Abstract
The network description is widely used to analyze the topology and the dynamics of complex systems. Residue interaction network (RIN) represents three-dimensional structure of protein as a set of nodes (residues) with their connections (edges). Calculated topological parameters from RIN correlate with various aspects of protein structure and function. Here, we reviewed the applications of RIN for the analysis and prediction of functionally important residues and ligand binding sites, protein–protein interactions , allosteric regulation , influence of point mutations on structure and dynamics of proteins.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Residue interaction network
- RIN
- Protein–protein interactions
- Allosteric regulation
- Scoring function
- Allosteric pathway
1 Introduction
Proteins play a vital role in biological systems and have numerous functions such as catalysts, transporters, regulators of signal transduction . They are linear heteropolymers folded into three-dimensional structures. The amino acid residues interact through various covalent and non-covalent bonds in a specific manner to obtain a particular three-dimensional structure, which determines their functions. Knowledge of the relationship between protein structure and its function is important in drug design , molecular medicine, and biotechnology.
Different computational methods have been used for investigations of protein structures and their functions, finding functionally important residues, prediction protein–protein interactions , discovering new biological active compounds. In the most approaches, the protein structures have been viewed as linear sequences of amino acid residues packed into 3D globules. In the last decade, an alternative view of proteins structures has emerged that describe the protein spatial structure as network of amino acids residues interaction.
Network analysis has successfully used in different fields, such as social networks [1], Internet networks [2], road networks [3]. In biology, this method is widely used for analysis of networks of gene regulation, protein–protein interaction , metabolites flow, prediction of drug side effects, etc., [4,5,6,7,8,9]. Applying network methodology for polypharmacology was reviewed in [10].
A network method is based on the graph theory and includes a set of entities (nodes) and of the relationships (edges) occurring among them. These nodes and edges can have various attributes. Depending on the object of the study, nodes can represent genes, proteins, small compounds, and edges connecting these nodes represent the physical interactions, genetic regulatory, or other properties linking the nodes. Edges can have additional information, such as weights, directions.
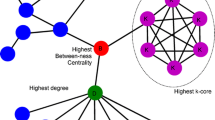
According to the structure of protein, every amino acid residue in it is considered to be a “node” or “vertex,” and the interaction of residues represents “edge” (Fig. 1). The existence of an edge between two nodes depends only on their spatial position in protein globule and has no relation to position in their primary sequence. The interaction can be represented as distance between Cα or any other atoms of amino acid residues, non-covalent interaction (electrostatic , hydrophobic , H-bonds) of the particular amino acids [11]. Additionally, in residue interaction network (RIN), the energy of interaction between residues can be used for weighting the edges [12, 13]. Proteins can be also modeled as subnetworks of amino acid residues having similar physiochemical properties. RIN method reduces spatial protein architectures to simple maps including nodes (residues) and edges (inter-residue interactions). Analysis of these graphs yields a characterization of the protein’s topology and network characteristics.
There are several names of the resultant intraprotein amino acid residue interaction networks . They are called residue interaction graphs [14], protein structure graphs [15, 16], protein residue networks [17], protein contact networks [18], protein energy networks [13], amino acid networks [19], protein structure networks [20], residue interaction networks [21]. In this review, we will use the residue interaction networks (RINs) to distinguish it from network of protein–protein interactions .
The application of RIN method in drug design is just at a beginning. RINs have been used to analyze protein stability and folding [22, 23], 3D structure modeling [19, 23], finding functionally important amino acid residues and sites [14, 24], analyzed protein–protein interactions [25], allosteric regulation [26], influence of amino acid mutations [27]. These studies showed that RIN method is valuable approaches allowed to improve the drug discovery process. Recently, several reviews on RINs have been published [28,29,30,31].
Herein, we aim to review the investigation of the construction, analysis, and application of RINs in fields related to drug design .
2 Graph Theory and Residue Interaction Network
Graph theory represents complex system as a set of elements (called vertices or nodes) with their connections (called edges). Each node can be connected to each other through multiple edges. Adding order of nodes in the graph, we get a directed graph, where edges are directed and usually represented as arrows. Introduction of the quantitative characteristics of the edges results in a weighted graph. Nodes with edges form a network. The network representation helps to analyze the interaction among individual elements and to characterize the whole system.
Residue interaction network is constructed on the base of the three-dimensional atomic coordinates of protein structure and consists of nodes and edges. Each node represents amino acid residue (or Cα atom) that is connected to the neighbor node. In the simplest variant, the edges are defined on the base of predefined cutoff of the distances in 3D structure between nodes. The values of distance may be varied based on nature of interactions (van der Waals , hydrophobic , electrostatic interactions, etc.). Frequently, the covalent backbones are included as edges in the networks. The edges can be weighed based on energy of interactions, knowledge-based potentials, or amino acid fluctuations in molecular dynamics simulation [30, 31]. The differential network (DDN) method was proposed where network formed by unique edges that are present only in one state but are absent in other ones [32].
Networks have several most common characteristics; some of them that frequently have been used for analysis of biological systems are listed below [28, 31, 33].
A degree of a node is a number of edges in a network that connect node with its neighbors. In a directed network, there might be two types of degrees, the in-degree, and the out-degree depending on the orientation of the edges. An average degree is the average number of connections that the nodes have in a network.
A connectivity represents a minimum number edges that need to be removed to make a disconnected graph. The connectivity structure and the degree of nodes analysis in RINs help to identify important residues, i.e., participating in ligand binding sites.
A shortest path is a path in which the two nodes are connected by the smallest number of intermediate nodes. A characteristic path length is defined as the number of edges in the shortest path between two nodes, averaged over all pairs of nodes. Residues with small shortest path lengths are often located in the active or ligand binding sites of proteins [17] and participate in allosteric pathways [34, 35].
A betweenness centrality of a node is the number of times that a node is included in the shortest path between each pair of nodes, normalized by the total number of pairs.
A closeness centrality of a node is the reciprocal of the average shortest path length.
The network concept is widely used to analyze and predict properties in different biological systems, from intramolecular interaction to whole cells and organisms. Biological networks are small worlds that means that two nodes are connected to each other via only a few other nodes [23, 30]. There are several network parameters for characterizing different aspects of biological networks.
A hub is defined as a node with a high degree or connectivity in a network. Hubs may play a structural role in proteins increasing the thermodynamic stability of proteins [14, 36].
A cluster is a set of nodes with the number of connections, which is higher than in the other nodes. Clusters often are equivalent to a domain of protein and participate in intramolecular interactions.
A clique is a set of nodes in which each node of graph is connected to every other node. Studies of cliques can help to understand ligand -induced population shift in protein [37].
There are several software packages, Web servers, and plug-ins available for construction and analyzing of RINs, such as Xpyder (http://xpyder.sourceforge.net/) [38], Network View [39], RING (http://protein.bio.unipd.it/ring/) [21, 40], RINalyzer (http://www.rinalyzer.de) [41], structureViz (http://www.cgl.ucsf.edu/cytoscape/structureViz/) [42].
Web server RING constructs physicochemically RINs from PDB files for subsequent visualization in the Cytoscape (software platform for the analysis and visualization of biological networks) (http://www.cytoscape.org) or Pymol (https://pymol.org/). Interactions (edges) are disulfide bonds, salt bridges, hydrogen bonds , aromatic interactions, and van der Waals contacts. Several features can be added to nodes and edges, such as secondary structure, solvent accessibility, energy score, sequence conservation. Subnetwork can be also constructed.
RINalyzer and structureViz are plug-ins for Cytoscape [43] that link Cytoscape with the molecular viewer UCSF Chimera (http://www.cgl.ucsf.edu/chimera/) [44]. They allow interactive structure analysis of RINs together with the corresponding 3D protein structure.
NetworkView plug-in for VMD (https://www.ks.uiuc.edu/Research/vmd/) allows to study allostery and signaling through network models. This plug-in can display the dynamical network representations.
3 RINs Application
3.1 Ligand Binding Sites
Identification of the ligand binding sites of proteins and functionally important residues is a crucial first step in drug design . However, it is a difficult task in the case of the absence of homologous proteins.
Several topological parameters of RINs may be used for the prediction of ligand binding sites. Several investigations showed that closeness and betweenness values of residues are correlated with ligand binding site residues [14, 34, 45,46,47,48]. The accuracy of prediction such residues may be improved by combining with such parameters as their solvent accessibility. So, Amitai et al. [14] could predict active site residues in 70% of the analyzed 178 enzymes proteins, using closeness centrality and solvent accessibility parameters. The similar result was obtained in [49]. The closeness centrality was used as parameter in machine learning methods for prediction of functionally important residues [50] or in score for docking [25].
However, for non-enzyme proteins correlation between closeness centrality and binding sites has not observed [34, 51]. In addition, global closeness centrality gave unsatisfactory result for non-globular and oligomer proteins. For such proteins, more tolerable prediction was obtained with local closeness [52]. It seems that the ligand binding sites in enzymes are correlated with centrality due to their typical location in cavities of the enzymes, whereas in oligomer proteins, the protein–protein interfaces are more flat [53], which reduces the centrality of their residues.
Coevolution residues networks, which include information about coevolved residues, were also used for predicting functionally important residues [54, 55]. RIN analysis was applied for prediction similarity of ligand binding sites in different proteins [56, 57].
The node-weighted RIN, called node-weighted amino acid contact energy network (NACEN) was developed for prediction hotspots, catalytic residues, and allosteric residues. Nodes were weighted based on structural, sequence, physicochemical and dynamic properties of the residues. SVM was used for design model to identify functionally important residues. The results revealed that parameters from node-weighted RIN have advantages over ones from unweighted network [58].
Poirrette et al. [56] designed RIN of the influenza sialidase binding site of Zanamivir and used it to predict proteins having the similar binding sites. Such an approach may be used for repurposing drugs or prediction of side effects.
3.2 Protein–Protein Interactions
Protein–protein interactions (PPIs) are crucial for many biological processes and functions; inhibition of PPIs with small molecules is a perspective way in drug design [53]. RIN method was used for analysis of protein–protein interfaces, prediction of hotspots, and selection of protein poses in the protein–protein docking .
Several investigations were done using RIN for analysis of protein–protein interfaces. They showed that hydrophobic and charged residues are predominant in the dimer interface and that arginine, histidine, glutamic acid, phenylalanine, and tyrosine are located in clusters at the interface [59, 60]. In those clusters, highly connected residues correlate with experimentally identified hotspots in the protein complexes [15, 16, 61, 62].
Correct prediction of protein–protein complexes using individual proteins by docking method is a big challenge, since the docking gives many false-positive solutions [63, 64]. Protein–protein complex formation may be viewed as combining of two RINs, where additional edges have appeared between nodes from different subunits. The interaction of residues occurs in accordance with their properties. Since native protein–protein complexes are far from random, the correct and incorrect poses have different topologies.
Chang et al. [65] designed hydrophobic and hydrophilic RINs of a protein–protein complex. Three terms based on these networks (degree, clustering coefficient, and characteristic path length) were calculated and used in network-based scoring function HPNet. Combining it with energy terms of RosettaDock [66] results in new combined scoring function HPNet-combine. It was found that HPNet-combine could improve the discrimination of the RosettaDock scoring function.
The similar methodology based on the construction of a hydrophobic and hydrophilic RINs of protein–protein complexes was used for the development NPPD scoring function [67]. Protein–protein docking , HoDock, and scoring function HPNCscore (hydrophobic , and polar network combined scoring function) were developed. It showed good results for several targets in Critical Assessment of PRedicted Interactions (CAPRI) rounds [68].
The weighed RINs were used for the development of Sn scoring function [69]. Two weighted parameters (strength and weighted average nearest neighbors’ degree) were introduced to develop a scoring function. The testing of this scoring function for 42 protein–protein complexes had shown a satisfied performance.
The scoring function based on the local network patterns, iScore, was proposed [70]. It achieved 83.6% specificity with 82% sensitivity for training set of ~1800 two domain proteins, homo- and heterodimers.
3.3 Allosteric Regulation
Allosteric regulation is a common mechanism to control the protein activities. The perturbation at the allosteric site results in transmission of signal through the protein structure to other sites leading to modification of catalytic activity, oligomerization, etc. [71, 72].
Allosteric sites became attractive target for drug design at last decade. Allosteric drugs have several potential benefits over orthosteric drugs . They may be more specific due to less similarity of allosteric sites comparing to active site in homologous proteins; they can increase or decrease the activity of enzymes and receptors; partially inhibiting by allosteric drugs may cause less side effects [73, 72].
Using allosteric sites for drug design, it is required to predict allosteric sites, residues involved in signal transduction pathways to the active sites. The search of allosteric sites by RIN method is similar to the other sites described above.
Allosteric pathways show how the signal may be transmitted over a long distance from allosteric to active sites within the protein. RIN is accurate and not time-consuming method for prediction such pathways.
Once the RIN constructed, several algorithms can be used to find allosteric pathways within the RINs. The common method is to find the shortest paths connecting the allosteric and active sites [34, 35, 74]. The shortest path may be determined by Floyd–Warshall algorithm. It was shown that many proteins may be considered as a set of modules (subgraphs with many interconnections and with few connections to other subgraphs). The residues involved in the interaction of such modules can participate in allosteric pathways [75]. It is proposed that such residues are conservative that also may be used for their prediction [76,77,78]. Proteins can have multiple allosteric pathways, which may preexist without effector binding at allosteric site [79]. Various pathways may be involved depending on the different changes in allosteric site.
However, RINs constructed based on a single structure do not take into account the structural changes in protein globule. Therefore, the combination of molecular dynamics simulation (MD) followed by RINs design frequently has been used to detect and to analyze allosteric pathways. In these cases, the edges in RINs are defined using various parameters obtained from MD. The edges may reflect the correlation of displacements of the residues [74, 80], the fluctuation of distances [81], interaction energy [82], etc.
Aminoacyl-tRNA synthetases are convenient objects for analysis of allosteric communication. The combination of MD with RIN was used for discovering pathways from anticodon region to the aminoacylation region for methionyl-tRNA synthetase [74, 83], glutaminyl-tRNA synthetase [84], cysteinyl-tRNA synthetase [35], and tryptophanyl-tRNA synthetase [85, 86]. Particularly, analysis of tryptophanyl-tRNA synthetase showed changes of flexibility around the active site induced by allosteric ligands binding and allowed to explain the molecular mechanism of half-of-the-sites reactivity (tryptophanyl-tRNA synthetase is a homodimer).
Another popular object is G protein-coupled receptors (GPCRs) [87,88,89]. It is a large family of membrane receptors, which have ligand binding site on the extracellular side of membrane and activation domain on its internal side. Using RIN method, several conservative residues participating in the signal transduction were discovered for the lutropin receptor [76] and A2A adenosine receptor [87] (Fig. 2).
3.4 Analyses of Mutations
RIN methods may be used for analysis and prediction of effects of amino acid mutation on protein properties, which may be useful for protein design , investigations of disease-associated single nucleotide polymorphisms, or mechanism of the drug resistance [27, 90,91,92].
Recently, we used RIN for investigation of the influence of several mutations on structure and flexibility of β-lactamase [93]. β-lactamases are class of enzymes responsible for bacteria resistant to β-lactam antibiotics . Besides, the key mutations, responsible for the extended spectrum β-lactamases or inhibitor resistance phenotype, secondary mutations , located far from active site and with a weak impact on the protein structure and enzyme activity, have been often appeared [94]. Analysis of MD trajectories showed that the secondary mutations , and the key mutations can exhibit opposite effect on the flexibility of the Ω-loop of β-lactamase that participate in antibiotic hydrolysis and transport in the active site [93]. Detailed analysis of RIN maps of proteins of consistent mutations from wild-type TEM-1 to TEM-72 (carrying two key mutations G238S and E240K and two secondary ones M182T and Q39K) showed that key mutations (responding for extended spectrum β-lactamases) lead to weakening interactions of the Ω-loop with protein globule. The appearance of secondary mutation M182T resulted in dramatic changing of conformation of R65, and this residue began to interact with the Ω-loop and fixed it near protein globule (manuscript submitted) (Fig. 3).
4 Conclusion
Herein, we have reviewed the development and current stage of RINs and their application for drug discovery.
RINs provide complex analysis of the proteins and their complexes. Residues are in tight contact with each other in protein globules, and RINs allowed to estimate their interdependence and to predict different properties and functionality of the individual residues and the whole proteins. In addition to topology, RINs allow to use chemicophysical properties of residues and energy of their interaction in RIN construction and analysis of proteins.
Besides, using RINs for investigation protein structure and functions, they may be applied in drug design in several ways.
Prediction of functionally important residues and sites can be helpful for understanding functions and regulation of uncharacterized proteins, finding active sites, allosteric and cryptic ligand binding sites. It may decrease the amount of “undruggable” protein, increasing field for drug design . On the other hand, many drug candidates fail in the late and costly stages of clinical trials [95]. Side effects are one of the main reasons for drug failure [96]. The detection of similarity in network topologies and interactions with ligands for several targets may indicate the promiscuity of drug candidates and possibly their side effects.
The development of inhibitors of protein–protein interactions is a perspective way in drug design , and RIN showed their applicability for this purpose. The analysis of networks may help to select correct poses in protein–protein docking that is important for the selection of inhibitor binding sites; incorporation of the terms from RINs may improve docking scoring functions.
Allosteric inhibitors are another mainstream in drug design in last decade. It is proposed that such inhibitors may regulate cellular processes more accurately. Allosteric regulation is the common property of protein, which may increase the number of druggable targets . RINs are convenient for finding allosteric sites, investigation of mechanism of intraprotein signal transmission. Prediction of the effect of amino acid mutations on protein structure and dynamics is crucial for the development drugs against diseases with a high probability of occurrence drug resistance , in particular antibacterial , antiviral , and anticancer drugs.
Nowadays, the application of RIN methods for drug discovery is at their early stage, but they already help to understand intimate properties of proteins and provide a new view for drug discovery.
Abbreviations
- CAPRI:
-
Critical assessment of predicted interactions
- DDN:
-
Differential network
- GPCR:
-
G protein-coupled receptor
- HPNCscore:
-
Hydrophobic and polar networks combined scoring function
- MD:
-
Molecular dynamics simulation
- NACEN:
-
Node-weighted amino acid contact energy network
- PPI:
-
Protein–protein interaction
- RIN:
-
Residue interaction network
- SVM:
-
Support vector machine
References
Otte E, Rousseau R (2002) Social network analysis: a powerful strategy, also for the information sciences. J Inform Sci 28:441–453
Meusel R, Vigna S, Lehmberg O, Bizer C (2015) The graph structure in the web – analyzed on different aggregation levels. J Web Sci 1:33–47
Bottinelli A, Louf R, Gherardi M (2017) Balancing building and maintenance costs in growing transport networks. Phys Rev E 96:032316
Murakami Y, Tripathi LP, Prathipati P, Mizuguchi K (2017) Network analysis and in silico prediction of protein-protein interactions with applications in drug discovery. Curr Opin Struct Biol 44:134–142
Zhao B, Wang J, Wu FX (2017) Computational methods to predict protein functions from protein-protein interaction networks. Curr Protein Pept Sci 18:1120–1131
Miryala SK, Anbarasu A, Ramaiah S (2018) Discerning molecular interactions: a comprehensive review on biomolecular interaction databases and network analysis tools. Gene 642:84–94
Laddach A, Ng JC, Chung SS, Fraternali F (2018) Genetic variants and protein-protein interactions: a multidimensional network-centric view. Curr Opin Struct Biol 50:82–90
Yao V, Wong AK, Troyanskaya OG (2018) Enabling precision medicine through integrative network models. J Mol Biol 430(18 Pt A):2913–2923
Xie L, Li J, Xie L, Bourne PE (2009) Drug discovery using chemical systems biology: identification of the protein-ligand binding network to explain the side effects of CETP inhibitors. PLoS Comput Biol 5:e1000387
Li P, Fu Y, Wang Y (2015) Network based approach to drug discovery: a mini review. Mini-Rev Med Chem 15:687–695
Aftabuddin M, Kundu S (2007) Hydrophobic, hydrophilic, and charged amino acid networks within protein. Biophys J 93:225–231
Bhattacharyya M, Vishveshwara S (2011) Probing the allosteric mechanism in pyrrolysyl-tRNA synthetase using energy-weighted network formalism. Biochemistry 50:6225–6236
Vijayabaskar MS, Vishveshwara S (2010) Interaction energy based protein structure networks. Biophys J J99:3704–3715
Amitai G, Shemesh A, Sitbon E, Shklar M, Netanely D, Venger I, Pietrokovski S (2004) Network analysis of protein structures identifies functional residues. J Mol Biol 344:1135–1146
Brinda KV, Vishveshwara S (2005) A network representation of protein structures: implications for protein stability. Biophys J J89:4159–4170
Brinda KV, Vishveshwara S (2005) Oligomeric protein structure networks: insights into protein-protein interactions. BMC Bioinform 6:296
Atilgan AR, Akan P, Baysal C (2004) Small-world communication of residues and significance for protein dynamics. Biophys J J86:85–91
Bagler G, Sinha S (2007) Assortative mixing in protein contact networks and protein folding kinetics. Bioinformatics 23:1760–1767
Zhou J, Yan W, Hu G, Shen B (2014) Amino acid network for the discrimination of native protein structures from decoys. Curr Protein Pept Sci 15:522–528
Hu G, Zhou J, Yan W, Chen J, Shen B (2013) The topology and dynamics of protein complexes: insights from intra–molecular network theory. Curr Protein Pept Sci 14:121–132
Martin AJ, Vidotto M, Boscariol F, Di Domenico T, Walsh I, Tosatto SC (2011) RING: networking interacting residues, evolutionary information and energetics in protein structures. Bioinformatics 27:2003–2005
Rao F, Caflisch A (2004) The protein folding network. J Mol Biol 342:299–306
Grewal RK, Roy S (2015) Modeling proteins as residue interaction networks. Protein Pept Lett 22:923–933
Zhou J, Yan W, Hu G, Shen B (2016) Amino acid network for prediction of catalytic residues in enzymes: a comparison survey. Curr Protein Pept Sci 17:41–51
Pons C, Glaser F, Fernandez-Recio J (2011) Prediction of protein-binding areas by small-world residue networks and application to docking. BMC Bioinform 12:378
Schueler-Furman O, Wodak SJ (2016) Computational approaches to investigating allostery. Curr Opin Struct Biol 41:159–171
Cheng TMK, Lu Y-E, Vendruscolo M, Lio P, Blundell TL (2008) Prediction by graph theoretic measures of structural effects in proteins arising from non-synonymous single nucleotide polymorphisms. PLoS Comput Biol 4:e1000135
Di Paola L, De Ruvo M, Paci P, Santoni D, Giuliani A (2013) Protein contact networks: an emerging paradigm in chemistry. Chem Rev 113:1598–1613
Csermely P, Korcsmáros T, Kiss HJ, London G, Nussinov R (2013) Structure and dynamics of molecular networks: a novel paradigm of drug discovery: a comprehensive review. Pharmacol Ther 138:333–408
Yan W, Zhou J, Sun M, Chen J, Hu G, Shen B (2014) The construction of an amino acid network for understanding protein structure and function. Amino Acids 46:1419–1439
Bhattacharyya M, Ghosh S, Vishveshwara S (2016) Protein structure and function: looking through the network of side-chain interactions. Curr Protein Pept Sci 17:4–25
Grewal RK, Mitra D, Roy S (2015) Mapping networks of light-dark transition in LOV photoreceptors. Bioinformatics 31:3608–3616
Doncheva NT, Assenov Y, Domingues FS, Albrecht M (2012) Topological analysis and interactive visualization of biological networks and protein structures. Nat Protoc 7:670–685
del Sol A, Fujihashi H, Amoros D, Nussinov R (2006) Residues crucial for maintaining short paths in network communication mediate signaling in proteins. Mol Syst Biol 2:0019
Ghosh A, Sakaguchi R, Liu C, Vishveshwara S, Hou YM (2011) Allosteric communication in cysteinyl tRNA synthetase: a network of direct and indirect readout. J Biol Chem 286:37721–37731
Estrada E (2010) Universality in protein residue networks. Biophys J 98:890–900
Ghosh A, Vishveshwara S (2008) Variations in clique and community patterns in protein structures during allosteric communication: investigation of dynamically equilibrated structures of methionyl tRNA synthetase complexes. Biochemistry 47:11398–11407
Pasi M, Tiberti M, Arrigoni A, Papaleo E (2012) xPyder: a PyMOL plugin to analyze coupled residues and their networks in protein structures. J Chem Inf Model 52:1865–1874
Eargle J, Luthey-Schulten Z (2012) NetworkView: 3D display and analysis of protein· RNA interaction networks. Bioinformatics 28:3000–3001
Piovesan D, Minervini G, Tosatto SC (2016) The RING 2.0 web server for high quality residue interaction networks. Nucleic Acids Res 44(W1):W367–W374
Doncheva NT, Klein K, Domingues FS, Albrecht M (2011) Analyzing and visualizing residue networks of protein structures. Trends Biochem Sci 36:179–182
Morris JH, Huang CC, Babbitt PC, Ferrin TE (2007) structureViz: linking Cytoscape and UCSF Chimera. Bioinformatics 23:2345–2347
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612
Yan Y, Zhang SG, Wu FX (2011) Applications of graph theory in protein structure identification. Proteome Sci 9(Suppl 1):S17
Thibert B, Bredesen DE, del Rio G (2005) Improved prediction of critical residues for protein function based on network and phylogenetic analyses. BMC Bioinform 6:213
Emerson IA, Gothandam KM (2012) Residue centrality in alpha helical polytopic transmembrane protein structures. J Theor Biol 309:78–87
Tse A, Verkhivker GM (2015) Molecular dynamics simulations and structural network analysis of c-Abl and c-Src kinase core proteins: capturing allosteric mechanisms and communication pathways from residue centrality. J Chem Inf Model 55:1645–1662
Chea E, Livesay DR (2007) How accurate and statistically robust are catalytic site predictions based on closeness centrality? BMC Bioinform 8:153
Tang YR, Sheng ZY, Chen YZ, Zhang Z (2008) An improved prediction of catalytic residues in enzyme structures. Protein Eng Des Sel 21:295–302
Sheftel S, Muratore K, Black M, Costanzi S (2013) Graph analysis of β2 adrenergic receptor structures: a “social network” of GPCR residues. Silico Pharmacol 1:16
Slama P, Filippis I, Lappe M (2008) Detection of protein catalytic residues at high precision using local network properties. BMC Bioinform 9:517
Veselovsky AV, Archakov AI (2007) Inhibitors of protein-protein interactions as potential drugs. Curr Comput-Aided Drug Des 3:51–58
Marino Buslje C, Teppa E, Di Domenico T, Delfino JM, Nielsen M (2010) Networks of high mutual information define the structural proximity of catalytic sites: implications for catalytic residue identification. PLoS Comput Biol 6:e1000978
Aguilar D, Oliva B, Buslje CM (2012) Mapping the mutual information network of enzymatic families in the protein structure to unveil functional features. PLoS ONE 7:e41430
Poirrette AR, Artymiuk PJ, Grindley HM, Rice DW, Willett P (1994) Structural similarity between binding sites in influenza sialidase and isocitrate dehydrogenase: implications for an alternative approach to rational drug design. Protein Sci 3:1128–1130
Liu ZP, Wu LY, Wang Y, Zhang XS, Chen L (2008) Analysis of protein surface patterns by pocket similarity network. Prot Pept Lett 15:448–455
Yan W, Hu G, Liang Z, Zhou J, Yang Y, Chen J, Shen B (2018) Node-weighted amino acid network strategy for characterization and identification of protein functional residues. J Chem Inf Model. (in press). https://doi.org/10.1021/acs.jcim.8b00146
Brinda KV, Kannan N, Vishveshwara S (2002) Analysis of homodimeric protein interfaces by graph-spectral methods. Protein Eng 15:265–277
Reichmann D, Rahat O, Albeck S, Meged R, Dym O, Schreiber G (2005) The modular architecture of protein–protein binding interfaces. PNAS 102:57–62
Brinda KV, Surolia A, Vishveshwara S (2005) Insights into the quaternary association of proteins through structure graphs: a case study of lectins. Biochem J J391:1–15
Kannan N, Chander P, Ghosh P, Vishveshwara S, Chatterji D (2001) Stabilizing interactions in the dimer interface of alpha-subunit in Escherichia coli RNA polymerase: a graph spectral and point mutation study. Protein Sci 10:46–54
Soni N, Madhusudhan MS (2017) Computational modeling of protein assemblies. Curr Opin Struct Biol 44:179–189
Zhang Q, Feng T, Xu L, Sun H, Pan P, Li Y, Li D, Hou T (2016) Recent advances in protein-protein docking. Curr Drug Targets 17:1586–1594
Chang S, Jiao X, Li CH, Gong XQ, Chen WZ, Wang CX (2008) Amino acid network and its scoring application in protein–protein docking. Biophys Chem 134:111–118
Gray JJ, Moughon S, Wang C, Schueler-Furman O, Kuhlman B, Rohl CA, Baker D (2003) Protein–protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J Mol Biol 33(1):281–299
Shih ESC, Hwang M-J (2015) NPPD: a protein-protein docking scoring function based on dyadic differences in networks of hydrophobic and hydrophilic amino acid residues. Biology 4:282–297
Gong X, Wang P, Yang F, Chang S, Liu B, He H, Cao L, Xu X, Li C, Chen W, Wang C (2010) Protein-protein docking with binding site patch prediction and network-based terms enhanced combinatorial scoring. Proteins 78:3150–3155
Jiao X, Chang S (2011) Scoring function based on weighted residue network. Int J Mol Sci 12:8773–8786
Luo Q, Hamer R, Reinert G, Deane CM (2013) Local network patterns in protein-protein interfaces. PLoS ONE 8:e57031
Greener JG, Sternberg MJ (2018) Structure-based prediction of protein allostery. Curr Opin Struct Biol 50:1–8
Nussinov R, Tsai CJ (2013) Allostery in disease and in drug discovery. Cell 153:293–305
Lu S, Li S, Zhang J (2014) Harnessing allostery: a novel approach to drug discovery. Med Res Rev 34:1242–1285
Ghosh A, Vishveshwara S (2007) A study of communication pathways in methionyl-tRNA synthetase by molecular dynamics simulations and structure network analysis. PNAS 104:15711–15716
del Sol A, Arauzo-Bravo MJ, Amoros D, Nussinov R (2007) Modular architecture of protein structures and allosteric communications: potential implications for signaling proteins and regulatory linkages. Genome Biol 8:R92
Angelova K, Felline A, Lee M, Patel M, Puett D, Fanelli F (2011) Conserved amino acids participate in the structure networks deputed to intramolecular communication in the lutropin receptor. Cell Mol Life Sci 68:1227–1239
Süel GM, Lockless SW, Wall MA, Ranganathan R (2003) Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat Struct Biol 10:59–69
Tang S, Liao JC, Dunn AR, Altman RB, Spudich JA, Schmidt JP (2007) Predicting allosteric communication in myosin via a pathway of conserved residues. J Mol Biol 373:1361–1373
del Sol A, Tsai CJ, Ma B, Nussinov R (2009) The origin of allosteric functional modulation: multiple pre-existing pathways. Structure 17:1042–1050
Sethi A, Eargle J, Black AA, Luthey-Schulten Z (2009) Dynamical networks in tRNA: protein complexes. PNAS 106:6620–6625
Dixit A, Verkhivker GM (2011) Computational modeling of allosteric communication reveals organizing principles of mutation-induced signaling in ABL and EGFR kinases. PLoS Comput Biol 7:e1002179
Kong Y, Karplus M (2009) Signaling pathways of PDZ2 domain: a molecular dynamics interaction correlation analysis. Proteins 74:145–154
Vishveshwara S, Ghosh A, Hansia P (2009) Intra and inter-molecular communications through protein structure network. Curr Protein Pept Sci 10:146–160
Sathyapriya R, Vishveshwara S (2007) Structure networks of E-coli glutaminyl-tRNA synthetase: effects of ligand binding. Proteins 68:541–550
Bhattacharyya M, Ghosh A, Hansia P, Vishveshwara S (2010) Allostery and conformational free energy changes in human tryptophanyl-tRNA synthetase from essential dynamics and structure networks. Proteins 78:506–517
Hansia P, Ghosh A, Vishveshwara S (2009) Ligand dependent intra and inter subunit communication in human tryptophanyl tRNA synthetase as deduced from the dynamics of structure networks. Mol Bio Syst 5:1860–1872
Fanelli F, Felline A (2011) Dimerization and ligand binding affect the structure network of A2A adenosine receptor. Biochim Biophys Acta 1808:1256–1266
Lee Y, Choi S, Hyeon C (2014) Mapping the intramolecular signal transduction of G-protein coupled receptors. Proteins 82:727–743
Miao Y, Nichols SE, Gasper PM, Metzger VT, McCammon JA (2013) Activation and dynamic network of the M2 muscarinic receptor. PNAS 110:10982–10987
Hu Z, Bowen D, Southerland WM, del Sol A, Pan Y, Nussinov R, Ma B (2007) Ligand binding and circular permutation modify residue interaction network in DHFR. PLoS Comput Biol 3:1097–1107
Li Y, Wen Z, Xiao J, Yin H, Yu L, Yang L, Li M (2011) Predicting disease-associated substitution of a single amino acid by analyzing residue interactions. BMC Bioinform 12:14
Tse A, Verkhivker GM (2015) Small-world networks of residue interactions in the Abl kinase complexes with cancer drugs: topology of allosteric communication pathways can determine drug resistance effects. Mol Biosyst 11:2082–2095
Shcherbinin DS, RubtsovaMYu Grigorenko VG, Uporov IV, Veselovsky AV, Egorov AM (2017) The study of the role of mutations M182T and Q39K in the TEM-72 β-lactamase structure by the molecular dynamics method. Biochem (Moscow), Suppl B: Biomed Chem 11:120–127
Grigorenko VG, RubtsovaMYu Uporov IV, Ishtubaev IV, Andreeva IP, Shcherbinin DS, Veselovsky AV, Egorov AM (2018) Bacterial TEM-type serine beta-lactamases: structure and analysis of mutations. biochemistry (Moscow). Suppl B: Biomed Chem 12:87–95
Nwaka S, Hudson A (2006) Innovative lead discovery strategies for tropical diseases. Nat Rev Drug Discov 5:941–955
Scheiber J, Chen B, Milik M, Sukuru SC, Bender A, Mikhailov D, Whitebread S, Hamon J, Azzaoui K, Urban L, Glick M, Davies JW, Jenkins JL (2009) Gaining insight into off-target mediated effects of drug candidates with a comprehensive systems chemical biology analysis. J Chem Inf Model 49:308–317
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Shcherbinin, D., Veselovsky, A. (2019). Analysis of Protein Structures Using Residue Interaction Networks. In: Mohan, C. (eds) Structural Bioinformatics: Applications in Preclinical Drug Discovery Process. Challenges and Advances in Computational Chemistry and Physics, vol 27. Springer, Cham. https://doi.org/10.1007/978-3-030-05282-9_3
Download citation
DOI: https://doi.org/10.1007/978-3-030-05282-9_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-05281-2
Online ISBN: 978-3-030-05282-9
eBook Packages: Chemistry and Materials ScienceChemistry and Material Science (R0)