal and oomycete pathogens challenge food security. Fungal Genet Biol 74:62–64
Ben Chaabane S, Liu R, Chinnusamy V, Kwon Y, Park JH, Kim SY, Zhu JK, Yang SW, Lee BH (2012) STA1, an Arabidopsis pre-mRNA processing factor 6 homolog, is a new player involved in miRNA biogenesis. Nucleic Acids Res 41(3):1984–1997
Berens ML, Berry HM, Mine A, Argueso CT, Tsuda K (2017) Evolution of hormone signaling networks in plant defense. Annu Rev Phytopathol 55:401–425
Bologna NG, Mateos JL, Bresso EG, Palatnik JF (2009) A loop-to-base processing mechanism underlies the biogenesis of plant microRNAs miR319 and miR159. EMBO J 28(23):3646–3656
Bollman KM, Aukerman MJ, Park MY, Hunter C, Berardini TZ, Poethig RS (2003) HASTY, the Arabidopsis ortholog of exportin 5/MSN5, regulates phase change and morphogenesis. Development 130(8):1493–1504
Burand JP, Hunter WB (2013) RNAi: future in insect management. J Invertebr Pathol 112:S68–S74
Cabrera J, Barcala M, García A, Rio-Machín A, Medina C, Jaubert-Possamai S, Favery B, Maizel A, Ruiz-Ferrer V, Fenoll C, Escobar C (2016) Differentially expressed small RNAs in Arabidopsis galls formed by Meloidogyne javanica: a functional role for miR390 and its TAS3-derived tasiRNAs. New Phytol 209(4):1625–1640
Cabrera J, Ruiz-Ferrer V, Fenoll C, Escobar C (2018) sRNAs involved in the regulation of plant developmental processes are altered during the root-knot nematode interaction for feeding site formation. Eur J Plant Pathol 152:945–955
Chen L, Ren Y, Zhang Y, Xu J, Zhang Z, Wang Y (2012) Genome-wide profiling of novel and conserved Populus microRNAs involved in pathogen stress response by deep sequencing. Planta 235(5):873–883
Chen Z, Hu L, Han N, Hu J, Yang Y, Xiang T, Zhang X, Wang L (2014) Overexpression of a miR393-resistant form of transport inhibitor response protein 1 (mTIR1) enhances salt tolerance by increased osmoregulation and Na+ exclusion in Arabidopsis thaliana. Plant Cell Physiol 56(1):73–83
Chi M, Bhagwat B, Lane WD, Tang G, Su Y, Sun R, Oomah BD, Wiersma PA, Xiang Y (2014) Reduced polyphenol oxidase gene expression and enzymatic browning in potato (Solanum tuberosum L.) with artificial microRNAs. BMC Plant Biol 14(1):62
Chinnusamy V, Zhu J-K (2003) Plant salt tolerance. In H Hirt, K Shinozaki Plant responses to abiotic stress. Springer, Berlin, pp 241–270
Chinnusamy V, Zhu JK (2009) Epigenetic regulation of stress responses in plants. Curr Opin Plant Biol 12(2):133–139
Chinnusamy V, Zhu J, Zhu JK (2006) Salt stress signaling and mechanisms of plant salt tolerance. In: Setlow JK (ed) Genetic engineering. Springer, Boston, MA, pp 141–177
Chinnusamy V, Zhu JK, Sunkar R (2010) Gene regulation during cold stress acclimation in plants. In: Sunkar R (ed) Plant stress tolerance. Humana Press, Totowa, NJ, pp 39–55
Clauw P, Coppens F, Korte A, Herman D, Slabbinck B, Dhondt S, Van Daele T, De Milde L, Vermeersch M, Maleux K, Maere S (2016) Leaf growth response to mild drought: natural variation in Arabidopsis sheds light on trait architecture. Plant Cell 28(10):2417–2434
Ding D, Zhang L, Wang H, Liu Z, Zhang Z, Zheng Y (2009) Differential expression of miRNAs in response to salt stress in maize roots. Ann Bot 103(1):29–38
Du T, Zamore PD (2005) microPrimer: the biogenesis and function of microRNA. Development 132(21):4645–4652
Eulalio A, Huntzinger E, Izaurralde E (2008) Getting to the root of miRNA-mediated gene silencing. Cell 132(1):9–14
Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, Carrington JC (2007) High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS One 2(2):e219
Feng J, Liu S, Wang M, Lang Q, Jin C (2014) Identification of microRNAs and their targets in tomato infected with Cucumber mosaic virus based on deep sequencing. Planta 240(6):1335–1352
Ferdous J, Hussain SS, Shi BJ (2015) Role of microRNAs in plant drought tolerance. Plant Biotechnol J 13(3):293–305
Guan Q, Lu X, Zeng H, Zhang Y, Zhu J (2013) Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. Plant J 74(5):840–851
Hajheidari M, Koncz C, Eick D (2013) Emerging roles for RNA polymerase II CTD in Arabidopsis. Trends Plant Sci 18(11):633–643
Hewezi T, Piya S, Qi M, Balasubramaniam M, Rice JH, Baum TJ (2016) Arabidopsis miR827 mediates post-transcriptional gene silencing of its ubiquitin E3 ligase target gene in the syncytium of the cyst nematode Heterodera schachtii to enhance susceptibility. Plant J 88(2):179–192
Jeyaraj A, Liu S, Zhang X, Zhang R, Shangguan M, Wei C (2017) Genome-wide identification of microRNAs responsive to Ectropis oblique feeding in tea plant (Camellia sinensis L.). Sci Rep 7(1):13634
Jodder J, Basak S, Das R, Kundu P (2017) Coherent regulation of miR167a biogenesis and expression of auxin signaling pathway genes during bacterial stress in tomato. Physiol Mol Plant Pathol 100:97–105
Kammerhofer N, Radakovic Z, Regis J, Dobrev P, Vankova R, Grundler FM, Siddique S, Hofmann J, Wieczorek K (2015) Role of stress-related hormones in plant defence during early infection of the cyst nematode Heterodera schachtii in Arabidopsis. New Phytol 207(3):778–789
Katiyar A, Smita S, Muthusamy SK, Chinnusamy V, Pandey DM, Bansal KC (2015) Identification of novel drought-responsive microRNAs and trans-acting siRNAs from Sorghum bicolor (L.) Moench by high-throughput sequencing analysis. Front Plant Sci 6:506
Khandal H, Parween S, Roy R, Meena MK, Chattopadhyay D (2017) MicroRNA profiling provides insights into post-transcriptional regulation of gene expression in chickpea root apex under salinity and water deficiency. Sci Rep 7(1):4632
Koc I, Filiz E, Tombuloglu H (2015) Assessment of miRNA expression profile and differential expression pattern of target genes in cold-tolerant and cold-sensitive tomato cultivars. Biotechnol Biotechnol Equip 29(5):851–860
Köhler A, Hurt E (2007) Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol 8(10):761
Kohli D, Joshi G, Deokar AA, Bhardwaj AR, Agarwal M, Katiyar-Agarwal S, Srinivasan R, Jain PK (2014) Identification and characterization of wilt and salt stress-responsive microRNAs in chickpea through high-throughput sequencing. PLoS One 9(10):e108851
Kundu A, Paul S, Dey A, Pal A (2017) High throughput sequencing reveals modulation of microRNAs in Vigna mungo upon Mungbean Yellow Mosaic India Virus inoculation highlighting stress regulation. Plant Sci 257:96–105
Lai EC (2003) microRNAs: runts of the genome assert themselves. Curr Biol 13(23):R925–R936
Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75(5):843–854
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, Kim VN (2003) The nuclear RNase III Drosha initiates microRNA processing. Nature 425(6956):415
Lenka SK, Muthusamy SK, Chinnusamy V, Bansal KC (2018) Ectopic expression of rice PYL3 enhances cold and drought tolerance in Arabidopsis thaliana. Mol Biotechnol 60:350–361
Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120(1):15–20
Li Y, Zhang Q, Zhang J, Wu L, Qi Y, Zhou JM (2010) Identification of microRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity. Plant Physiol 152(4):2222–2231
Li B, Duan H, Li J, Deng XW, Yin W, Xia X (2013) Global identification of miRNAs and targets in Populus euphratica under salt stress. Plant Mol Biol 81(6):525–539
Li Y, Lu YG, Shi Y, Wu L, Xu YJ, Huang F, Guo XY, Zhang Y, Fan J, Zhao JQ, Zhang HY (2014) Multiple rice microRNAs are involved in immunity against the blast fungus Magnaporthe oryzae. Plant Physiol 164(2):1077–1092
Li H, Dong Y, Chang J, He J, Chen H, Liu Q, Wei C, Ma J, Zhang Y, Yang J, Zhang X (2016) High-throughput microRNA and mRNA sequencing reveals that microRNAs may be involved in melatonin-mediated cold tolerance in Citrullus lanatus L. Front Plant Sci 7:1231
Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP (2003) Vertebrate microRNA genes. Science 299(5612):1540–1540
Liu Q, Chen YQ (2009) Insights into the mechanism of plant development: interactions of miRNAs pathway with phytohormone response. Biochem Biophys Res Commun 384(1):1–5
Liu HH, Tian X, Li YJ, Wu CA, Zheng CC (2008) Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA 14(5):836–843
Liu M, Yu H, Zhao G, Huang Q, Lu Y, Ouyang B (2017) Profiling of drought-responsive microRNA and mRNA in tomato using high-throughput sequencing. BMC Genomics 18(1):481
López C, Pérez-Quintero A (2012) The micromics revolution: microRNA-mediated approaches to develop stress-resistant crops. In: Tuteja N, Gill SS, Tiburcio AF, Tuteja R (eds) Improving crop resistance to abiotic stress, vol 1 and 2. Wiley, Chichester, pp 559–590
Mangrauthia SK, Agarwal S, Sailaja B, Madhav MS, Voleti SR (2013) MicroRNAs and their role in salt stress response in plants. In: Ahmad P, Azooz MM, Prasad MNV (eds) Salt stress in plants. Springer, New York, NY, pp 15–46
Mangrauthia SK, Bhogireddy S, Agarwal S, Prasanth VV, Voleti SR, Neelamraju S, Subrahmanyam D (2017) Genome-wide changes in microRNA expression during short and prolonged heat stress and recovery in contrasting rice cultivars. J Exp Bot 68(9):2399–2412
Martinez J, Tuschl T (2004) RISC is a 5′ phosphomonoester-producing RNA endonuclease. Genes Dev 18(9):975–980
Megha S, Basu U, Kav NN (2018) Regulation of low temperature stress in plants by microRNAs. Plant Cell Environ 41(1):1–15
Megraw M, Baev V, Rusinov V, Jensen ST, Kalantidis K, Hatzigeorgiou AG (2006) MicroRNA promoter element discovery in Arabidopsis. RNA 12(9):1612–1619
Mondal TK, Ganie SA (2014) Identification and characterization of salt responsive miRNA-SSR markers in rice (Oryza sativa). Gene 535(2):204–209
Muthusamy SK, Dalal M, Chinnusamy V, Bansal KC (2016) Differential regulation of genes coding for organelle and cytosolic ClpATPases under biotic and abiotic stresses in wheat. Front Plant Sci 7:929
Muthusamy SK, Dalal M, Chinnusamy V, Bansal KC (2017) Genome-wide identification and analysis of biotic and abiotic stress regulation of small heat shock protein (HSP20) family genes in bread wheat. J Plant Physiol 211:100–113
Nowara D, Gay A, Lacomme C, Shaw J, Ridout C, Douchkov D, Hensel G, Kumlehn J, Schweizer P (2010) HIGS: host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell 22(9):3130–3141
Pacheco R, García-Marcos A, Barajas D, Martiáñez J, Tenllado F (2012) PVX–potyvirus synergistic infections differentially alter microRNA accumulation in Nicotiana benthamiana. Virus Res 165(2):231–235
Pradhan B, Naqvi AR, Saraf S, Mukherjee SK, Dey N (2015) Prediction and characterization of Tomato leaf curl New Delhi virus (ToLCNDV) responsive novel microRNAs in Solanum lycopersicum. Virus Res 195:183–195
Ramesh SV, Sahu PP, Prasad M, Praveen S, Pappu HR (2017) Geminiviruses and plant hosts: a closer examination of the molecular arms race. Viruses 9(9):256
Reyes CA, Ocolotobiche EE, Marmisollé FE, Robles Luna G, Borniego MB, Bazzini AA, Asurmendi S, García ML (2016) Citrus psorosis virus 24K protein interacts with citrus miRNA precursors, affects their processing and subsequent miRNA accumulation and target expression. Mol Plant Pathol 17(3):317–329
Rogers K, Chen X (2013) Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25(7):2383–2399
Salvador-Guirao R, Baldrich P, Weigel D, Rubio-Somoza I, San Segundo B (2017) The microRNA miR773 is involved in the Arabidopsis immune response to fungal pathogens. Mol Plant-Microbe Interact 31(2):249–259
Schwessinger B, Zipfel C (2008) News from the frontline: recent insights into PAMP-triggered immunity in plants. Curr Opin Plant Biol 11(4):389–395
Song G, Zhang R, Zhang S, Li Y, Gao J, Han X, Chen M, Wang J, Li W, Li G (2017) Response of microRNAs to cold treatment in the young spikes of common wheat. BMC Genomics 18(1):212
Snyman MC, Solofoharivelo MC, Souza-Richards R, Stephan D, Murray S, Burger JT (2017) The use of high-throuhput small RNA sequencing reveals differentially expressed microRNAs in response to aster yellows phytoplasma-infection in Vitis vinifera cv. ‘Chardonnay’. PLoS One 12(8):e0182629
Stepien A, Knop K, Dolata J, Taube M, Bajczyk M, Barciszewska-Pacak M, Pacak A, Jarmolowski A, Szweykowska-Kulinska Z (2017) Posttranscriptional coordination of splicing and miRNA biogenesis in plants. Wiley Interdiscip Rev RNA 8(3). https://doi.org/10.1002/wrna.1403
Sun X, Xu L, Wang Y, Yu R, Zhu X, Luo X, Gong Y, Wang R, Limera C, Zhang K, Liu L (2015) Identification of novel and salt-responsive miRNAs to explore miRNA-mediated regulatory network of salt stress response in radish (Raphanus sativus L.). BMC Genomics 16(1):197
Sun Y, Niu X, Fan M (2017) Genome-wide identification of cucumber green mottle mosaic virus-responsive microRNAs in watermelon. Arch Virol 162(9):2591–2602
Sunkar R, Zhu JK (2004) Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16(8):2001–2019
Tamaoki D, Seo S, Yamada S, Kano A, Miyamoto A, Shishido H, Miyoshi S, Taniguchi S, Akimitsu K, Gomi K (2013) Jasmonic acid and salicylic acid activate a common defense system in rice. Plant Signal Behav 8(6):e24260
Tian B, Wang S, Todd TC, Johnson CD, Tang G, Trick HN (2017) Genome-wide identification of soybean microRNA responsive to soybean cyst nematodes infection by deep sequencing. BMC Genomics 18(1):572
Tran LT, Taylor JS, Constabel CP (2012) The polyphenol oxidase gene family in land plants: lineage-specific duplication and expansion. BMC Genomics 13(1):395
Treiber T, Treiber N, Meister G (2012) Regulation of microRNA biogenesis and function. Thromb Haemost 108(04):605–610
Wu J, Yang Z, Wang Y, Zheng L, Ye R, Ji Y, Zhao S et al (2015) Viral-inducible Argonaute18 confers broad-spectrum virus resistance in rice by sequestering a host microRNA. elife 4:e05733
Xia X, Shao Y, Jiang J, Du X, Sheng L, Chen F, Fang W, Guan Z, Chen S (2015) MicroRNA expression profile during aphid feeding in chrysanthemum (Chrysanthemum morifolium). PLoS One 10(12):e0143720
Xie Z, Allen E, Fahlgren N, Calamar A, Givan SA, Carrington JC (2005) Expression of Arabidopsis MIRNA genes. Plant Physiol 138(4):2145–2154
Xu HJ, Chen T, Ma XF, Xue J, Pan PL, Zhang XC, Cheng JA, Zhang CX (2013) Genome-wide screening for components of small interfering RNA (siRNA) and micro-RNA (miRNA) pathways in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae). Insect Mol Biol 22(6):635–647
Yang X, Ren W, Zhao Q, Zhang P, Wu F, He Y (2014) Homodimerization of HYL1 ensures the correct selection of cleavage sites in primary miRNA. Nucleic Acids Res 42(19):12224–12236
Yin X, Wang J, Cheng H, Wang X, Yu D (2013) Detection and evolutionary analysis of soybean miRNAs responsive to soybean mosaic virus. Planta 237(5):1213–1225
Yu B, Bi L, Zheng B, Ji L, Chevalier D, Agarwal M, Ramachandran V, Li W, Lagrange T, Walker JC, Chen X (2008) The FHA domain proteins DAWDLE in Arabidopsis and SNIP1 in humans act in small RNA biogenesis. Proc Natl Acad Sci U S A 105(29):10073–10078
Yu X, Hou Y, Chen W, Wang S, Wang P, Qu S (2017) Malus hupehensis miR168 targets to ARGONAUTE1 and contributes to the resistance against Botryosphaeria dothidea infection by altering defense responses. Plant Cell Physiol 58(9):1541–1557
Zhang B, Pan X, Cobb GP, Anderson TA (2006) Plant microRNA: a small regulatory molecule with big impact. Dev Biol 289(1):3–16
Zhang W, Gao S, Zhou X, Chellappan P, Chen Z, Zhou X, Zhang X, Fromuth N, Coutino G, Coffey M, Jin H (2011) Bacteria-responsive microRNAs regulate plant innate immunity by modulating plant hormone networks. Plant Mol Biol 75(1–2):93–105
Zhou X, Wang G, Sutoh K, Zhu JK, Zhang W (2008) Identification of cold-inducible microRNAs in plants by transcriptome analysis. Biochim Biophys Acta 1779(11):780–788
Zhou L, Liu Y, Liu Z, Kong D, Duan M, Luo L (2010) Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa. J Exp Bot 61(15):4157–4168
Zhou M, Li D, Li Z, Hu Q, Yang C, Zhu L, Luo H (2013) Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bentgrass. Plant Physiol 161(3):1375–1391
Zhu H, Zhou Y, Castillo-González C, Lu A, Ge C, Zhao YT, Duan L, Li Z, Axtell MJ, Wang XJ, Zhang X (2013) Bidirectional processing of pri-miRNAs with branched terminal loops by Arabidopsis Dicer-like1. Nat Struct Mol Biol 20(9):1106
Zuo JH, Wang YX, Liu HP, Ma YZ, Ju Z, Zhai BQ, Fu DQ, Zhu Y, Luo YB, Zhu BZ (2011) MicroRNAs in tomato plants. Sci China Life Sci 54(7):599–605
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Jagannadham, P.T.K., Muthusamy, S.K., Chidambaranathan, P. (2019). Micromics: A Novel Approach to Understand the Molecular Mechanisms in Plant Stress Tolerance. In: Wani, S. (eds) Recent Approaches in Omics for Plant Resilience to Climate Change. Springer, Cham. https://doi.org/10.1007/978-3-030-21687-0_5
Download citation
DOI: https://doi.org/10.1007/978-3-030-21687-0_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-21686-3
Online ISBN: 978-3-030-21687-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)