Abstract
The cultivated pear is a major fruit crop in Eurasia that underpins many local economies. However, its origin and domestication history, as well as the diversity of wild pears in natural ecosystems, are at the early stages of exploration. In this chapter, we provide an overview of the described diversity and genetic relationships among wild and cultivated Pyrus species. Non-discriminatory morphological characters, poor diagnostic genetic tools, and lack of access to samples scattered throughout worldwide genebank collections make it difficult to definitively elucidate relationships of pear species and more generally Pyrus diversification and domestication. High-throughput sequencing is providing advancements in our understanding of the domestication process of the pear, and of biogeography, taxonomy, and ecology of wild pears. This knowledge will be crucial for future breeding programs focused on improving quality and production traits.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
3.1 Introduction: Assessing Pyrus Diversity
Cultivated pears are produced throughout temperate regions on both a commercial scale and for local household use; however, their origin and domestication history are at the early stages of exploration. Over the past 4000 years, pear cultivation has led to the identification and/or development of a vast number of landraces and recent cultivars through natural and artificial hybridization. Vegetative propagation by grafting has allowed interesting and/or desirable phenotypes to be maintained and spread (Zohary and Spiegel-Roy 1975). As a result, cultivated pears exhibit a wide range of desirable traits, including fruit attractiveness, flavor, size, and shape. Numerous molecular studies, primarily based mostly on a few marker loci, have been used to characterize the diversity of pear cultivars and the origin of this diversity in wild species. However, the genetics underlying key agronomic traits are just beginning to be understood.
Assessments of pear species diversity and distribution are usually determined using regional inventory and census counts. These records are often not collected using standardized techniques and have gaps with respect to coverage. In addition, recurrent hybridizations and resulting introgressions among species have made it difficult to differentiate species. Consequently, it is difficult to identify the geographical range of wild Pyrus species.
Wild relatives of cultivated pears offer novel allelic diversity and allelic combinations that can provide sources of resistance and tolerance to abiotic and biotic stresses for pear breeding programs. Pyrus wild species such as P. communis spp. pyraster, P. calleryana, P. ussuriensis, P. pyrifolia, P. fauriei, P. dimorphoylla, P. betulifolia, and P. × nivalis have desirable levels of disease resistance to various pathogens, including pear leaf spot (Entomosporium mespili (DC.) Sacc.), fire blight (Erwinia amylovora (Burr.) Winslow et al.), and pear psylla (Cacopsylla pyricola (Foerster)) (van der Zwet et al. 1983; Bell 1992; Bell and Itai 2011). These species can be used as parents in breeding programs, as providers of specific alleles for introgression, or as rootstocks. Many wild pear species, including P. pashia, P. korshinskyi, P. syriaca, P. × hopiensis, P. gharbiana, P. betulifolia, P. calleryana, P. cossonii. P. dimorphophylla, P. fauriei, P. pyrifolia, P. ussuriensis, and P. xerophila, are recognized for their desirable rootstock traits, providing tolerance to extreme heat, humidity, and cold, as well as disease resistance (Ercisli 2004; Bao et al. 2008; Zong et al. 2014b; U.S. Department of Agriculture 2017).
This chapter focuses on the measured diversity of wild Pyrus species and described relationships between wild species and cultivated forms. The life history traits of pears, with long lifespans and high levels of gene flow among populations and species, combined with their ancient origin, render Pyrus as a valuable model for studying fruit tree species diversification. Expanded knowledge of pear genetic diversity and evolution will also assist in pinpointing sources of allelic variation in the wild useful for future breeding programs. Such studies are particularly timely, as wild gene pools may be sources of alleles for resistance to biotic and abiotic stresses (van der Zwet et al. 1983; Bell 1992; Bell and Itai 2011), and these are currently under threat of fragmentation in their centers of origin.
3.2 Diversification of Wild Pears
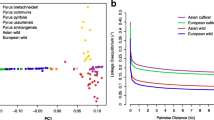
The genus Pyrus is presumed to have originated during the Tertiary Period (65–55 million years ago [Mya]) (Silva et al. 2014), or in particular in the Oligocene Epoque, 33–25 Mya (Korotkova et al. 2018) in the mountainous regions of Western China or Asia Minor. Microsatellite or simple sequence repeat (SSR) markers, as well as genomic studies, have revealed strong genetic differentiation between two main genetic groups, an Occidental (European/Central Asian) and an Asian (East Asia), which diverged between 6.6 and 3.3 Mya (Fig. 3.1) (Liu et al. 2015; Volk et al. 2019; Wu et al. 2018). Two non-coding regions of the cpDNA and one low copy nuclear gene have also demonstrated the differentiation between wild Asian and Occidental pear groups (Zheng et al. 2014). Altogether, this suggests spatial dispersal events to eastern and northern Eurasia, whereby Asian wild species have diversified, and to western Eurasia, whereby Occidental wild species have diversified (Figs. 3.2 and 3.3).
Generalized diagram of network relationships and shared haplotypes of Pyrus species, from Volk et al. (2019). North African Pyrus species include P. cossonii, P. gharbiana, and P. mamorensis, while West Asian Pyrus species include P. elaegrifolia, P. glabra P. korshinskyi, P. sachokiana, P. salicifolia, P. spinosa, and P. syriaca
The use of classical microsatellite genetic markers has shed light on the genetic diversity of some wild pear species. Nuclear microsatellite data demonstrated that the genetic variation of wild populations of P. calleryana, P. communis subsp. pyraster, P. pashia, and P. ussuriensis is higher within (ranging from 80 to 96%) than among populations (ranging from 4 to 20%) (Liu et al. 2012; Wolko et al. 2015; Zong et al. 2014a; Wuyun et al. 2015) (Table 3.1). This observed wide range across wild Pyrus species may be in part due to physical sampling methods used; e.g., distances between sites and familial relationships among individuals. The heterozygosity of these populations ranges from 0.48 for P. ussuriensis (Wuyun et al. 2015) to 0.76 for P. communis subsp. caucasica and P. communis subsp. pyraster (Table 3.2; Asanidze et al. 2014; Wolko et al. 2015). Hereafter, we review the literature on specific diversity and evolution of Asian (pea pear and large-fruited) and Occidental pears.
3.2.1 Genetic Diversity of Asian Wild Pears
Asian wild pears are often described as belonging to either the “pea pear” or the “large-fruited pear” groups. Pea pears, including P. betulifolia, P. calleryana, P. dimorphophylla, P. fauriei, and P. koehnei, produce fruits that are less than 1 cm in diameter with two carpels (Jiang et al. 2016). In contrast, large-fruited Asian pear species include, among others, P. pashia, P. pyrifolia, P. ussuriensis, P. xerophyla, and P. hondoensis (Challice and Westwood 1973). It has been difficult to genetically differentiate between “pea” and “large-fruited” pears (Jiang et al. 2016; Zheng et al. 2014). Genetic diversity assessments of Asian wild pears have focused primarily on differences/relatedness of either within species or between wild species and cultivated forms.
Molecular genetic markers have facilitated identification of basal species and hybrids in the Asian wild Pyrus group. Sequence-specific amplification polymorphism (SSAP) suggest that P. betulifolia, P. pashia, P. pyrifolia, and P. ussuriensis are primitive genepools of wild Asian species (Jiang et al. 2016). Other original wild Asian species include P. koehnei and P. fauriei (Zheng et al. 2014). Pyrus species of ambiguous identities or origins include P. dimorphophylla (sometimes classified as a variety of P. calleryana), P. calleryana (with leaf shape similar to P. pashia and fruit similar to P. betulifolia), and P. × bretschneideri (genetically similar to P. ussuriensis). Asian wild pear species resulting from hybridizations between wild pear species include P. xerophila (P. pashia, × P. ussuriensis × Occidental), P. sinkiangensis (P. pyrifolia × Occidental), P. phaeocarpa (P. betulifolia × P. ussuriensis × P. pyrifolia), P. hondoensis (P. dimophophylla × P. ussuriensis), P. neoserrulata and P. serrulata (P. calleryana × P. pyrifolia), and P. hopeiensis (P. ussuriensis × [P. × phaeocarpa or P. betulifolia]) (Jiang et al. 2016; U.S. Department of Agriculture 2017).
3.2.1.1 Genetic Diversity Within the Asian Pea Pear Species
The following Pyrus pea pear taxa, P. betulifolia, P. calleryana, P. dimorphophylla, P. fauriei, and P. koehnei are native to China, Japan, and the Korean peninsula (Fig. 3.2). Pyrus betulifolia is described as an ancient pear species that shares some traits with both Asian and Occidental pear types (Zong et al. 2014b, 2017). Diversity of this species, as measured using chloroplast intergenic fragments and microsatellite genetic markers (SSRs), has revealed that the Taihang Mountains are natural genetic barriers, and that range expansion and contraction events must have occurred during and between glacial periods (Zong et al. 2014b, 2017). Furthermore, populations within P. betulifolia are more easily distinguishable using chloroplast markers rather than nuclear SSRs as pollen-mediated gene flow has likely homogenized genetic diversity at the nuclear level (Zong et al. 2017). Future work using additional markers, such as single nucleotide polymorphisms (SNPs), will provide more insights into the population structure of P. betulifolia.
On the other hand, P. calleryana, native to southern China, Japan, and the Korean Peninsula, is classified as a wild pea pear that shares some similarities with both P. pashia and P. betulifolia (Jiang et al. 2016). In Southern China, the range of native species of P. calleryana, P. pashia, and P. betulifolia is found to overlap (Liu et al. 2012; Jiang et al. 2016). Using both nuclear microsatellite and chloroplast sequence markers, two genepools are identified in eight populations of P. calleryana in the Zhejiang Province in China (Liu et al. 2012). These genepools correspond to two geographic regions, with one located in the northeast and the other located in the southwest.
3.2.1.2 Genetic Diversity Within the Large-Fruited Asian Pear Species
The wild large-fruited Asian pear species include P. pashia, P. pyrifolia, P. ussuriensis, P. xerophyla, and P. hondoensis. Pyrus ussuriensis is native to northeastern and north-central Chinese provinces, as well as to Japan (Fig. 3.2; Katayama et al. 2016). Each of chloroplast sequences, SSAPs, and SSRs has been used to assess genetic variations among and within P. ussuriensis populations throughout its native range. These genetic studies have revealed existence of a spatial genetic structure across sampling regions. Furthermore, within-population diversity is found to be high, likely due to self-incompatibility, while between-population differentiation is weak, except for those genetically distant populations from Inner Mongolia (Wuyun et al. 2015). It is reported that Inner Mongolian populations may have experienced some bottleneck effects due to their demographic decline (Wuyun et al. 2015).
As P. pashia is another ancient species, it may be intermediate between Asian and Occidental pear groups. Whereas, P. pashia is native to Southwest China and to the Himalayan region (Fig. 3.2; Zong et al. 2014a). Due to high levels of within-site diversity, based on SSR profiles, Zong et al. (2014a) have proposed that some of the sampled populations may have likely served as sources for range expansions during interglacial periods. Liu et al. (2013) have used chloroplast sequence data to assess the diversity of individuals within 22 populations. As with other wild pear species, a high level of genetic variation is detected within populations. Range expansions may explain lack of correlations between genetic and geographic distances across the range of P. pashia (Liu et al. 2013).
3.2.2 Genetic Diversity in the Occidental Pear Species
Occidental pear species are likely to have radiated westward from China and currently occupy overlapping ranges (Fig. 3.3). Chloroplast and nuclear genes have been used to reconstruct the phylogeny of Occidental Pyrus species using 50 accessions representing the following 11 species: P. communis, P. nivalis, P. cordata, P. eleagrifolia, P. spinosa, P. regelii, P. salacifolia, P. syriaca, P. cossonii, P. gharbiana, and P. mamorensis (Zheng et al. 2014). It is found that all Occidental species, except for P. regelii and P. gharbiana, have shared haplotypes. Moreover, it appears that P. regelii, the most easterly West Asian species, must have diversified early, becoming isolated, and it is the only west Asian species P. regelii that is monophyletic (Fig. 3.1; Zheng et al. 2014; Volk et al. 2019). In addition, P. regelii has an ancestral phenotype with dissected adult leaves and ovaries with few locules (Zheng et al. 2014).
Based on a phylogenetic dendrogram, accessions of some Occidental species, including P. spinosa, P. cossonii, P. regelii, P. gharbiana, and P. mamorensis, are located on distinct branches (Zheng et al. 2014). In contrast, P. eleagrifolia, P. nivalis, and P. salicifolia are spread throughout the phylogenetic dendrogram (Zheng et al. 2014). Recently, Volk and co-authors (2019) have observed lower levels of differentiation among Occidental species using chloroplast sequence data (Fig. 3.1). Furthermore, P. spinosa, native to Turkey, Southeastern Europe, France, and Spain, has primitive characters, suggesting that it may be yet another ancient species; whereas, P. salicifolia and P. nivalis have overlapping phenotypes with regard to leaf shape (lanceolate or elliptical leaves) and level of hairiness (Zheng et al. 2014; Paganová 2003). Wild P. communis subsp. pyraster in Poland and Germany have high levels of diversity within populations, as well as weak correlations between genetic and geographical distance (Wolko et al. 2015; Reim et al. 2017). Recent genomic sequencing data reveal that many pear accessions assigned to Occidental species may be highly admixed (Wu et al. 2018).
3.3 Domestication
Pyrus communis subsp. communis is a European pear known for its soft and juicy flesh, and includes cultivars such as ‘Bartlett’ and ‘Anjou’. In contrast, P. pyrifolia, the Asian pear, has a crisp and juicy texture. Asian pears include a number of types of cultivated pears, including Chinese white pear cultivars (such as ‘Ya Li’ and ‘Tse Li’) and Japanese pears (such as ‘Kosui’, ‘Hosui’, and ‘Nijisseki’). Genetic markers have been developed and used to reconstruct the domestication process that has resulted in the evolution of European, Chinese white, and Japanese pear cultivars, as well as various Asian landraces that include Chinese sand pears, Ussurian pears, and Xinjiang pears. Recently, SNP data have elucidated this dichotomy between Occidental and Asian cultivated pears (Kumar et al. 2017). These two pear types, from Europe and Asia, respectively, originated from different wild pear relatives specific to their regions of origin (Fig. 3.4). This suggests two independent domestication events, one in Europe and one in Asia from distinct wild species, which was recently confirmed by fully sequenced genomes of a large collection of wild and cultivated pears (Wu et al. 2018). Specifically, P. communis subsp. communis is derived from P. pyraster, and P. pyrifolia is derived from the wild P. pyrifolia (Wu et al. 2018).
3.3.1 The Chinese White, Japanese, and Chinese Sand Pear Cultivar Groups
The cultivated Chinese white pears, Japanese pears, and Chinese sand pears share a common ancestor, P. pyrifolia (Fig. 3.5a; Bao et al. 2007; Jiang et al. 2016).
The Chinese white pear is the most commonly grown pear in northern China, and it is found at the intersection of the native species ranges of P. ussuriensis and P. pyrifolia (Bao et al. 2007). The Chinese white pears, grown in northern China, may have originated from a gene pool whereby P. ussuriensis has hybridized with P. pyrifolia (Jiang et al. 2016). Pyrus × bretschneideri is a hybrid species (sometimes considered to be P. pyrifolia) between P. ussuriensis and P. pyrifolia. This hybrid species, P. × bretschneideri, is considered as the source species for Chinese white pears (Liu et al. 2015).
The Japanese pear is the most commonly grown commercial pear in Japan. Nishio and co-authors (2016) have used microsatellite markers to assess the genetic diversity and ancestry of modern Japanese pear cultivars. These cultivars are genetically similar to local cultivars from the Kanto region of Japan. Iketani et al. (2010) have found that these local Japanese cultivars are more similar to P. pyrifolia of China than P. ussuriensis of Japan.
Chinese sand pears are primarily local cultivars grown in Sichuan Province, along the Yangtze River, and in southern regions of China (Song et al. 2014). Chinese white and Japanese pears have fewer numbers of haplotypes than those of Chinese sand pears, suggesting that Chinese sand pears have higher levels of diversity, and are likely to be more basal than other cultivars derived from P. pyrifolia (Teng et al. 2015). Although Chinese sand pears may have been derived primarily from P. pyrifolia (Jiang et al. 2009), there is some SSAP marker evidence suggesting that Chinese sand and Japanese pears may have resulted from introgressive hybridizations between P. pyrifolia and P. pashia in Southern China (Jiang et al. 2016).
Zangli pears are yet another Asian pear landrace, native to Eastern Tibet, Western Sichuan, and Northwestern Yunnan provinces. Cultivars of Zangli pears are resistant to bitter cold, dry air, and high winds (Xue et al. 2017). Microsatellite markers have revealed that Zangli pears are genetically similar to Chinese sand pears and that they may have been introduced north from Yunnan and west from Sichuan (Xue et al. 2017).
3.3.2 The Ussurian Cultivated Pear
Ussurian pear cultivars are native to the southern area of northeastern China, as well as to Hebei, Shanxi, and Gansu provinces (Fig. 3.2). Domesticated Ussurian pears are genetically and phenotypically distinct from wild P. ussuriensis (Wuyun et al. 2015). Cultivated Ussurian pears are known for their strong cold resistance, and they can endure up to −52 °C (Katayama et al. 2016). The domesticated Ussurian pears have lineages from the following two species, P. ussuriensis and P. pyrifolia (Fig. 3.5b; Jiang et al. 2016; Yu et al. 2016). It is likely that P. ussuriensis and P. pyrifolia have also hybridized in the northern part of Japan, where the two species overlap (Katayama et al. 2016). Recently, full-sequencing genome data have revealed that cultivated P. ussuriensis is derived from the wild P. ussuriensis (Wu et al. 2018). Various samples selected for genomic and genetic analyses may have affected conclusions obtained from the different studies.
3.3.3 The Xinjiang Pear
Xinjiang pear cultivars are derived from hybridizations between P. pyrifolia (possibly Chinese white pears) and Occidentals (Fig. 3.5c; Jiang et al. 2016). It is presumed that Occidental pears have been introduced from abroad in the Xinjiang Province in China (Chang et al. 2017). It has been reported that the ‘Korla’ pear, the most famous Xinjiang pear cultivar, shares chloroplast haplotypes with Chinese white pears, as well as with other Xinjiang cultivated pear accessions (Chang et al. 2017).
3.3.4 The Cultivated European Pear
The European pear, P. communis subsp. communis, is commercially grown, and is thought to have originated from smaller fruited P. communis subsp. pyraster, a subspecies native to Eastern Europe, and P. communis subsp. caucasica, a subspecies native to the Caucasus Mountains of Russia, Crimea, Armenia, and Georgia (Fig. 3.5d; Volk et al. 2006). Microsatellite markers have successfully differentiated P. communis subsp. pyraster and P. communis subsp. caucasica, from P. communis cultivars (Volk et al. 2006). In a later study, Asanidze and co-authors (2014) have compared local Georgian pear cultivars to wild species of P. communis subsp. caucasica, P. balansae, P. salicifolia, P demetrii, P. syriaca, P. ketzkhovelii, and P. sachokiana found in Georgia. Based on microsatellite marker relationships and morphological similarities, it is likely that P. communis subsp. caucasica and P. balansae (sometimes considered to be P. communis; U.S. Department of Agriculture 2017) are progenitors of local Georgian pear cultivars (Asanidze et al. 2011, 2014).
3.4 Conclusions
Altogether, studies based on genetic data, mainly of SSRs and chloroplast sequences, provide a first glimpse of the genetic diversity and evolution of the Pyrus genus. Population genetic studies have revealed that within-population variation and gene flow among populations of Pyrus species are high, as well as between-species hybridizations recurrent. This adds to the taxonomic complexity of differentiating Pyrus species, either based on morphological or genetic traits. Yet, many of the current findings are based on relatively few numbers of markers—nuclear or chloroplast microsatellite or sequence data. The use of genome-wide SNP data using high-throughput sequencing technologies holds promise in reducing costs per marker and per sample (see Kumar et al. 2017; Wu et al. 2018). This research will be limited, however, based on the availability of true-to-type reference materials and access to wild populations of Pyrus species within the native range.
Genebanks currently offer reference materials and some collections of wild species material with detailed passport information (collection site, georeferencing, and half-sib relationships, among others) that can serve as sources of such population genomic studies. Efforts to identify markers that are associated with traits of physiological and agronomic significance will facilitate measurement of “useful” variation within species, thus opening the door to exploring effects of specific allelic diversity within breeding programs.
3.5 Future Directions
Future efforts that unify taxonomic descriptions, based on morphological and genetic characters, of Pyrus genetic resources within worldwide genebanks will facilitate access to and use of genebank materials. In addition, further work is required to unravel the large-scale evolutionary history of the Pyrus genus, and in particular the origin of edible pears. We must re-assess pear diversity in terms of species and genetic diversity in Europe, Central Asia, and Eastern Asia using genomic tools such as genotyping-by-sequencing, whole-genome sequencing, or SNP arrays (Montanari et al. 2013; Kumar et al. 2017; Xue et al. 2017). The recent release of reference genomes for P. × bretschneideri (Wu et al. 2013) and for the European pear P. communis (Chagné et al. 2014), together with new population-level genetic frameworks designed to search for molecular signatures of evolutionary processes and to infer complex demographic histories (Beichman et al. 2018; Csilléry et al. 2010; Gutenkunst et al. 2010), has rendered studies of genomic consequences of pear domestication timely. Recent resequencing of both wild and cultivated pears has revealed demographic history and genomic signatures of adaptation during pear domestication (Wu et al. 2018). The combination of these genomic approaches is providing us with a more precise picture of the genomic diversity and evolution of the Pyrus genus and, more generally, of processes of adaptation in perennials.
References
Asanidze Z, Akhalkatsi M, Gvritishvili M (2011) Comparative morphometric study and relationships between the Caucasian species of wild pear (Pyrus spp.) and local cultivars in Georgia. Flora 206:974–986
Asanidze Z, Akhalkatsi M, Henk AD, Richards CM, Volk GM (2014) Genetic relationships between wild progenitor pear (Pyrus L.) species and local cultivars native to Georgia, South Caucasus. Flora 209:504–512
Bao L, Chen K, Zhang D, Cao Y, Yamamoto Y, Teng Y (2007) Genetic diversity and similarity of pear (Pyrus L.) cultivars native to East Asia revealed by SSR (simple sequence repeat) markers. Genet Resour Crop Evol 54:959–971
Bao L, Chen K, Zhang D, Li X, Teng Y (2008) An assessment of genetic variability and relationships within Asian pears based on AFLP (amplified fragment length polymorphism) markers. Scient Hort 116:374–380
Beichman AC, Huerta-Sanchez E, Lohmueller KE (2018) Using genomic data to infer historic population dynamics of nonmodel organisms. Ann Rev Ecol Evol System 49. https://www.annualreviews.org/doi/abs/10.1146/annurev-ecolsys-110617–62431
Bell RL (1992) Additional East European Pyrus germplasm with resistance to pear psylla nymphal feeding. HortScience 27:412–413
Bell RL, Itai A (2011) Pyrus. In: Kole C (ed) Wild crop relatives: genomic and breeding resources, temperate fruits. Springer, Berlin, pp 147–176
Chagné D, Crowhurst RN, Pindo M, Thrimawithana A, Deng C et al (2014) The draft genome sequence of European pear (Pyrus communis L. ‘Bartlett’). PLoS ONE 9(4):e92644
Challice JS, Westwood MN (1973) Numerical taxonomic studies of the genus Pyrus using both chemical and botanical characters. Bot J Linn Soc 67:121–148
Chang Y-J, Cao Y-F, Zhang J-M, Tian L-M, Dong X-G, Zhang Y, Qi D, X-s Zhang (2017) Study on chloroplast DNA diversity of cultivated and wild pears (Pyrus L.) in Northern China. Tree Genet Genomes 13:44. https://doi.org/10.1007/s11295-017-1126-z
Csilléry K, Blum MG, Gaggiotti OE, François O (2010) Approximate Bayesian computation (ABC) in practice. Trends Ecol Evol 25(7):410–418
Ercisli S (2004) A short review of the fruit germplasm resources of Turkey. Genet Resour Crop Evol 51:419–435
Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD (2010) Diffusion approximations for demographic inference: DaDi. Nature Preced hdl:10101/npre.2010.4594.1
Iketani H, Yamamoto T, Katayama H, Uematsu C, Mase N, Sato Y (2010) Introgression between native and prehistorically naturalized (archaeophytic) wild pear (Pyrus spp.) populations in Northern Tohoku, Northeast Japan. Conserv Genet 11:115–126
Jiang Z, Tang F, Huang H, Hu H, Chen Q (2009) Assessment of genetic diversity of Chinese sand pear landraces (Pyrus pyrifolia Nakai) using simple sequence repeat markers. HortScience 44:619–626
Jiang S, Zheng X, Yu P, Yue X, Ahmed M, Cai D, Teng Y (2016) Primitive genepools of Asian pears and their complex hybrid origins inferred from fluorescent sequence-specific amplification polymorphism (SSAP) markers based on LTR retrotransposons. PLoS ONE 11(2):e0149192. https://doi.org/10.1371/journal.pone.0149192
Katayama H, Amo H, Wuyun T, Uematsu C, Iketani H (2016) Genetic structure and diversity of the wild Ussurian pear in East Asia. Breed Sci 66:90–99
Korotkova N, Parolly G, Khachatryan A, Ghulikyan L, Sargsyan H, Akopian J, Borsch T, Gruenstaeudl M (2018) Towards resolving the evolutionary history of Caucasian pear Pyrus, Rosaceae)—Phylogenetic relationships, divergence times and leaf trait evolution. J Systemat Evol 56:35–47
Kumar S, Kirk C, Wiedow C, Knaebel M, Brewer L (2017) Genotyping-by-sequencing of pear (Pyrus spp.) accessions unravels novel patterns of genetic diversity and selection footprints. Hort Res 4:17015
Liu J, Zheng X, Potter D, Hu C, Teng Y (2012) Genetic diversity and population structure of Pyrus calleryana (Rosaceae) in Zhejiang province, China. Biochem System Ecol 45:69–78
Liu J, Sun P, Zheng X, Potter D, Li K, Hu C, Teng Y (2013) Genetic structure and phylogeography of Pyrus pashia L. (Rosaceae) in Yunnan Province, China, revealed by chloroplast DNA analyses. Tree Genet Genomes 9:433–441
Liu Q, Song Y, Liu L, Zhang M, Sun J, Zhang S, Wu J (2015) Genetic diversity and population structure of pear (Pyrus spp.) collections revealed by a set of core genome-wide SSR markers. Tree Genet Genomes 11:128. https://doi.org/10.1007/211295-015-0953-z
Montanari S, Saeed M, Knäbel M, Kim YK, Troggio M, Malnoy M, Velasco R, Fontana P, Won KH, Durel C-E, Perchepied L, Schaffer R, Wiedow C, Bus V, Brewer L, Gardiner SE, Crowhurst RN, Chagné D (2013) Identification of Pyrus single nucleotide polymorphisms (SNPs) and evaluation for genetic mapping in European pear and interspecific hybrids. PLoS ONE 8(10):e77022
Nishio S, Takada N, Saito T, Yamamoto T, Iketana H (2016) Estimation of loss of genetic diversity in modern Japanese cultivars by comparison of diverse genetic resources in Asian pear (Pyrus spp.). BMC Genet 17:81. https://doi.org/10.1186/212863-016-0380-7
Paganová V (2003) Taxonomic reliability of leaf and fruit morphological characteristics of the Pyrus L. taxa in Slovakia. Hort Sci (Prague) 30:98–107
Reim S, Lochschmidt F, Proft A, Wolf H, Wolf H (2017) Species delimitation, genetic diversity and structure of the European indigenous wild pear (Pyrus pyraster) in Saxony, Germany. Genet Resour Crop Evol 64:1075–1085
Richards CM, Volk GM, Reilley AA, Henk AD, Lockwood DR, Reeves PA, Forsline PL (2009) Genetic diversity and population structure in Malus sieversii, a wild progenitor species of the domesticated apple. Tree Genet Genomes 5(2):339–347
Silva, GJ, Medeiros Souza T, Lía Barbieri R, Costa de Oliveira A (2014) Origin, domestication, and dispersing of pear (Pyrus spp.). Adv Agric 2014:541097
Song Y, Fan L, Chen H, Zhang M, Ma Q, Zhang S, Wu J (2014) Identifying genetic diversity and a preliminary core collection of Pyrus pyrifolia cultivars by a genome-wide set of SSR markers. Scientia Hort 167:5–16
Teng Y, Yue X, Zheng X, Cai D (2015) Genetic clue to the origin of cultivated Asian pears inferred from cpDNA haplotypes. Acta Hort 1094:31–39
U.S. Department of Agriculture (2017) Germplasm Resources Information Network (GRIN-Global). GRIN Taxonomy. https://npgsweb.ars-grin.gov/gringlobal/taxon/taxonomyquery.aspx. Accessed 7 Dec 2017
van der Zwet T, Stankovic D, Cociu V (1983) Collecting Pyrus germplasm in Eastern Europe and its significance to the USDA pear breeding program. Acta Hortic 140:43–45
Volk GM, Richards CM, Henk AD, Reilley AA, Bassil NV, Postman JD (2006) Diversity of wild Pyrus communis based on microsatellite analyses. J Amer Soc Hort Sci 131:408–417
Volk GM, Henk AD, Richards CM, Bassil NV, Postman J (2019) Chloroplast sequence data differentiate Maleae, and specifically Pyrus, species in the USDA-ARS National Plant Germplasm System. Genet Resour Crop Evol 66(1):5–15
Wolko Ł, Bocianowski J, Antkowiak W, Słomski R (2015) Genetic diversity and population structure of wild pear (Pyrus pyraster (L.) Burgsd.) in Poland. Open Life Sci 10:19–29
Wu J, Wang Z, Shi Z, Zhang S, Ming R, Zhu S, Khan MA, Tao S, Korban SS, Wang H, Chen NJ, Nishio T, Xu X, Cong L, Qi K, Huang X, Wang Y, Zhao X, Wu J, Deng C, Gou C, Zhou W, Yin H, Qin G, Sha Y, Tao Y, Chen H, Yang Y, Song Y, Zhan D, Wang J, Li L, Dai M, Gu C, Wang Y, Shi D, Wang X, Zhang H, Zeng L, Zheng D, Wang C, Chen M, Wang G, Xie L, Sovero V, Sha S, Huang W, Zhang S, Zhang M, Sun J, Xu L, Li Y, Liu X, Li Q, Shen J, Wang J, Paull RE, Bennetzen JL, Wang J, Zhang S (2013) The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res 23(2):396–408
Wu J, Wang Y, Xu J, Korban SS, Fei Z, Tao S, Ming R, Tai S, Khan MA, Postman JD, Gu C, Yin H, Zheng D, Qi K, Li Y, Wang R, Deng CH, Kumar S, Chagné D, Li X, Wu J, Huang X, Zhang H, Xie Z, Li X, Zhang M, Li Y, Yue Z, Fang X, Li J, Li L, Jin C, Qin M, Zhang J, Wu X, Ke Y, Wang J, Yang H, Zhang S (2018) Diversification and independent domestication of Asian and European pears. Genome Biol 19:77. https://doi.org/10.1186/s13059-018-1452-y
Wuyun T, Amo H, Xu J, Ma T, Uematsu C, Katayama H (2015) Population structure of and conservation strategies for wild Pyrus ussuriensis Maxim. in China. PLoSOne 10(8):e013368. https://doi.org/10.1371/journal.pone.0133686
Xue L, Liu Q, Qin M, Zhang M, Wu X, Wu J (2017) Genetic variation and population structure of “Zangli” pear landraces in Tibet revealed by SSR markers. Tree Genet Genomes 13:26. https://doi.org/10.1007/s11295-017-1110-7
Yu P, Jiang S, Wang X, Bai S, Teng Y (2016) Retrotransposon-based sequence-specific amplification polymorphism markers reveal that cultivated Pyrus ussuriensis originated from an interspecific hybridization. Eur J Hort Sci 81:264–272
Zheng X, Cai D, Potter D, Postman J, Liu J, Teng Y (2014) Phylogeny and evolutionary histories of Pyrus L. revealed by phylogenetic trees and networks based on data from multiple DNA sequences. Mol Phylogen Evol 80:54–65
Zohary D, Spiegel-Roy P (1975) Beginnings of fruit growing in the old world. Science 187(4174):319–327
Zong Y, Sun P, Liu J, Yue Z, Li K, Teng Y (2014a) Genetic diversity and population structure of seedling populations of Pyrus pashia. Plant Mol Biol Rep 32:644–651
Zong Y, Sun P, Liu J, Yue X, Niu Q, Teng Y (2014b) Chloroplast DNA-based genetic diversity and phylogeography of Pyrus betulaefolia (Rosaceae) in Northern China. Tree Genet Genomes 10:739–749
Zong Y, Sun P, Yue X, Niu Q, Teng Y (2017) Variation in microsatellite loci reveals a natural boundary of genetic differentiation among Pyrus betulaefolia populations in Northern China. J Am Soc Hort Sci 142:319–329
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 This is a U.S. government work and not under copyright protection in the U.S.; foreign copyright protection may apply
About this chapter
Cite this chapter
Volk, G.M., Cornille, A. (2019). Genetic Diversity and Domestication History in Pyrus. In: Korban, S. (eds) The Pear Genome. Compendium of Plant Genomes. Springer, Cham. https://doi.org/10.1007/978-3-030-11048-2_3
Download citation
DOI: https://doi.org/10.1007/978-3-030-11048-2_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-11047-5
Online ISBN: 978-3-030-11048-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)