Abstract
Bioinformatics plays an indispensable role and has a number of applications in today’s plant and crop science. Powerful tools and methods are required to organize the huge data and extend our ability to analyse complex biological systems. At the same time, many researchers in plant biology are unacquainted with available bioinformatics methods, tools and databases, which could lead to unexploited opportunities or misinterpretation of the information. Omics has allowed us to come to terms with vast amounts of biological data being generated by sequencing projects and has made considerable progress in plant breeding by providing scientists and breeders access to genomic information. In plant biology, these omics tools (proteomics, genomics, transcriptomics) are helpful in improving the quality of traditional medicinal plants; improving crop nutrition quality, single gene analysis, sequence similarity, modelling of protein, crop breeding and insect resistance; improving nutritional quality; and development of drought-resistant varieties. Thus, plant genomics has assisted in the efficient exploitation of plants as biological resources. Further, the progress made in molecular plant breeding, genetics, genomic selection and genome editing has contributed to a more comprehensive understanding of molecular markers, provided deeper insights into the diversity available for crops and greatly complemented breeding stratagems. In this chapter, we describe the main bioinformatics approaches in the area of next-generation sequencing (NGS) for its impact on crop improvement and breeding programmes. We also cover some molecular and genetic marker databases, which find its application in breeding programmes. Finally, we explore a few emerging research topics in this innovative field of research.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Agricultural bioinformatics
- Omics technologies
- Crop improvement
- Crop breeding
- Fruit breeding
- Molecular markers
- Genetic markers
2.1 Introduction
Crops play a significant and diverse role in our economy, environment and feeding the increasing world population. Increased demand for biofuel crops, population explosion and global climate change have become a challenge for current plant biotechnology, and sustainable agricultural production is an urgent issue in this response (Brown and Funk 2008; Ozturk 2010; Hakeem et al. 2012). Climate change will severely influence the world’s food supply, and it is predicted to have immense negative effects on both the yield and the quality of crop plants (Kumar 2016), unless steps are taken to increase crop resilience. Plant genomics is a potentially powerful defence against this looming threat. Now to solve these issues and increase crop yields, breeding of novel crops and adaptation of current crops to the new environment based on a better molecular understanding of gene function, and on the regulatory mechanisms involved in crop production (Takeda and Matsuoka 2008), have become a primary necessity. Crop yields have increased during the past century and will continue due to enhanced breeding and new biotechnological-engineered strategies. Some of the important gene sequences and their function have been designated, many of which are related to crop yields (production), crop quality, tolerance to biotic and abiotic stresses and development of molecular markers (De Filippis 2012). One vital tool of bioinformatics is “genomics”, which is commonly used to identify genotypic and phenotypic changes in plants, and this information helps in improving the overall performance of crop plants (Ahmad et al. 2011).
Modern technologies of bioinformatics have enhanced the study of plant biology to a higher level than before and have assisted in unravelling genetic and molecular networks (Schuster 2007). After a rapid surge in genome sequencing through innovative high-throughput methods, scientists have an opportunity to exploit the structure of the plant genetic material at the molecular level which is known as “plant genomics” (Govindaraj et al. 2015). Some of the latest applications of bioinformatics in plant science research field (Fig. 2.1) are as follows:
-
Integrated “omics” strategies clarify the molecular system of the plant which is used to improve the plant productivity. Innovations in omics-based research improve plant-based research.
-
Genomics strategy, especially comparative genomics, helps in understanding the genes and their functions and the biological properties of each species.
-
Bioinformatics databases are also used in designing new techniques and experiments for increased plant production (Mochida and Shinozaki 2010).
-
Advancement in the bioinformatics tools has enabled us in providing information about the genes present in the genome of microorganisms (role in agriculture). These tools have also made it possible to predict the function of different genes and factors affecting these genes, and this information is used by scientists to produce enhanced species of plants which have a drought, herbicide and pesticide resistance in them (Mochida and Shinozaki 2010).
-
Nowadays, genomics provides breeders with a new set of tools and techniques that allow the study of the whole genome, and which represents a paradigm shift, by facilitating the direct study of the genotype and its relationship with the phenotype (Tester and Langridge 2010). The present genomics is leading to a new revolution in plant breeding at the beginning of the twenty-first century.
At the most fundamental level, the advances in genomics will greatly accelerate the acquisition of knowledge and that, in turn, will directly affect many aspects of the processes associated with plant improvement. Bioinformatics information and databases have become ready-to-use tools for crop scientists and breeders in gene data mining and linking this knowledge to its biological significance (Mochida and Shinozaki 2010). Knowledge of the function of all plant genes, in conjunction with the further development of tools for modifying and interrogating genomes, will lead to the development of a genuine genetic engineering paradigm in which rational changes can be designed and modelled from the first principles. Bioinformatics, when combined with genomics, has the potential to help maintain food security in the face of climate change through the accelerated production of climate-ready crops (Batley and Edwards 2016). Based on these understandings, this chapter focuses on challenges and opportunities, which knowledge and skills in bioinformatics can bring to plant scientists in present plant genomics era as well as future aspects in critical need for effective tools to facilitate the translation of knowledge from new sequencing data to the improvement of plant productivity. This chapter emphasizes on a number of applications of bioinformatics in agriculture in view of crop improvement, breeding programmes, fruit breeding, overviewing the main bioinformatics strategies and challenges, as well as perspectives in this field and various bioinformatics tools/databases important for breeders and plant biotechnologists.
2.2 Role of Bioinformatics in Crop Improvement
To understand and unravel the genetic and molecular basis of all biological processes in plants have become a key objective for plant and crop biologists. This understanding helps in the practical utilization of plants as biological resources in the development of new cultivars with improved quality and at reduced economic costs (Schlueter et al. 2003). Since climate change and population explosion have increased pressure on our ability to produce sufficient food, the breeding of novel crops and the adaptation of current crops to the new environment are required to ensure continued food production. At the most fundamental level, advances in genomics have accelerated the acquisition of knowledge and that, in turn, has helped in providing rational annotation of genes, proteins and phenotypes, and this omics data can now be envisioned as a highly important tool for plant improvement (Fig. 2.1). Several new gene-finding tools are tailored for applications to plant genomic sequences, which have resulted in enhancing the nutritional quality and composition of food crops, increasing agricultural production for food, feed and energy (Schlueter et al. 2003; Van Emon 2016). This amalgam of bioinformatics with genomic tools has potential to maintain food security in the face of climate change through the accelerated production of new cultivars with improved quality and reduced economic and environmental cost (Batley and Edwards 2016).
The onset of research in the field of sequence analysis and genome annotation has played a significant role in the area of crop improvement. Whole-genome sequencing of several species permits to define their organization and provides the starting point for understanding their functionality (Ellegren 2014), therefore favouring human agriculture practice. An extremely large amount of genomics data is available from plants due to the tremendous improvements in the field of omics (Fig. 2.1), and nowadays function of different genes in the plant and the factors affecting these genes can be predicted (Morrell et al. 2012). This information has helped scientists to generate plant species resistant to abiotic and biotic stresses, herbicides and pesticides. In recent years, a number of latest sequencing technologies, which are adaptations of already existing pyro-sequencing methods (Ansorge 2009), have provided us with new opportunities to be addressed at the entire genome level in the fields of comparative genomics, meta-genomics and evolutionary genomics (Varshney et al. 2009). Indeed, the contribution of genomics to agriculture spans the identification and the manipulation of genes linked to specific phenotypic traits (Zhang et al. 2014) as well as genomics breeding by marker-assisted selection of variants (Organization 2005). Efforts addressed to the achievement of appropriate knowledge of associated molecular information, such as the one arising from transcriptome, metabolome and proteome sequencing (Fig. 2.2), are also essential to better depict the gene content of a genome and its main functionalities.
Current advances in genomics and bioinformatics provide opportunities for accelerating crop improvement (Fig. 2.2) in the following areas:
-
“Gene finding” refers to the prediction of introns and exons in a segment of DNA sequence. Bioinformatics has aided in genome sequencing, and it has shown its success in locating the genes, in phylogenetic comparison and in the detection of transcription factor binding sites of the genes. Such an approach to identify key genes and understand their function will result in a “quantum leap” in quantitative and qualitative trait improvement in commercially important crops (Morrell et al. 2012).
-
“Comparative genetics” (model and non-model plant) with computational tools can reveal an organization of agronomically important genes with respect to each other which can be further used for transferring information from the model crop systems to other food crops. Species-specific nucleotide sequences are now providing information related to phenotypic characters, even when based on genome comparative analyses from the few model plants available (Cogburn et al. 2007; Paterson 2008).
-
“Cheminformatics” for designing of agrochemicals is based on an analysis of the components of signal perception and transduction pathways to select targets and to identify potential compounds that can be used as herbicides, pesticides or insecticides, thereby improving plant quality and quantity (Bennetzen et al. 1998).
-
“Agricultural genomics” leads to the global understanding of plant and pathogen biology, and its application would be beneficial for agriculture and in providing massive information to improve the crop phenotype. Further, whole-genome sequencing in plants allows chromosome-scale genetic comparisons, thereby identifying conserved genetic areas, which can facilitate identification and documentation of similar genomic sequences in related plant species (Haas et al. 2004; De Bodt et al. 2005).
-
“Microarray technology” has been widely adopted in gene expression analysis in crop plants to clarify the function of key genes and uncover the regulation mechanism through dissecting regulatory elements and the interaction of responsible genes. These gene expression studies allow us to understand how plants respond to and interact with the physical environment and management practices. These data may become a crucial tool of future breeding decision management systems (Langridge and Fleury 2011).
-
“Full-length cDNA libraries” serve as primary sequence resources for designing microarray probes and as clone resources for genetic engineering to improve crop efficiency (Futamura et al. 2008). These libraries have been used to identify biological features through comparisons of target sequences with those of model organisms.
-
“Multiple alignments” provide a method to estimate the number of genes in the gene families and in the identification of the previously undescribed genes. The multiple alignment information helps in studying the gene expression pattern in plants.
-
“Mutant analysis” is an effective approach for the investigation of gene function (Stanford et al. 2001). Comprehensive collections of mutant lines are also essential bioresources for radically accelerating forward and reverse genetics.
-
“Gene pyramiding” or gene stacking implies multiple desirable genes are assembled from different parent crops to enhance trait and develop elite lines and varieties. It is mainly used in improving existing elite cultivars for a few unsatisfactory traits, for which genes with large positive effects are identified (Malav et al. 2016).
-
“Molecular DNA marker” identification and location have contributed significantly to marker-assisted studies and selection (MAS) in plant breeding, and in a wider range of research, including species identification and evolution (Feltus et al. 2004; Varshney et al. 2005).
-
“Genetic markers” constructed to cover the complete genome may allow identification of individual genes associated with complex traits by QTL (quantitative trait loci) analysis and the identification of genetic diversity and induced variations (Feltus et al. 2004; Varshney et al. 2005).
-
In silico genomics technology has made it easier for researchers (working on plant-pathogen interactions) to identify defence/disease-resistant gene-enzyme with their promoter region and transcription factor which help to enhance the immunity and defence mechanisms (Pandey and Somssich 2009).
-
Bioinformatics has also enabled scientists to improve the nutritional quality of the plants by making changes in its genome. Researchers have been successful in inserting genes in the genome of rice to increase vitamin A levels. The genetically modified rice contains more vitamin A (essential to maintain healthy eyes) that has helped in reducing the blindness rate worldwide (Ye et al. 2000).
-
Bioinformatics tools are also indispensable to agriculture and horticulture from the climate change perspective. Some varieties of cereal have been modified to be drought/submergence resistant and enhanced to grow in infertile soils.
-
“Host-pathogen interactions” help in understanding the disease genetics and pathogenicity factor of a pathogen, which ultimately helped in designing best management options. Metagenomics and transcriptomics approaches are used to understand the genetic architecture of microorganism and pathogens to check how these microbes affect the host plant so that pathogen−/insect-resistant crop is generated and in the identification of host beneficial microbes (Schenk et al. 2012).
-
“Insect genomics” helps in the identification of resistance mechanisms and finding the novel target sites (Cory and Hoover 2006). By mapping the genome for Bacillus thuringiensis (bacteria that increases soil fertility and protects the plants from pests), scientists were able to incorporate these genes into the plant (e.g. cotton, maize and potato), which made them insect resistant. This resulted in a decrease in insecticide usage, enhancing productivity and nutritional value of crops.
-
Improving crops through breeding is a sustainable approach to increase yield and yield stability without intensifying the use of fertilizers and pesticides. Third-generation sequencing technologies are assisting to overcome challenges in plant genome assembly caused by polyploidy and frequent repetitive elements. As a result, high-quality crop reference genomes are increasingly available, benefitting downstream analyses such as variant calling and association mapping that identify breeding targets in the genome (Hu et al. 2018).
-
Machine learning also helps identify genomic regions of agronomic value by facilitating functional annotation of genomes and enabling real-time high-throughput phenotyping of agronomic traits in the glasshouse and in the field.
-
Crop databases integrate the growing volume of genotype and phenotype data, providing a valuable resource for breeders and an opportunity for data mining approaches to uncover novel trait-associated candidate genes (Hu et al. 2018).
2.3 Crop Breeding: Bioinformatics and Preparing for Climate Change
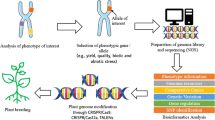
Plant breeding has been practised for thousands of years, since near the beginning of human civilization (Kingsbury 2009), “plant breeding is the art and science of changing the genetic structure of plants in order to produce desired characteristics” (Sleper and Poehlbman 2006). Bioinformatics has been involved in different aspects of sciences including plant breeding, and a large portion of these tools and techniques are related to the omics category (Barh et al. 2013). From the past few years, plant breeding has been extended through development and deployment of a large number of methods and tools with respect to specific objectives (Al-Khayri et al. 2015). With the use of omics, the consistency and predictability of plant breeding programmes have been improved, reducing the time and the expense of stress-tolerant varieties (Van Emon 2016). The field of genomics and its application to plant breeding are developing very quickly, and this boom in plant breeding has started after genome sequencing of Arabidopsis and rice (Kaul et al. 2000; Matsumoto et al. 2005), followed by many genome sequencing projects of different plant species (Skuse and Du 2008). The combination of conventional breeding techniques with genomic tools and approaches is leading to a new genomics-based plant breeding (Fig. 2.3). A fully assembled and well-annotated genome will allow breeders to discover genes related to agronomic traits, determine their location and function as well as develop genome-wide molecular markers (Hu et al. 2018).
Reaping benefits of omics in crop breeding. Discovery of the genes and the genetic architecture by different omics underlying critical traits provides insights for crop improvement. Identification of genes and quantitative trait loci (QTL) and genome-wide association study (GWAS) enhances rice yield, quality and stress tolerance in a wide range of environments. Genetic maps help to locate genes and provide molecular/genetic markers for selection. Gene discovery provides knowledge of genetic mechanisms and interactions. Databases provided (barcoded) sample tracking and breeding history (pedigrees). Phenotypes (trait measurements) are stored with experimental design and environmental data and can be connected to individual and genotype (marker). All these constitute a toolbox for plant breeders
One of the most substantial transformation of bioinformatics’ techniques in plant breeding is that it had replaced the conventional molecular marker technology with high-throughput DNA sequencing technologies and has developed a number of databases (Mochida and Shinozaki 2010) (Table 2.1). These and other technical revolutions provide genome-wide molecular tools for breeders (large collections of markers, high-throughput genotyping strategies, high-density genetic maps, etc.) that can be incorporated into existing breeding methods (Tester and Langridge 2010; Lorenz et al. 2011). With the progress of genome sequencing and large-scale EST analysis in various species, these sequence datasets have become quite efficient sequence resources for designing molecular markers covering the entire genomes (Feltus et al. 2004). Recent advances in genomics are producing new plant breeding methodologies, improving and accelerating the breeding process in many ways (e.g. association mapping, marker-assisted selection, “breeding by design”, gene pyramiding, genomic selection, etc.) (Lorenz et al. 2011). Some of these molecular and genetic markers, which have played a significant role in improving plant breeding, are as follows:
-
1.
Crop breeders have known the complexity of multiple alleles for decades. However, with the advent of molecular markers, genetic diversity and other forms of genetic structure in breeding populations are possible. For high-throughput genotyping, a number of platforms have been developed that have been applied to genetic map construction, marker-assisted selection and QTL cloning using multiple segregation populations (Hori et al. 2007). Such genotyping systems have also been used in post-genome sequencing projects such as genotyping of genetic resources, accessions to evaluate population structure and association studies to identify genetic loci involved in phenotypic changes of species. Listed in Table 2.1 are the most important web-based sites for DNA markers.
-
2.
Molecular DNA markers have contributed significantly to marker-assisted studies and selection (MAS) in plant breeding and in a wider range of research, including species identification and evolution (Feltus et al. 2004).
-
3.
Genetic markers designed to cover a genome extensively allow not only identification of individual genes associated with complex traits by QTL analysis but also the exploration of genetic diversity with regard to natural variations (Feltus et al. 2004; Varshney et al. 2005).
-
4.
A number of attempts to design polymorphic markers from accumulated sequence datasets have been made for various species, e.g., genome-wide rice (Oryza sativa) DNA polymorphism datasets have been constructed based on alignment between japonica and indica rice genomes (Han and Xue 2003; Shen et al. 2004).
-
5.
The most important database EST (expressed sequence tag) consists of ESTs drawn from the multiple cDNA. Large-scale EST datasets are also important resources for the discovery of sequence polymorphisms, especially for allocating expressed genes onto a genetic map (Heesacker et al. 2008).
-
6.
The Illumina GoldenGate Assay allows the simultaneous analysis of up to 1536 SNPs in 96 samples and has been used to analyse genotypes of segregation populations in order to construct genetic maps allocating SNP (single nucleotide polymorphism) markers in crops (Hyten et al. 2008).
-
7.
Diversity Arrays Technology (DArT) is a high-throughput genotyping system that was developed based on a microarray platform (Wenzl et al. 2007). These DArT markers have been used together with conventional molecular markers to construct denser genetic maps and/or to perform association studies in various crop species.
-
8.
Affymetrix Gene Chip Arrays have been used to discover nucleotide polymorphisms as single-feature polymorphisms based on the differential hybridization of Gene Chip probes in barley and wheat (Rostoks et al. 2005; Bernardo et al. 2009).
-
9.
Transcriptomics subcategories of omics attract a large number of biologists, especially in plant breeding area (Hakeem et al. 2016).
-
10.
The most powerful application of third-generation sequencing for breeding is the assembly of improved contiguous crop genomes (Hu et al. 2018).
Further, as the resolution of genetic maps in the major crops increases, and as the molecular basis for specific traits or physiological responses becomes better elucidated, it will be increasingly possible to associate candidate genes, discovered in model species, with corresponding loci in crop plants (Fig. 2.3). Appropriate relational databases will make it possible to freely associate across genomes with respect to the gene sequence, putative function and genetic map position. Once such tools have been implemented, the distinction between breeding and molecular genetics will fade away. Breeders will routinely use computer models as toolbox to formulate predictive hypotheses to create phenotypes of interest from complex allele combinations (Fig. 2.3) and then construct those combinations by scoring large populations for very large numbers of genetic markers (Walsh 2001).
2.3.1 Informative Bioinformatics Databases/Tools for Crop Breeders
Crop breeding has long relied on cycles of phenotypic selection and crossing, which generate superior genotypes through genetic recombination. When genome sequences are available, all genes and genetic variants contributing to agronomic traits can be identified, and changes made during breeding processes can be assessed at the genotype level (Hu et al. 2018). Availability of ready-to-go genomic data for breeders today plays an increasingly important role in all aspects of crop breeding, such as quantitative trait loci (QTL) mapping and genome-wide association studies (GWAS), where genomic sequencing of crop populations can allow gene-level resolution of agronomic variation. The progress made in genomics-based breeding has even assisted in identification of genetic variation in crop species, which can be applied to produce climate-resilient varieties (Mousavi-Derazmahalleh et al. 2018; Dwivedi et al. 2017).
GWAS (comparative genomic analysis, phylogenomics, evolutionary analysis and genome-wide association study) is presently a favourable tool to explore the allelic variation in a broader scope for extensive phenotypic diversity and higher resolution of QTL mapping. GWAS is an alternative to overcome the disadvantages of existing classical crop breeding methods, e.g. a biparental cross-mapping method for genetic dissections of the agronomically important traits (Myles et al. 2009). GWAS has a powerful application in plant breeding for identifying phenotypic diversity in trait-associated loci, as well as allelic variation in candidate genes addressing quantitative and complex traits (Kumar et al. 2013). GWAS has been successfully applied to study Arabidopsis thaliana, where more than 1300 distinct accessions have been genotyped for 250,000 SNP (Kozlov et al. 2015) phenotypes. A few rice genes having large effects in controlling traits are involved in determining yield, morphology and stress tolerance, and nutritional quality was also identified (Famoso et al. 2011). GWAS has been widely used to dissect complex traits in some other major crops, e.g. maize and soya bean (Li et al. 2013; Hwang et al. 2014). Several bioinformatics approaches have been introduced as GWAS acceleration tools (Table 2.1).
Advances in genomics offer the potential to accelerate the genomics-based breeding of crop plants (Fig. 2.3). However, relating genomic data to climate-related agronomic traits for use in breeding remains a huge challenge and one which will require coordination of diverse skills and expertise. Bioinformatics, when combined with genomics, has the potential to help maintain food security in the face of climate change through the accelerated production of climate-ready crops (Batley and Edwards 2016). The vast breeding knowledge gathered over the last several decades will become directly linked to basic plant biology and enhance the ability to elucidate gene function in model organisms (Hospital et al. 2002). The expected dramatic improvements in phenotypes of commercial interest include both the improvement of factors that traditionally limit agronomic performance (input traits) and the alteration of the amount and kinds of materials that crops produce (output traits). Examples include:
-
Abiotic stress tolerance
-
Biotic stress tolerance
-
Improving nutrient use efficiency
-
Manipulation of plant architecture and development (size, organ shape, number and position, the timing of development and senescence)
-
Metabolite partitioning (redirecting of carbon flow among existing pathways or shunting into new pathways)
Appropriate relational databases will make it possible to freely associate across genomes with respect to the gene sequence, putative function or genetic map position. Once such tools have been implemented, the distinction between breeding and molecular genetics will fade away. Breeders will routinely use computer models to formulate predictive hypotheses to create phenotypes of interest from complex allele combinations and then construct those combinations by scoring large populations for very large numbers of genetic markers (Walsh 2001; Deckers and Hospital 2002).
2.4 Application of Bioinformatics in Fruit Breeding
During the last three decades, the world has witnessed a rapid increase in the knowledge about the plant genome sequences and the physiological and molecular roles of various plant genes, which have revolutionized the molecular genetics and its efficiency in plant breeding programmes. Since bioinformatics has application in every field of science, genome programme can now be envisioned as a highly important tool for fruit breeding. Identifying key genes and understanding their function will result in a “quantum leap” in improving fruit quality and quantity (Meyer and Mewes 2002). The revolution in life sciences brought on by genomics dramatically increases the scale and scope of our experimental enquiry and applications in fruit plant breeding. The scale and high-resolution power of genomics make possible a broad and detailed genetic understanding of plant performance at multiple levels of aggregation (Meyer and Mewes 2002).
The primary goal of fruit plant genomics is to understand the genetic and molecular basis of all biological processes in plants that are relevant to the species. This understanding is fundamental to allow efficient exploitation of fruit plants as biological resources in the development of new cultivars of improved quality and reduced economic and environmental costs (Fig. 2.1). This knowledge is also vital for the development of new diagnostic tools and traits of primary interest like pathogen resistance and abiotic stress, fruit quality and yield. Moreover, gene expression analysis will allow us to understand how fruit plants respond to and interact with the physical environment and management practices. This information, in conjunction with appropriate technology, may provide predictive measures of plant health and fruit quality and become part of future breeding decision management systems. Current genome programmes generate a large amount of data that will require processing, storage and distribution to the international research community. The data include not only sequence information but also information on mutations, markers, maps and functional discoveries.
The key objectives for fruit plant bioinformatics include:
-
Integrating phenotypes, genomics and bioinformatics tools and resources in public and private breeding pipelines will address this challenge and help deliver breeding targets
-
Providing rational annotation of genes, proteins and phenotypes.
-
Elaborating relationships both within the data on individual fruits and between fruits and other organisms.
2.5 Future Prospects
Bioinformatics era and high-throughput sequencing (HTS) are revolutionizing the experimental design in molecular biology, strikingly contributing to increasing scientific knowledge while affecting relevant applications in many different aspects of agriculture. Bioinformatics plays a significant role in the development of the agricultural sector, agro-based industries, agricultural by-product utilization and better management of the environment. With the increase of sequencing projects, bioinformatics continues to make considerable progress in biology by providing scientists with access to the genomic information and plays a big role to analyse the data properly. Recent wealth of plant genomic resources, along with advances in bioinformatics, have enabled plant researchers to achieve a fundamental and systematic understanding of economically important plants and plant processes, critical for advancing crop improvement. The scale and high-resolution power of genomics enable to achieve a broad as well as a detailed genetic understanding of plant performance at multiple levels of aggregation. Advances in genomics are providing breeders with new tools and methodologies that allow a great leap forwards in plant breeding, including the “super domestication” of crops and the genetic dissection and breeding for complex traits. The ability to represent high-resolution physical and genetic maps of crops has been one of the paramount implications of bioinformatics. Plant scientists have an opportunity to use these resources to the full, to ensure that bench work, both in the present and in the future, can be combined with bioinformatics to fully reap the rewards of the genomics revolution. By applying novel technologies and methods in concert, future plant breeding can achieve the crop improvement rate required to ensure food security. Despite these exciting achievements, there remains a critical need for effective tools and methodologies to advance plant biotechnology, to tackle questions that are hardly solved using current approaches and to facilitate the translation of this newly discovered knowledge to improve plant productivity. Overall, due to great impact of plant breeding in order to provide the world food security through improving current staple food crops and also overcome the current harsh environmental situation (as a result of climate change), it is necessary to assess the role and achievements of bioinformatics in breeding science of crop plants.
References
Ahmad P, Ashraf M, Younis M, Hu X, Kumar A, Akram NA, Al-Qurainy F (2011) Role of transgenic plants in agriculture and biopharming. Biotechnol Adv 30:525–540
Al-Khayri JM, Jain SM, Johnson DV (2015) Advances in plant breeding strategies: breeding, biotechnology and molecular tools. Springer International Publishing
Ansorge WJ (2009) Next-generation DNA sequencing techniques. Nat Biotechnol 25:195–203
Barh D, Zambare V, Azevedo V (2013) Omics: applications in biomedical, agricultural, and environmental sciences. CRC Press
Batley J, Edwards D (2016) The application of genomics and bioinformatics to accelerate crop improvement in a changing climate. Curr Opin Plant Biol 30:78–81
Bennetzen JL et al (1998) A plant genome initiative. Plant Cell 10:488–493
Bernardo AN, Bradbury PJ, Ma H, Hu S, Bowden RL, Buckler ES et al (2009) Discovery and mapping of single feature polymorphisms in wheat using Affymetrix arrays. BMC Genomics 10:251
Blenda A, Scheffl er J, Scheffl er B, Palmer M, Lacape JM et al (2006) CMD: a cotton microsatellite database resource for Gossypium genomics. BMC Genomics 7:132
Bombarely A, Menda N, Tecle IY, Buels RM, Strickler S et al (2011) The sol genomics network (solgenomics.Net): growing tomatoes using Perl. Nucleic Acids Res 39:D1149–D1155
Brown M, Funk CC (2008) Climate. Food security under climate change. Science 319:580–581
Canaran P, Stein L, Ware D (2006) LookAlign: an interactive web-based multiple sequence alignment viewer with polymorphism analysis support. Bioinformatics 22:885–886
Carollo V, Matthews DE, Lazo GR, Blake TK, Hummel DD, Lui N et al (2005) Grain genes 2.0. An improved resource for the smallgrains community. Plant Physiol 139:643–651
Cogburn LA, Porter TE, Duclos MJ, Simon J, Burgess SC, Zhu JJ et al (2007) Functional genomics of the chicken-a model organism. Poult Sci 86:2059–2094
Cory JS, Hoover K (2006) Plant-mediated effects in insect-pathogen interactions. Trends Ecol Evol 21:278–286
De Bodt S, Maere S, Van de Peer Y (2005) Genome duplication and the origin of angiosperms. Trends Ecol Evol 20:591–597
De Filippis LF (2012) Breeding for biotic stress tolerance in plants. In: Asharaf M, Ozturk M, Ahmad MSA, Aksoy A (eds) Crop production for agricultural improvement. Springer Science
Deckers J, Hospital F (2002) The use of molecular genetics in the improvement of agricultural populations. Nat Rev Genet 3:22–32
Dwivedi SL, Scheben A, Edwards D, Spillane C, Ortiz R (2017) Assessing and exploiting functional diversity in germplasm pools to enhance abiotic stress adaptation and yield in cereals and food legumes. Front Plant Sci 8:1461
Ellegren H (2014) Genome sequencing and population genomics in nonmodel organisms. Trends Ecol Evol 29(1):51–63
Famoso AN, Zhao K, Clark RT, Tung CW, Wright MH, Bustamante C, Kochian LV, McCouch SR (2011) Genetic architecture of aluminum tolerance in rice (Oryza sativa) determined through genome-wide association analysis and QTL mapping. PLoS Genet 7(8):e1002221
Feltus FA, Wan J, Schulze SR, Estill JC, Jiang N, Paterson AH (2004) An SNP resource for rice genetics and breeding based on subspecies indica and japonica genome alignments. Genome Res 14:1812–1819
Futamura N, Totoki Y, Toyoda A, Igasaki T, Nanjo T, Seki M et al (2008) Characterization of expressed sequence tags from a full-length enriched cDNA library of Cryptomeria japonica male strobili. BMC Genomics 9:383
Govindaraj M, Vetriventhan M, Srinivasan M (2015) Importance of genetic diversity assessment in crop plants and its recent advances: an overview of its analytical perspectives. Genet Res Int 20(15):431–487
Grant D, Nelson RT, Cannon SB, Shoemaker RC (2010) SoyBase, the USDA-ARS soybean genetics and genomics database. Nucleic Acids Res 38:D843–D846
Haas BJ, Delcher AL, Wortman JR, Salzberg SL (2004) DAGchainer: a tool for mining segmental genome duplications and synteny. Bioinformatics 20:3643–3646
Hakeem K, Ozturk M, Memon AR (2012) Biotechnology as an aid for crop improvement to overcome food shortage. In: Ashraf M et al (eds) Crop production for agricultural improvement. Springer
Hakeem KR, Tombuloğlu H, Tombuloğlu G (2016) Plant omics: trends and applications. Springer International Publishing, pp 109–136
Han B, Xue Y (2003) Genome-wide intraspecific DNA-sequence variations in rice. Curr Opin Plant Biol 6:134–138
Heesacker A, Kishore VK, Gao W, Tang S, Kolkman JM, Gingle A et al (2008) SSRs and INDELs mined from the sunflower EST database: abundance, polymorphisms, and cross-taxa utility. Theor Appl Genet 117:1021–1029
Hori K, Sato K, Takeda K (2007) Detection of seed dormancy QTL in multiple mapping populations derived from crosses involving novel barley germplasm. Theor Appl Genet 115:869–876
Hospital F, Bouchez A, Lecomete L, Causse M, Charcosset A (2002) Use of markers in plant breeding: lessons from genotype building experiments. 7th WCGALP, Montpellier, pp 22–25
Hu H, Scheben A, Edwards D (2018) Advances in integrating genomics and bioinformatics in the plant breeding pipeline. Agriculture 8(6):75
Huang M (2015) In Biogpu: a high performance computing tool for genome-wide association studies, Plant and Animal Genome XXIII conference, Plant and Animal Genome
Hwang EY, Song Q, Jia G, Specht JE, Hyten DL, Costa J, Cregan PB (2014) A genome-wide association study of seed protein and oil content in soybean. BMC Genomics 15(1):1
Hyten DL, Song Q, Choi IY, Yoon MS, Specht JE, Matukumalli LK et al (2008) High-throughput genotyping with the GoldenGate assay in the complex genome of soybean. Theor Appl Genet 116:945–952
Jayashree B, Buhariwalla HK, Shinde S, Crouch JH (2005) A legume genomics resource: the chickpea root expressed sequence tag database. Electron J Biotechnol 8:128–133
Kaul S, Koo HL, Jenkins J, Rizzo M, Rooney T et al (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408(6814):796–815
Kingsbury N (2009) Hybrid: the history and science of plant breeding. University of Chicago Press
Kobayashi M (2015) In heap: a SNPs detection tool for NGS data with special reference to GWAS and genomic prediction, Plant and Animal Genome XXIII conference, Plant and Animal Genome
Kozlov AM, Aberer AJ, Stamatakis A (2015) ExaML version 3: a tool for phylogenomic analyses on supercomputers. Bioinformatics 31(15):2577–2579
Kumar S (2016) Crop breeding: bioinformatics and preparing for climate change. Apple Academic Press
Kumar S, Garrick DJ, Bink MC, Whitworth C, Chagné D, Volz RK (2013) Novel genomic approaches unravel genetic architecture of complex traits in apple. BMC Genomics 14(1):393
Lawrence CJ, Schaeffer ML, Seigfried TE, Campbell DA, Harper LC (2007) MaizeGDB’s new data types, resources and activities. Nucleic Acids Res 35:D895–D900
Li H, Peng Z, Yang X, Wang W, Fu J, Wang J, Han Y, Chai Y, Guo T, Yang N (2013) Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat Genet 45(1):43–50
Liang C, Jaiswal P, Hebbard C, Avraham S, Buckler ES, Casstevens T et al (2008) Gramene: a growing plant comparative genomics resource. Nucleic Acids Res 36:D947–D953
Lorenz AJ, Chao S, Asoro FG, Heffner EL, Hayashi T et al (2011) Genomic selection in plant breeding: knowledge and prospects. Adv Agron 110:77–123
Malav AK, Indu, Chandrawat KS (2016) Gene pyramiding: an overview. Int J Curr Res Biosci Plant Biol 3(7):22–28
Matsumoto T, Wu JZ, Kanamori H, Katayose Y, Fujisawa M et al (2005) The map-based sequence of the rice genome. Nature 436(7052):793–800
Meyer K, Mewes HW (2002) How can we deliver the large plant genomes? Strategies and perspectives. Curr Opin Plant Biol 5:173–177
Mochida K, Shinozaki K (2010) Genomics and bioinformatics resources for crop improvement. Plant Cell Physiol 51:497–523
Mochida K, Saisho D, Yoshida T, Sakurai T, Shinozaki K (2008) TriMEDB: a database to integrate transcribed markers and facilitate genetic studies of the tribe Triticeae. BMC Plant Biol 8:72
Morrell PL, Buckler ES, Ross-Ibarra J (2012) Crop genomics: advances and applications. Nat Rev Genet 13(2):85–96
Mousavi-Derazmahalleh M, Bayer PE, Hane JK, Babu V, Nguyen HT et al (2018) Adapting legume crops to climate change using genomic approaches. Plant Cell Environ 42(1):6–19
Myles S, Peiffer J, Brown PJ, Ersoz ES, Zhang Z, Costich DE, Buckler ES (2009) Association mapping: critical considerations shift from genotyping to experimental design. Plant Cell 21(8):2194–2202
Organization EPS (2005) European plant science: a field of opportunities. J Exp Bot 56(417):1699–1709
Ozturk M (2010) Agricultural residues and their role in bioenergy production. Proceedings-second consultation AgroResidues-Second expert consultation ‘The utilization of agricultural residues with special emphasis on utilization of agricultural residues as biofuel’, pp 31–43
Pandey SP, Somssich IE (2009) The role of WRKY transcription factors in plant immunity. Plant Physiol 150:1648–1655
Paterson AH (2008) Genomics of sorghum. Int J Plant Genomics 200:362–451
Plechakova O, Tranchant-Dubreuil C, Benedet F, Couderc M, Tinaut A et al (2009) MoccaDB – an integrative database for functional, comparative and diversity studies in the Rubiaceae family. BMC Plant Biol 9:123
Robinson AJ, Love CG, Batley J, Barker G, Edwards D (2004) Simple sequence repeat marker loci discovery using SSR primer. Bioinformatics 20:1475–1476
Rostoks N, Borevitz JO, Hedley PE, Russell J, Mudie S, Morris J et al (2005) Single-feature polymorphism discovery in the barley transcriptome. Genome Biol 6:R54
Schenk PM, Carvalhais LC, Kazan K (2012) Unraveling plant-microbe interactions: can multispecies transcriptomics help? Trends Biotechnol 30:177–184
Schlueter SD, Dong Q, Brendel V (2003) GeneSeqer@PlantGDB: gene structure prediction. Nucleic Acids Res 32:D354–D359
Schuster SC (2007) Next-generation sequencing transforms today’s biology. Nature 200(8):16–18
Shen YJ, Jiang H, Jin JP, Zhang ZB, Xi B, He YY et al (2004) Development of genome-wide DNA polymorphism database for map-based cloning of rice genes. Plant Physiol 135:1198–1205
Skuse GR, Du C (2008) Bioinformatics tools for plant genomics. Int J Plant Genomics 2008:910474
Sleper DA, Poehlman JM (2006) Breeding field crops. Blackwell Publishing, Oxford, p 424
Stanford WL, Cohn JB, Cordes SP (2001) Gene-trap mutagenesis: past, present and beyond. Nat Rev Genet 2:756–768
Steinbach D (2015) In GnpIS-Asso: a generic database for managing and exploiting plant genetic association studies results using high throughput genotyping and phenotyping data, Plant and Animal Genome XXIII conference, Plant and Animal Genome
Takeda S, Matsuoka M (2008) Genetic approaches to crop improvement: responding to environmental and population changes. Nat Rev Genet 9:444–457
Tester M, Langridge P (2010) Breeding technologies to increase crop production in a changing world. Science 327:818–822
Van Emon JM (2016) The omics revolution in agricultural research. J Agric Food Chem 64(1):36–44
Varshney RK, Graner A, Sorrells ME (2005) Genomics-assisted breeding for crop improvement. Trends Plant Sci 10:621–630
Varshney RK, Nayak SN, May GD, Jackson SA (2009) Next-generation sequencing technologies and their implications for crop genetics and breeding. Trends Biotechnol 27:522–530
Walsh B (2001) Quantitative genetics in the age of genomics. Theor Popul Biol 59:175–184
Wang J (2015) In A Bayesian model for detection of high-order interactions among genetic variants in genome-wide association studies and its application on soybean oil/protein traits, Plant and Animal Genome XXIII conference, Plant and Animal Genome. p 159–162
Wenzl P, Raman H, Wang J, Zhou M, Huttner E, Kilian A (2007) A DArT platform for quantitative bulked segregant analysis. BMC Genomics 8:196
Ye X, Al-Babili S, Klöti A et al (2000) Engineering the provitamin A (β-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 287(5451):303–305
Zhang Z, Ober U, Erbe M, Zhang H, Gao N, He J, Li J, Simianer H (2014) Improving the accuracy of whole genome prediction for complex traits using the results of genome wide association studies. PLoS One 9(3):e93017
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Shafi, A., Zahoor, I., Haq, E., Fazili, K.M. (2019). Impact of Bioinformatics on Plant Science Research and Crop Improvement. In: Hakeem, K., Shaik, N., Banaganapalli, B., Elango, R. (eds) Essentials of Bioinformatics, Volume III. Springer, Cham. https://doi.org/10.1007/978-3-030-19318-8_2
Download citation
DOI: https://doi.org/10.1007/978-3-030-19318-8_2
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-19317-1
Online ISBN: 978-3-030-19318-8
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)