Abstract
Plants have four main types of photoreceptor proteins: phytochromes (up to 5 different types with partly different functions), cryptochromes (2 types), phototropins (2 types), and UVR8. The signal transduction from these is not simple chains, they form a network, and excitation of one photoreceptor may enhance or inhibit the effect of another in a very complex manner. This chapter gives an account of these pathways and interactions.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
14.1 Introduction
Plants have evolved the capacity to sense and interpret diverse light signals to modulate their development. In the extensively studied plant Arabidopsis thaliana, four distinct families of photoreceptors (phytochromes, cryptochromes, phototropins, and UV-B photoreceptor) have been well characterized. In this chapter, we give a general description on molecular mechanism of photoreceptors and basic structures of different light signaling. We then dedicate to the signaling cross talk under the control of various kinds of photoreceptors, including (1) signaling interaction between light and UV-B; (2) physical interactions among phytochrome, phototropin, and cryptochrome; and (3) signaling integration of phytochrome and cryptochrome.
14.2 Photoreceptor Regulation and Activity: An Overview
Photoreceptors have the ability to sense and distinguish light of different spectral composition, delivering the signal to the downstream components, initiating transcription of various genes. This finally results in multiple developmental processes, including seed germination, seedling photomorphogenesis, phototropism, gravitropism, chloroplast movement, shade avoidance, circadian rhythms, and flower induction (Quail 2002). Therefore, the photoreceptors play the central role in plant life. After germination, seedlings follow one of two developmental patterns. Skotomorphogenesis (or etiolation) in the dark is characterized by long hypocotyl, closed cotyledons in Arabidopsis, and the development of proplastids into etioplasts. By contrast, growth in the light leads to photomorphogenesis (or de-etiolation) characterized by short hypocotyls, expanded open cotyledons, and the development of mature chloroplasts (Jiao et al. 2007). A wide spectrum of light can induce photomorphogenesis; however, for clarification, we here define the far red-, red-, blue-, and UV-A-induced photomorphogenesis as light photomorphogenesis, while UV-B-induced one as UV-B photomorphogenesis. In Arabidopsis, there are five phytochromes (PHYA to PHYE) that perceive red/far-red light, three cryptochromes (CRY1 to CRY3) and two phototropins (PHOT1 and PHOT2) that sense blue/UV-A light, and one known UV-B photoreceptor (UVR8) that senses UV-B (280-215 nm).
Phytochromes (phys) are homodimeric photoreceptors of approximately 120 kDa monomers that bear a single linear tetrapyrrole chromophore, which exists in two photoconvertible forms: Pr (red light-absorbing phy) and Pfr (far-red light-absorbing phy) (Vierstra and Zhang 2011). The photoconversion results in their translocation from the cytoplasm into the nucleus (Quail 2002). This translocation is caused by the phosphorylation of PHYs, which leads to alter its stability and affinity toward downstream signal components (Kim et al. 2004; Ryu et al. 2005). Photoactivated phytochrome has kinase activity, inducing phosphorylation of PIF3 (PHYTOCHROME-INTERACTING PROTEIN 3) and PKS1, initiating the downstream signaling (Al-Sady et al. 2006; Fankhauser et al. 1999) (Fig. 14.1).
The structure of various kinds of photoreceptors. The phytochromes are dimeric chromoproteins. Each polypeptide consists of an N-terminal photosensory domain that covalently binds a single bilin chromophore (PΦB), followed by a C-terminal domain. Phototropins have a photosensory N-terminal half and a C-terminal half with serine/threonine kinase function. The N terminus contains two flavin mononucleotide (FMN) chromophore-binding LOV domains (LOV1 and LOV2). Cryptochromes 1 and 2 have an N-terminal photolyase-related (PHR) domain (CNT) and a less conserved, intrinsically unstructured C-terminal DAS domain (CCT), which is not present in CRY3. The PHR domain non-covalently binds to two chromophores, a flavin adenine dinucleotide (FAD), and a pterin. UVR8 is a seven-bladed β-propeller protein. Arginine (Arg) residues at positions 286 and 338 facilitate hydrogen bonds that hold the homodimer together, while tryptophan (Trp) residues at positions 285 and 233 serve as chromophores. C27 is a protein interaction domain, which is responsible for the interaction with RUPs and COP1
Cryptochromes (crys) are photolyase-like blue light/UV-A receptors that bind a flavin adenine dinucleotide chromophore (Yu et al. 2010). Arabidopsis has three members of this family, CRY1, CRY2, and CRY3, which show significant homology to microbial photolyase, but lack photorepair activity. CRY1 is nuclear localized in the dark but largely cytoplasmic under light, while CRY2 is constitutively nuclear (Lin and Shalitin 2003). A more divergent CRY3 does not have the photolyase-related (PHR) domain and less conserved DAS domain (CCT), which are present in CRY1 and CRY2 Partch et al. 2005). However, CRY3 has a dual function in regulating its transport to chloroplast and mitochondria (Kim et al. 2004). Light can induce the autophosphorylation of CRY1 and CRY2, resulting from the conformational change in the C-terminus CCT domain by the activation of the N-terminus CNT domain. Phosphorylated CRYs can be dimerized and possibly interact with downstream factors (Sang et al. 2005).
Phototropins (phots) are the plant-specific blue light receptors with serine/threonine protein kinase activity. They bear two LOV domains that bind flavin mononucleotide chromophores (Christie 2007). Several transient developments, including phototropism, chloroplast movement, and stomatal opening, are controlled by them. Arabidopsis PHOT1 and PHOT2 are mainly associated with the plasma membrane, but only a fraction of PHOT1 is relocated to the cytoplasm (Christie 2007). Blue light triggers the autophosphorylation of PHOT1 and PHOT2, initiating the transduction of the light signal (Christie 2007).
UV-B Photoreceptor: UVR8 (UV RESISTANCE LOCUS 8) is the recently identified UV-B-specific photoreceptor, with two tryptophans (W233 and W285) as the chromophores. In the absence of UV-B radiation, dimeric UVR8 is localized in the cytoplasm. Upon exposure to UV-B, it monomerizes and migrates to the nucleus. This change is essential for the following signaling transduction.
14.3 Simplified Light and UV-B Signaling
14.3.1 Introduction
Light signaling is one of the most well-elucidated pathways in Arabidopsis, which more than a hundred regulators have been found to play a role in response to light. Several reviews have been published to summarize the complicated signaling (Quail 2002; Jiao et al. 2007), and here we give a simplified signaling model in regard of seedling morphogenesis and will focus on the interconnections shortly.
14.3.2 Light Signaling Pathway
In the dark, a seedling undergoes skotomorphogenesis, in the absence of photoreceptor activation. In this process, there are two groups of proteins, namely, COP/DET/FUS (CONSTITUTIVE PHOTOMORPHOGENIC/DE-ETIOLATED/FUSCA) and PIFs, to maintain the status of skotomorphogenesis. The COP/DET/FUS complex is a member of the ubiquitin system, which is responsible for protein degradation (Sullivan et al. 2003). Because they work in concert to degrade a number of photomorphogenesis-promoting transcription factors, such as HY5 (LONG HYPOCOTYL 5), COP/DET/FUS are the central repressors of photomorphogenesis (Saijo et al. 2003; Osterlund et al. 2000). COP1 is one of the proteins in the complex and has the activity of Ub E3 ligase by interacting with many of the photomorphogenesis-promoting transcription factors. It has been shown that the ubiquitination is promoted by SPA (SUPPRESSOR OF PHYA-105) family proteins (Osterlund et al. 2000; Laubinger et al. 2004). After light illumination, the activated photoreceptors (PHYA, PHYB, CRY1, and CRY2) suppress COP1 function by translocating from nucleus to cytoplasm, resulting in the photomorphogenesis-promoting transcription factors, which initiate the expression of a large number of light-responsive genes to promote photomorphogenesis (Osterlund et al. 2000).
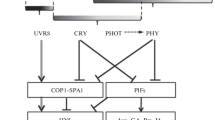
Contrasting to the repressed function of COP/DET/FUS by inactivate photomorphogenesis-promoting transcription factors, another group of proteins called PIFs directly regulate gene expression to promote skotomorphogenesis (Leivar et al. 2008). PIFs, the basic helix-loop-helix (bHLH) transcription factors, include PIF1/PIL5 (PIF3-LIKE 5), PIF3, PIF4, PIF5/PIL6, and PIF6/PIL2, and they physically interact with phytochromes (Castillon et al. 2007). In the dark, PIFs activate skotomorphogenesis-promoted genes (Leivar et al. 2008), but they are inactivated by light resulting in repression of photomorphogenesis. This inactivation is due to the phosphorylation by photoactivated Pfr form of phytochromes. The phosphorylated PIFs are subsequently degraded by proteasome, and its stability is dependent on COP1 and SPA protein (Al-Sady et al. 2006; Shen et al. 2007). Briefly, the complicated signaling pathway downstream of light photoreceptors is mainly classified into two above branches, COP1-HY5 and PIFs pathways, and recently research revealed that these key components play an important role in light signaling cross talk (Fig. 14.2).
A simplified light signaling pathway in Arabidopsis. Under light, the photoreceptors suppress two main branches of light signaling, COP1-HY5 and PIFs. COP1, which is repressed by phytochromes and cryptochromes, is an ubiquitin E3 ligase, and it directly targets HY5 for degradation. HY5 is a bZIP transcription factor that promotes photomorphogenesis in all light conditions (Fig. 14.1). PIFs are bHLH transcription factors that are required for skotomorphogenesis. Phytochromes directly interact with PIFs, resulting in PIFs’ degradation, while COP1 positively affects PIF’s protein level. Note: between two proteins, solid lines indicate a direct effect, while dotted lines represent an indirect regulation
14.3.3 UV-B Signaling Pathway
Compared to the complicated signaling pathways in light responses, the pathways activated by UV-B radiation is much simpler to chart. With the recently identified UV-B receptor UVR8, this signaling pathway can be structurally described. Upon UV-B irradiation, UVR8 is activated by converting from dimers to monomers (Rizzini et al. 2011). The monomerized UVR8 further interacts with COP1 to promote its accumulation (Favory et al 2009). COP1 then stabilizes the central transcription factors HY5 and its homolog, HYH, resulting in the initiation of UV-B-responsive genes to promote UV-B photomorphogenesis (Ulm et al. 2004). However, not all of these genes are UVR8 dependent. To fine-tune this linear pathway, several components have been identified to form feedback regulations. STO/BBX24 (SALT TOLERANCE/B-BOX DOMAIN PROTEIN 24) interacts with both HY5 and COP1 to involve in a negative feedback loop by impinging on HY5 (Jiang et al. 2012). FHY3 (FAR-RED ELONGATED HYPOCOTYL 3) and HY5 work together to modulate the expression of COP1 as the members of a positive feedback loop (Huang et al. 2012). RUP1/2 (REPRESSOR UV-B PHOTOMORPHOGENESIS 1/2) belong to the WD40-repeat protein superfamily, interacting both with COP1 and UVR8 to act as negative feedback regulators (Gruber et al. 2010). It has been reported that RUP1/2 mediate the redimerization of UVR8, leading to the disruption of UVR8-COP1 interaction to attenuate the signaling, and this regulation is independent of COP1 (Heijde and Ulm 2013). For the detailed information on UV-B signaling, please refer to the reviews by Jenkins (2009), Heijde and Ulm (2012), Jiang et al. (2012) and Tilbrook et al. (2013) (Fig. 14.3).
UV-B signaling pathway in Arabidopsis. The UV-B radiation is specifically perceived by the UV-B photoreceptor UVR8, resulting in the rapid accumulation of COP1 and its interaction with UVR8. COP1-UVR8 interaction presumably stabilizes and activates HY5 for its transcriptional activity, leading to UV-B-regulated gene expression and photomorphogenesis (Fig. 14.3). This UV-B signaling cascade is feedback regulated by a few negative regulators, including BBX24 and RUP1/RUP2. While RUP1/RUP2 negatively regulates UV-B signaling by interacting with UVR8, the BBX24 protein fine-tunes the UV-B responses by impinging on HY5. FHY3 and HY5 have the ability to bind to COP1 promoter to control the expression of COP1
14.4 Signaling Cross Talk Between Light and UV-B
14.4.1 Introduction
Before the identification of UVR8, many attempts have been made in pursuit of the distinct UV-B signal transduction pathway. The light-induced CHS gene is found to be upregulated by all kinds of light and controlled by photoreceptors. For instance, UV-A and blue light-induced CHS expression is modulated by cry1, but cry1 is not required for UV-B-induced expression. In addition, UV-A and blue light each act synergistically with UV-B to stimulate CHS, and this synergism is retained in cry1-deficient mutant (Fuglevand et al. 1996). These results support the notion that there might be a considerable complexity in both photoreception and signal transduction in regulating CHS by UV-B and UV-A/blue light in Arabidopsis.
Stomatal opening is one of many physiological responses of plants regulated by blue light receptors, PHOT1 and PHOT2 (Kinoshita et al. 2001; Boccalandro et al. 2012), but it can also be modulated by UV-B (Eisinger et al. 2000). The blue light- and UV-B-induced stomatal opening is reversed by green light (Eisinger et al. 2003). However, phot1/phot2 double mutants show normal stomatal opening in response to UV-B, and green light fails to antagonize this process (Eisinger et al. 2003), indicating that phot1 and phot2 are required for the inhibition of UV-B effects by green light. The UV-B bending response of Arabidopsis hypocotyls during phototropism appears to be mediated by phototropins (Eisinger et al. 2003). The above evidence give a clear picture that UV-B is governed by a distinct photoreceptor and deeply cross-talked with other light photoreceptors. However, the above phenotypes are difficult to identify in large genetic screening. The phenotypes with shorter hypocotyl and higher pigment accumulation under low-fluence rate of UV-B are easier to screen by the eyes. Thus, all the regulators in UV-B signaling are identified by these two phenotypes. Moreover, hypocotyl growth inhibition and pigment accumulation are also featured in light photomorphogenesis. With the identification of UVR8, several main regulators in UV-B signaling have been revealed. Not surprisingly, most of them have other functions in other light signaling.
14.4.2 COP1: A Negative Factor in Light Signaling but a Positive Factor in UV-B Signaling
COP1 encodes a RING finger E3 ubiquitin ligase, which consists of three functional domains: a RING finger required for ligase activity, a coiled-coil domain for dimerization, and a WD40-repeat domain for target binding (Yi and Deng 2005). In the dark, COP1 function as an E3 ligase to target the light-responsive transcription factors (such as HY5) to repress the light photomorphogenesis. While, in the presence of light, COP1 is inactivated, resulting in the stabilization of HY5 to unleash the light responses (Osterlund et al. 2000). It is believed that early inactivation of COP1 by visible light occurs through direct interaction with phytochromes and cryptochromes (Wang et al. 2001; Seo et al. 2004), and the nuclear exclusion of COP1 is a rate-limiting step for photomorphogenic development (Subramanian et al. 2004). Moreover, the E3 ligase activity requires plant-specific SPA proteins, which is physically interacting with COP1 through the coiled-coil domain (Laubinger et al. 2004).
The cop1 mutants display light-grown phenotypes even in darkness, including short hypocotyls, open cotyledons, and elevated pigment levels. These constitutive light and UV-B photomorphogenic phenotypes make them very difficult to investigate under UV-B, but after prolonged UV-B treatment, cop1–4 mutant shows chlorosis, suggesting COP1 may plant a positive role in UV-B responses (Oravecz et al. 2006). Surprisingly, a large number of UV-B-responsive genes are inactivated in the cop1–4 mutant (Oravecz et al. 2006). In contrast to the relocation to cytoplasm in light, COP1 is stabilized and accumulates in the nucleus (Oravecz et al. 2006). This accumulation is caused by direct gene upregulation by UV-B (Huang et al. 2012). The rapid interaction of UVR8 and COP1 is believed to account for the stabilization (Favory et al. 2009), but the molecular mechanism is still unknown. COP1 is also crucial to UV-B-induced HY5 activation, and they both accumulate in the nucleus in response to UV-B (Oravecz et al. 2006). This indicates that COP1 may not have an E3 ligase activity in UV-B signaling, and the notion is further supported by the evidence that SPA is not required for the proper function of COP1 in response to UV-B (Oravecz et al. 2006). Thus this protein has dual opposite functions in response to two different spectra of light. One might ask what the function is under solar light, which contains both UV-B and visible light. Is COP1 located in the cytoplasm or in the nucleus? Is the amount of COP1 decreased or increased? These are questions to be elucidated in the field condition.
14.4.3 HY5 Has the Leading Role in Both Light and UV-B Signaling
Distinct light qualities, which are mediated through different photoreceptors, have similar effects on the transcriptomes during photomorphogenesis (Ma et al. 2001). Therefore, it is reasonable to suggest that one or more integration points exist for light signaling. A few light-induced transcription factors have been identified as key regulators during seedling morphogenesis. One of them is HY5, which is a target of COP1 for 26S proteasome-mediated degradation (Osterlund et al. 2000).
HY5 encodes a bZIP (basic leucine zipper) transcription factor, which could be the most important transcription factor that not only controls photomorphogenesis (Osterlund et al. 2000) but also governs other kinds of development, such as plant hormone responses (Lau and Deng 2010). Genome-enabled high-throughput analysis of immunoprecipitated chromatin (ChIP-chip or ChIP-seq) have revealed that HY5 binds directly to the promoter regions of light-responsive genes (Lee et al. 2007) and controls ~20 % of the light-regulated genes in Arabidopsis (Ma et al. 2002). Meanwhile, HY5 also guides the posttranscriptional regulation systems. For example, HY5 regulates eight miRNA (micro RNA) genes that in turn control the transcript abundance of specific target genes. Overexpressing HY5-targeted miR408 resulted in phenotypes that are opposite to the hy5 mutants (Zhang et al. 2011). Therefore, HY5 is likely to play a hierarchical role in photomorphogenesis. Recently developed large-scale analysis tools have enabled us to gain more insight into the mechanisms of how HY5 functions as a central transcription factor. More than 10,000 genes are found to be regulated by HY5 (Zhang et al. 2011), which is three times larger than previously reported (Lee et al. 2007), owing to the development of the resolution of ChIP-seq. As a transcription factor, HY5 has its own binding characters, which is aimed at 1 kb upstream of the transcription start site (TSS) and 5′ untranslated regions (UTRs) (Zhang et al. 2011). Three binding motifs, G-box, C-box, and hybrid CG-boxes, have been found to occur at a higher frequency in the identified HY5-binding regions (Zhang et al. 2011). Most surprisingly, histone modification is involved in HY5-mediated gene regulation. Comparing the pattern between wild-type and hy5 seedlings revealed that the level of permissive H3 histone modifications (acetylated H3K9 and tri-methylated H3K4) was increased, while that of the inhibitory histone H3 modification (di-methylated H3K9) was substantially decreased in HY5-bound regions for genes positively regulated by HY5 (Zhang et al. 2011). Another important feature is that HY5 can be a candidate in a feedback regulation loop, and we will talk about this later.
The hy5 mutant shows aberrant light-mediated phenotypes in Arabidopsis, including an elongated hypocotyl and reduced chlorophyll/anthocyanin accumulation (Oyama et al. 1997). But this kind of disturbance is slightly reduced by photomorphogenic UV-B radiation (Ulm et al. 2004), and the hypersensitive phenotype is further extended to UV-B stress (Brown et al. 2005). Genome-wide expression analysis reveals that a large number of UV-B-inducible genes are regulated by HY5, including the genes involved in UV protection (Ulm et al. 2004). Therefore, similar to the role in light signaling, HY5 is also a central transcription factor in UV-B-responsive pathway. However, the function of HY5 in UV-B signaling is under the control of UVR8, rather than other photoreceptors, because UV-B-induced upregulation of HY5 does not show any changes in various photoreceptor mutants (Ulm et al. 2004). UV-B-induced HY5-dependent genes have a large overlap with UVR8- and COP1-dependent genes, suggesting these three components work together to control the UV-B responses (Brown and Jenkins 2008; Oravecz et al. 2006). Interestingly, UVR8 itself is proposed to associate with chromatin in the vicinity of the HY5 genomic locus regardless of UV-B (Brown et al. 2005), but this is not the mechanism that UV-B-induced HY5 expression is tightly regulated by UVR8. In contrast to the COP1-mediated degradation of HY5 in darkness, COP1 is required for the HY5 expression under UV-B (Oravecz et al. 2006). In turn, HY5 is found to bind to the ACGT-containing elements (ACEs) within the COP1 promoter to upregulate COP1 (Huang et al. 2012). Thus, HY5 is involved in a positive feedback loop to fine-tune the UV-B responses.
14.4.4 BBX24/STO Negatively Modulates Light and UV-B Signaling
STO is a B-box-type zinc finger protein with sequence similarities to CONSTANS (Griffiths et al. 2003). It was identified through a screening from a series of yeast calcineurin mutants, where STO protein can rescue the yeast salt-sensitive phenotype caused by the deficient of the catalytic subunit genes (cna1cna2) or the regulatory subunit gene (cnb1) (Lippuner et al. 1996). However, Arabidopsis overexpressed with STO has not shown typical salt tolerance phenotypes, and STO gene itself cannot be regulated by salt treatment (Nagaoka and Takano 2003). Even more surprisingly, STO is found to have a negative role in light signaling (Indorf et al. 2007). STO also called BBX24 according to the nomenclature for the B-box transcription factors (Khanna et al. 2009).
Despite the ambiguous phenotypes in salt treatment and sto/bbx24 mutant and its overexpression, Arabidopsis have the obvious responses to red, far-red, and blue lights. Mutant has a shorter hypocotyl length under these light conditions, indicating STO/BBX24 is a negative regulator in light pathways (Indorf et al. 2007). The transcript is also controlled by lights and circadian clock, which is supported by the evidence that the regulation is through PHYA and PHYB, not PHOTs or CRYs (Indorf et al. 2007). STO/BBX24 interacts with COP1 in a yeast two-hybrid system (Holm et al. 2001), and both of them are co-localized in vivo in darkness (Indorf et al. 2007; Yan et al. 2011). It is assumed that COP1 is responsible for the degradation of STO/BBX24 in darkness and the proper function in light (Indorf et al. 2007; Yan et al. 2011). After accumulation of STO/BBX24 in light, it negatively modulates the light-responsive genes, such as CHS (Indorf et al. 2007).
A recent study has also extended the negative function of STO/BBX24 to the UV-B signaling, which is in agreement with the hypersensitive phenotypes under UV-B, such as short hypocotyls and enhanced anthocyanin accumulation (Jiang et al. 2012). Both as a negative factor in two kinds of signaling, is there any difference on the mechanism? STO/BBX24 can interact with COP1 and HY5 in vivo, and the interaction with COP1 is dependent on the presence of UV-B (Jiang et al. 2012). This interaction bears the most different mechanism involved in these two signaling, where COP1 functions as an E3 ligase to degrade STO/BBX24 in dark, and COP1 disarms the interaction to stabilize STO/BBX24 in light (Indorf et al. 2007; Yan et al. 2011), while COP1 is required for the accumulation of STO/BBX24 under UV-B (Jiang et al. 2012). This indicates STO/BBX24 acts downstream of UVR8, because UVR8-COP1 interaction is the prerequisite for the stabilization of COP1 under UV-B. However, no evidence shows that STO/BBX24 has a direct physical interaction with UVR8 (Jiang et al. 2012). As a central transcription factor in both light and UV-B signaling, HY5-STO/BBX24 interaction seems essential to the negative function of STO/BBX24. The UV-B-induced accumulation of HY5 is reduced by STO/BBX24, and even the transcriptional activity of HY5 is also repressed by STO/BBX24, resulting in the downregulation of most of the UV-B-responsive genes (Jiang et al. 2012). Interestingly, HY5-STO/BBX24 interaction seems reduced by UV-B (Jiang et al. 2012). It is not clear whether the negative function of STO/BBX24 in light signaling achieves through HY5, but one should keep in mind that STO/BBX24 is also a transcription factor. The targets of STO/BBX24 remain to be further elucidated.
14.4.5 FHY3: A Dual Function in Light and UV-B Signaling
As we mentioned before that phytochrome is activated to a Pfr form by red and far-red light, and this activation results in the translocation from cytoplasm and nucleus. Genetic studies have identified two pairs of homologous genes essential for PHYA signaling: FHY1 (FAR-RED ELONGATED HYPOCOTYL 1) and FHL (FHY1-LIKE), and FAR1 (FAR-RED-IMPAIRED RESPONSE 1) and FHY3 (FAR-RED ELONGATED HYPOCOTYL 3) (Hiltbrunner et al. 2005, 2006; Lin et al. 2007). FHY1 and FHL are essential for light-regulated PHYA nuclear accumulation and subsequent light responses (Hiltbrunner et al. 2005, 2006; Rosler et al. 2007). However, the expression of FHY1/FHL is activated by FAR1/FHY3, transposase-derived transcription factors, through directly binding to the promoter region of FHY1/FHL (Lin et al. 2007). Meanwhile, the expression of FAR/FHY3 is in turn negatively regulated by PHYA signaling (Lin et al. 2007). Thus, FAR/FHY3 act at a focal point of a feedback loop that maintains the homeostasis of PHYA signaling. However, this feedback regulation does not act alone, because HY5 is reported to bind to the FHY1/FHL promoters and the binding region is very near to the FAR/FHY3 binding region (Li et al. 2010). Further, it is found that HY5 interferes with FHY3 for binding to the FHY1 promoter (Li et al. 2010). Moreover, HY5 has a physical interaction with FAR/FHY3, but HY5 is a repressor for FHY1/FHL, while FAR/FHY3 is an activator (Li et al. 2010). These provide a complicated mechanism that HY5 works together with other cofactors to fine-tune the PHYA signaling.
Surprisingly, FHY3 shows a positive function in UV-B signaling (Huang et al. 2012). FHY3 is repressed by far-red light (Li et al. 2010) but is activated by UV-B and is believed to be regulated at a posttranscriptional level as the protein level is eventual downregulated under UV-B (Huang et al. 2012). FHY3 also has the ability to bind to COP1 promoter to further positively regulate UV-B signaling (Huang et al. 2012). But its homolog, FAR, which has a function in PHYA signaling, is not essential in photomorphogenic UV-B response, and FHY1, a target of FHY3 in PHYA signaling, shows no transcriptional response to UV-B (Huang et al. 2012). The different transcriptional behaviors of FHY3, FAR, and FHY1 indicate a fine-tuning of light signals that distinguish the PHYA signaling from UV-B pathway. UV-B can attenuate the interaction of HY5 with FHY3 (Huang et al. 2012), which is very similar to the HY5-STO/BBX24 interaction. As mentioned before, HY5 can also bind to COP1 promoter (Huang et al. 2012), suggesting HY5-FHY3 interaction modulates the UV-B-induced COP1 expression in concert. Therefore, the working model of HY5-FHY3 interaction is absolutely contrasting from PHYA signaling to UV-B signaling, where HY5 and FHY3 work antagonistically to bind to FHY1/FHL to regulate PHYA activities in far-red light, but they function synergistically to bind to COP1 to modulate UV-B responses.
The main regulators described above are all involved in both light and UV-B signaling, despite two negative factors, RUP1 and RUP2. Expression of both RUP1 and RUP2 is induced by UV-B in a UVR8-, COP1-, and HY5-dependent manner (Gruber et al. 2010). RUPs interact with UVR8 resulting in modulating UVR8 redimerization (Heijde and Ulm 2013). Therefore, RUPs play an important role in a negative feedback regulation. Interestingly, in another study that described RUP1 and RUP2 as EFO1 and EFO2 (EARLY FLOWERING BY OVEREXPRESSION 1 and 2), RUP1/EFO1 and RUP2/EFO2 expression is seen to be gated by the circadian clock, with expression levels peaking at daybreak and gradually subsiding to their lowest level at the onset of the night period (Wang et al. 2011). However, UV-B-induced RUPs are not likely to have the rhythmical expression type, but UV-B is indeed entertaining signal for circadian clock. The expression of select clock genes is UV-B responsive, indicating that UV-B entrains the plant clock via transcriptional activation. Moreover, UV-B induction of clock gene expression is gated by the clock. UVR8 and COP1 are essential to this biological process, and HY5 and HYH are dispensable (Feher et al. 2011). However, like RUPs, UV-B-dependent induction of HY5 expression does not follow a circadian rhythm (Feher et al. 2011). It can be deduced that temporal restriction of UV-B-specific responses by the clock may limit metabolic energy costs without compromising UV-B protection. However, the exact role of clock-regulated UV-B gene expression induction is still unknown.
14.5 Physical Interaction Between Photoreceptors
14.5.1 Introduction
With the identification of UVR8 as a UV-B receptor, the photomorphogenic UV-B signaling is getting better to be understood. But UVR8 is likely the “strangest” photoreceptor when compared with others (i.e., intrinsic tryptophan vs. bounded cofactor as a chromophore, dimer to monomer vs. monomer to dimer, or phosphorylation to activation). Meanwhile, UVR8 is not evidenced to physically interact with other photoreceptors, suggesting UVR8-mediated UV-B signaling is independent of other photoreceptors, though the critical regulators are mostly shared by others. In this section, we will focus on the physical interaction between PHYs, CRYs, and PHOTs.
14.5.2 Phytochrome-Phototropin Interaction
In higher plants, phytochrome is believed to enter the nucleus upon light activation and regulate transcription, whereas phototropins are considered to be member associated. These two kinds of photoreceptors govern almost different developmental processes, suggesting they are not associated together. However, some evidence is challenging this notion. Arabidopsis fhl/fhy1 mutants do not show light-dependent phyA nuclear translocation, but phyA-mediated responses still can be found, implying that a cytoplasmic signal also exists (Rosler et al. 2007). Phytochrome is surprisingly associated with the plasma membrane as derived from a fluorescence correlation microscopy study in moss Ceratodon, implying that phytochrome assembled with phycoerythrobilin is less mobile at the cell periphery than in the cytoplasm (Böse et al. 2004). After all, this challenging evidence is difficult to understand unless the finding of neochrome in ferns and algae. Neochrome is a chimeric molecular that is capable of phytochrome function in a phototropin (Nozue et al. 1998; Suetsugu et al. 2005). Further, a finding suggests that phytochrome is associated physically with phototropin at the plasma membrane in moss Physcomitrella patens (Jaedicke et al. 2012).
Phytochrome 4 (Pp.phy4) is the predominate photoreceptor responsible for vectorial light sensing in Physcomitrella (Mittmann et al. 2004). By using various kinds of protein interaction assay, such as yeast two hybrid, bimolecular fluorescence complementation, and co-immunoprecipitation, Pp.phy4 is confirmed to be physically associated with phototropins at the plasma membrane, and light has no effect on their association (Mittmann et al. 2004). Meanwhile, Arabidopsis phyA and phot1 also interact at the plasma membrane in onion epidermis cells, but only a small fraction of the phytochrome is associated with phototropin (Mittmann et al. 2004). This is in agreement with the evidence that a small proportion of phyA always retains in the cytoplasm irrespective of the light conditions (Hisada et al. 2000). However, Arabidopsis phyA-phot1 interaction cannot be detectable in a yeast two-hybrid assay (Hisada et al. 2000), probably because of lack of some important factors, such as PKS1. All these indicate that there might be a phytochrome cytoplasmic signaling, suggesting signals from plant photoreceptors do not always end up in transcriptional changes. And the downregulation of protochlorophyllide oxidoreductase A (PORA) by phytochrome is controlled via cytoplasmic Pfr (Paik et al. 2012).
14.5.3 Phytochrome-Cryptochrome Interaction
Pretreatment of plant tissue with red light (which specifically activates the phytochrome photoreceptor) can significantly enhance cryptochrome-dependent responses to blue light, whereas far-red light (which converts phytochrome to the inactive Pr form) reduces responsiveness to blue light (Mohr 1994). Genetic experiments with severely phytochrome-deficient single and double mutants demonstrate that a minimal level of active phytochrome is necessary for full cryptochrome activity; Arabidopsis phyA/phyB double mutant is impaired in cry1-mediated inhibition of hypocotyl elongation and anthocyanin accumulation (Casal and Boccalandro 1995; Ahmad and Cashmore 1997). It seems that phytochrome is required for the full activity of cryptochrome. As mentioned before, the photoactivated phytochrome has the kinase activity, and surprisingly cryptochromes are its target (Ahmad et al. 1998). In vitro kinase assay demonstrated that cry1 is phosphorylated in a phytochrome-dependent manner, but red or blue light does not affect this in vitro phosphorylation. However, in vivo assay exhibits that the phosphorylation of cry1 is enhanced by red light but compromised by supplemental with far-red light (Ahmad et al. 1998). Meanwhile, phytochrome is mainly targeting the C-terminal of cry1 (Ahmad et al. 1998). The phyA-cry1 interaction is further confirmed in the yeast two-hybrid assay, where both photoreceptors form a better conformational activity than in vitro purification (Ahmad et al. 1998).
Loss of cry1 activity in turn impairs the phytochrome responses. The time to flowering under short-day conditions is dramatically reduced in phyB mutant (Goto et al. 1991), lacking the phyB signal transduction pathway (Ahmad and Cashmore 1996). However, hy4 mutant, deficient in cry1 protein and showing reduced responsiveness to red light, flowered significantly earlier than the wild-type parent (Ahmad and Cashmore 1996). This result indicates a direct interaction of the mutant alleles of CRY1 with phyB resulting in impaired function.
14.6 Phytochrome and Cryptochrome Signal Integration
To date, there is no evidence showing cryptochromes could interact with phototropins or UVR8 could associate with other photoreceptors. Different light colors that selectively activate different photoreceptors activate a highly overlapping set of genes. Therefore, the signaling responded to different spectrum of light is sharing some components downstream from the photoreceptors. These include the negative regulator of the DET/COP/FUS class and the positive regulator HY5 (Quail 2002), which are already discussed above. In particular, COP1 is responsible for the degradation of phyA, cry2, and HY5 (Holm et al. 2001; Seo et al. 2004; Shalitin et al. 2002). In this section, we are going to discuss three signaling components which act downstream of both phyA and cryptochrome and are required for a subset of light responses.
OBP3 (OBF4-binding protein 3) belongs to a Dof (DNA binding with one finger) transcription factor (Kang and Singh 2000), and the function in light signaling is identified through an activation-tagging mutagenesis of phyB (Ward et al. 2005). A gain-of-function mutant, sob1-D (suppressor of phyB-4 dominant), suppresses the long hypocotyl phenotype of phyB, which is caused by the overexpression of OBP3 (Ward et al. 2005). OBP3-RNAi transgenic lines show reduced responsive to red light in terms of hypocotyl length, and this aberrant phenotype requires functional phyB, indicating OBP3 is a positive regulator of phyB-mediated inhibition of hypocotyl elongation (Ward et al. 2005). Furthermore, OBP3-RNAi lines are found to have larger cotyledons and reach the most dramatic size under blue light. The OBP3-mediated cotyledon expansion requires cry1 in blue light. These suggest that OBP3 is a negative regulator of cry1-mediated cotyledon expansion (Ward et al. 2005). Thus, OBP3 is a component in phyB and cry1 signaling pathway, acting as a positive and negative role, respectively. Ward et al. suggest that OBP3 might also work as a general inhibitor of tissue expansion with phyB and cry1. But these provide the solid evidence that OBP3 represents a connection between phyB and cry1 signal transduction.
HFR1 (long hypocotyl in far-red 1) is a bHLH transcription factor, which is a positive regulator of phyA-mediated light responses (Fairchild et al. 2000). phyA/hfr1 double mutant shows enhanced hypocotyl growth phenotype and each of the single parental mutant, indicating HFR1 has the function independent of phyA (Fairchild et al. 2000). When grown in blue light, however, hfr1 mutants exhibit reduced de-etiolation responses, including hypocotyl growth, cotyledon opening, and anthocyanin accumulation (Duek and Fankhauser 2003). Moreover, the functional cry1, not cry2, is required for the hyposensitive to blue light, but HFR1 is not important for the normal accumulation of cry1 (Duek and Fankhauser 2003). Genetic analysis further reveals that HFR1 is downstream of phyA and cry1 (Duek and Fankhauser 2003), providing a model that HFR1 is a positive integrator for both phyA and cry1 signaling.
SUB1 (short under blue light 1), encoding a Ca2+-binding protein, is first identified by exaggerative short hypocotyl under blue light but later is found to have the same response to far-red not to red light (Guo et al. 2001). sub1 is epistatic to cry2 in blue light, and phyA is epistatic to sub1 in both far-red and blue light, suggesting SUB1 functions both in phyA and cry2 signal transduction pathways (Guo et al. 2001). Moreover, blue- and far-red-induced CHS and CHI are significantly enhanced in sub1 mutant, and this enhancement is also observed in light-induced HY5 protein accumulation (Guo et al. 2001). Therefore, SUB1 defines a point of cross talk between cryptochrome and PHYA signaling.
At molecular level, it has been shown that the interaction between phytochromes and cryptochromes occurs at the level of photoreceptor. With the advanced of technique, especially the large scale of expression analysis, the presence of multiple levels of signal integration has been suggested. HY5 and COP1 represent two typical integrators, which master the signal transduction under all photoreceptors. More specifically, here we discussed three components integrating the phytochrome and cryptochrome signaling. OBP3 has a dual function, HFR1 is a positive regulator, and SUB1 negatively modulates both phytochrome and cryptochrome responses. However, there is evidence that phototropin signaling is cross-talked with phytochrome or cryptochrome. For instance, phototropism and chloroplast movement are primarily controlled by phototropin, but the amplitude of the response is modulated by both phytochrome and cryptochrome (DeBlasio et al. 2003; Ohgishi et al. 2004). To date, the underlying molecular mechanisms are still unknown.
References
Ahmad M, Cashmore AR (1996) The pef mutants of Arabidopsis thaliana define lesions early in the phytochrome signaling pathway. Plant J 10:1103–1110
Ahmad M, Cashmore AR (1997) The blue-light receptor cryptochrome 1 shows functional dependence on phytochrome A or phytochrome B in Arabidopsis thaliana. Plant J 11:421–427
Ahmad M, Jarillo JA, Smirnova O, Cashmore AR (1998) The CRY1 blue light photoreceptor of Arabidopsis interacts with phytochrome A in vitro. Mol Cell 1:939–948
Al-Sady B, Ni W, Kircher S, Schäfer E, Quail PH (2006) Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell 23:439–446
Boccalandro HE, Giordano CV, Ploschuk EL, Piccoli PN, Bottini R, Casal JJ (2012) Phototropins but not cryptochromes mediate the blue light-specific promotion of stomatal conductance, while both enhance photosynthesis and transpiration under full sunlight. Plant Physiol 158:1475–1484
Böse G, Schwille P, Lamparter T (2004) The mobility of phytochrome within protonemal tip cells of the moss Ceratodon purpureus, monitored by fluorescence correlation spectroscopy. Biophys J 87:2013–2021
Brown BA, Jenkins GI (2008) UV-B signaling pathways with different fluence-rate response profiles are distinguished in mature Arabidopsis leaf tissue by requirement for UVR8, HY5, and HYH. Plant Physiol 146:576–588
Brown BA, Cloix C, Jiang GH, Kaiserli E, Herzyk P, Kliebenstein DJ, Jenkins GI (2005) A UV-B-specific signaling component orchestrates plant UV protection. Proc Natl Acad Sci U S A 102:18225–18230
Casal JJ, Boccalandro H (1995) Co-action between phytochrome B and HY4 in Arabidopsis thaliana. Planta 197:213–218
Castillon A, Shen H, Huq E (2007) Phytochrome interacting factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci 12:514–521
Christie JM (2007) Phototropin blue-light receptors. Annu Rev Plant Biol 58:21–45
DeBlasio SL, Mullen JL, Luesse DR, Hangarter RP (2003) Phytochrome modulation of blue light-induced chloroplast movements in Arabidopsis. Plant Physiol 133:1471–1479
Duek PD, Fankhauser C (2003) HFR1, a putative bHLH transcription factor, mediates both phytochrome A and cryptochrome signaling. Plant J 34:827–836
Eisinger W, Swartz T, Bogonolni R, Taiz L (2000) The ultraviolet action spectrum for stomatal opening in broad bean. Plant Physiol 122:99–105
Eisinger WR, Bogomolni RA, Taiz L (2003) Interactions between a blue-green reversible photoreceptor and a separate UV-B receptor in stomatal guard cells. Am J Bot. 90:1560–1566
Fairchild CD, Schumaker MA, Quail PH (2000) HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev 14:2377–2391
Fankhauser C, Yeh K-C, Lagarias JC, Zhang H, Elich TD, Chory J (1999) PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 284:1539–1541
Favory JJ, Stec A, Gruber H, Rizzini L, Oravecz A, Funk M (2009) Interaction of COP1 and UVR8 regulates UV-B induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J 28:591–601
Feher B, Kozma-Bognar L, Kevei E, Hajdu A, Binkert M, Davis SJ, Schäfer E, Ulm R, Nagy F (2011) Functional interaction of the circadian clock and UV RESISTANCE LOCUS 8-controlled UV-B signaling pathways in Arabidopsis thaliana. Plant J 67:37–48
Fuglevand G, Jackson JA, Jenkins GI (1996) UV-B, UV-A, and blue light signal transduction pathways interact synergistically to regulate chalcone synthase gene expression in Arabidopsis. Plant Cell 8:2347–2357
Goto N, Kumagai T, Koornneef M (1991) Flowering responses to light-breaks in photomorphogenic mutants of Arabidopsis thaliana. Physiol Plant 83:209–215
Griffiths S, Dunford RP, Coupland G, Laurie DA (2003) The evolution of CONSTANS-like gene families in barley, rice, and Arabidopsis. Plant Physiol 131:1855–1867
Gruber H, Heijde M, Heller W, Albert A, Seidlitz HK, Ulm R (2010) Negative feedback regulation of UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. Proc Natl Acad Sci U S A 107:20132–20137
Guo HW, Mockler T, Duong H, Lin C (2001) SUB1, an Arabidopsis Ca2+-binding protein involved in cryptochrome and phytochrome coaction. Science 291:487–490
Heijde M, Ulm R (2012) UV-B photoreceptor-mediated signaling in plants. Trends Plant Sci 17:230–237
Heijde M, Ulm R (2013) Reversion of the Arabidopsis UV-B photoreceptor UVR8 to the homodimeric ground state. Proc Natl Acad Sci U S A 110:1113–1118
Hiltbrunner A, Tscheuschler A, Viczian A, Kunkel T, Kircher S, Toth R, Honsberger A, Nagy F, Fankhauser C, Schäfer E (2005) Nuclear accumulation of the phytochrome A photoreceptor requires FHY1. Curr Biol 15:2125–2130
Hiltbrunner A, Tscheuschler A, Viczian A, Kunkel T, Kircher S, Schäfer E (2006) FHY1 and FHL act together to mediate nuclear accumulation of the phytochrome A photoreceptor. Plant Cell Physiol 47:1023–1034
Hisada A, Hanzawa H, Weller JL, Nagatani A, Reid JB, Furuya M (2000) Light-induced nuclear translocation of endogenous pea phytochrome A visualized by immunocytochemical procedures. Plant Cell 12(7):1063–1078
Holm M, Hardtke CS, Gaudet R, Deng XW (2001) Identification of a structural motif that confers specific interaction with the WD40 repeat domain of Arabidopsis COP1. EMBO J 20:118–127
Huang X, Ouyang X, Yang P, Lau OS, Li G, Li J, Chen H, Deng XW (2012) Arabidopsis FHY3 and HY5 positively mediate induction of COP1 transcription in response to photomorphogenic UV-B light. Plant Cell 24:4590–4606
Indorf M, Cordero J, Neuhaus G, Rodriuez-Franco M (2007) Salt tolerance (STO), a stress-related protein, has a major role in light signalling. Plant J 51:563–574
Jaedicke K, Lichtenthaler AL, Meyberg R, Zeidler M, Hughes J (2012) A phytochrome-phototropin light signaling complex at the plasma membrane. Proc Natl Acad Sci U S A 109:12231–12236
Jenkins G (2009) Signal transduction in response to UV-B radiation. Annu Rev Plant Biol 60:407–431
Jiang L, Wang Y, Björn LO, He JX, Li SS (2012) Sensing of UV-B radiation by plants. Plant Sig Behav 7:1–5
Jiao Y, Lau OS, Deng XW (2007) Light-regulated transcriptional networks in higher plants. Nat Rev Genet 8:217–230
Kang HG, Singh KB (2000) Characterization of salicylic acid-responsive, Arabidopsis Dof domain proteins: Overexpression of OBP3 leads to growth defects. Plant J 21:329–339
Khanna R, Kronmiller B, Maszle DR, Coupland G, Holm M, Mizuno T, Wu SH (2009) The Arabidopsis B-Box Zinc Finger Family. Plant Cell 21:3416–3420
Kim JI, Shen Y, Han YJ, Park JE, Kirchenbauer D, Soh MS, Nagy F, Schafer E, Song PS (2004) Phytochrome phosphorylation modulates light signaling by influencing the protein-protein interaction. Plant Cell 16:2629–2640.
Kinoshita T, Doi M, Suetsugu N, Kagawa T, Wada M, Shimazaki K (2001) Phot1 and phot2 mediate blue light regulation of stomatal opening. Nature 414:656–660
Lau OS, Deng XW (2010) Plant hormone signaling lightens up: integrators of light and hormones. Curr Opin Plant Biol 13:571–577
Laubinger S, Fittinghoff K, Hoecker U (2004) The SPA quartet: a family of WD-repeat proteins with a central role in suppression of photomorphogenesis in Arabidopsis. Plant Cell 16:2293–2306
Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19:731–749
Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH (2008) Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol 18:1815–1823
Li J, Li G, Gao S, Martinez C, He G, Zhou Z, Huang X, Lee J-H, Zhang H, Shen Y, Wang H, Deng XW (2010) Arabidopsis transcription factor ELONGATED HYPOCOTYL5 plays a role in feedback regulation of phytochrome A signaling. Plant Cell 22:3634–3649
Lin C, Shalitin D (2003) Cryptochrome structure and signal transduction. Annu Rev Plant Biol. 54:469–496
Lin R, Ding L, Casola C, Ripoll DR, Feschotte C, Wang H (2007) Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318:1302–1305
Lippuner V, Cyert MS, Gasser CS (1996) Two classes of plant cDNA clones differentially complement yeast calcineurin mutants and increase salt tolerance of wild-type yeast. J Biol Chem 271:12859–12866
Ma L, Li J, Qu L, Hager J, Chen Z, Zhao H, Deng XW (2001) Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13:2589–2607
Ma L, Gao Y, Qu L, Chen Z, Li J, Zhao H, Deng XW (2002) Genomic evidence for COP1 as a repressor of light-regulated gene expression and development in Arabidopsis. Plant Cell 14:2383–2398
Mittmann F, Brückern G, Zeidler M, Repp A, Abts T, Hartmann E, Hughes J (2004) Targeted knockout in Physcomitrella reveals direct actions of phytochrome in the cytoplasm. Proc Natl Acad Sci U S A 101:13939–13944
Mohr H (1994) Coaction between pigment systems. In: Kendrick RE, Kronenberg GHM (eds) Photomorphogenesis in plants, 2nd edn. Kluwer Academic Publishers, Dordrecht, pp 353–372
Nagaoka S, Takano T (2003) Salt tolerance-related protein STO binds to a Myb transcription factor homologue and confers salt tolerance in Arabidopsis. J Exp Bot 54:2231–2237
Nozue K, Kanegae T, Imaizumi T, Fukuda S, Okamoto H, Yeh KC, Lagarias JC, Wada M (1998) A phytochrome from the fern Adiantum with features of the putative photoreceptor NPH1. Proc Natl Acad Sci U S A 95:15826–15830
Ohgishi M, Saji K, Okada K, Sakai T (2004) Functional analysis of each blue light receptor, cry1, cry2, phot1, and phot2, by using combinatorial multiple mutants in Arabidopsis. Proc Natl Acad Sci U S A 101:2223–2228
Oravecz A, Baumann A, Máté Z, Brzezinska A, Molinier J, Oakeley EJ, Adam E, Schäfer E, Nagy F, Ulm R (2006) CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell 18:1975–1990
Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Target destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405:462–466
Oyama T, Shimura Y, Okada K (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev 11:2983–2995
Paik I, Yang S, Choi G (2012) Phytochrome regulates translation of mRNA in the cytosol. Proc Natl Acad Sci U S A 109:1335–1340
Partch CL, Sancar A (2005) Photochemistry and photobiology of cryptochrome blue-light photopigments: the search for a photocycle. Photochem Photobiol. 81:1291–1304
Quail PH (2002) Phytochrome photosensory signalling networks. Nat Rev Mol Cell Biol 3:85–93
Rizzini L, Favory JJ, Cloix C, Faggionato D, O’Hara A, Kaiserli E, Baumeister R, Schäfer E, Nagy F, Jenkins GI, Ulm R (2011) Perception of UV-B by the Arabidopsis UVR8 protein. Science 332:103–106
Rosler J, Klein I, Zeidler M (2007) Arabidopsis fhl/fhy1 double mutant reveals a distinct cytoplasmic action of phytochrome A. Proc Natl Acad Sci U S A 104:10737–10742
Ryu JS, Kim JI, Kunkel T, Kim BC, Cho DS, Hong SH, Kim SH, Fernández AP, Kim Y, Alonso JM, Ecker JR, Nagy F, Lim PO, Song PS, Schäfer E, Nam HG (2005) Phytochrome-specific type 5 phosphatase controls light signal flux by enhancing phytochrome stability and affinity for a signal transducer. Cell 120:395–406
Saijo Y, Sullivan JA, Wang H, Yang J, Shen Y, Rubio V, Ma L, Hoecker U, Deng XW (2003) The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev 17:2642–2647
Sang Y, Li QH, Rubio V, Zhang YC, Mao J, Deng XW, Yang HQ (2005) N-terminal domain-mediated homodimerization is required for photoreceptor activity of Arabidopsis CRYPTOCHROME 1. Plant Cell 17:1569–1584
Seo HS, Watanabe E, Tokutomi S, Nagatani A, Chua NH (2004) Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev 18:617–622
Shalitin D, Yang H, Mockler TC, Maymon M, Guo H, Whitelam GC, Lin C (2002) Regulation of Arabidopsis cryptochrome 2 by blue- light-dependent phosphorylation. Nature 417:763–767
Shen Y, Khanna R, Carle CM, Quail PH (2007) Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol 145:1043–1051
Subramanian C, Kin BH, Lyssenko NN, Xu X, Johnson CH, von Arnim AG (2004) The Arabidopsis repressor of light signaling, COP1, is regulated by nuclear exclusion: Mutational analysis by bioluminescence resonance energy transfer. Proc Natl Acad Sci U S A 101:6798–6802
Suetsugu N, Mittmann F, Wagner G, Hughes J, Wada M (2005) A chimeric photoreceptor gene, NEOCHROME, has arisen twice during plant evolution. Proc Natl Acad Sci U S A 102:13705–13709
Sullivan JA, Shirasu K, Deng XW (2003) The diverse roles of ubiquitin and the 26S proteasome in the life of plants. Nat Rev Genet 4:948–958
Tilbrook K, Arongaus AB, Binkert M, Heijde M, Yin R, Ulm R (2013) The UVR8 UV-B photoreceptor, perception, signaling and response. Arabidopsis Book 11:e0164
Ulm R, Baumann A, Oravecz A, Mate Z, Adam E, Oakeley EJ, Schäfer E, Nagy F (2004) Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc Natl Acad Sci U S A 101:1397–1402
Vierstra RD, Zhang J (2011) Phytochrome signaling: solving the Gordian knot with microbial relatives. Trends Plant Sci 16:417–426
Wang H, Ma LG, Li JM, Zhao HY, Deng XW (2001) Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294:154–158
Wang W, Yang D, Feldmann KA (2011) EFO1 and EFO2, encoding putative WD-domain proteins, have overlapping and distinct roles in the regulation of vegetative development and flowering of Arabidopsis. J Exp Bot 62:1077–1088
Ward JM, Cufr CA, Denzel MA, Neff MM (2005) The Dof transcription factor OBP3 modulates phytochrome and cryptochrome signaling in Arabidopsis. Plant Cell 17:475–485
Yan H, Marquardt K, Indorf M, Jutt D, Kircher S, Neuhaus G, Rodriguez-Franco M (2011) Nuclear localization and interaction with COP1 are required for STO/BBX24 function during photomorphogenesis. Plant Physiol 156:1772–1782
Yi C, Deng XW (2005) COP1 – From plant photomorphogenesis to mammalian tumorigenesis. Trends Plant Sci 15:618–625
Yu X, Liu H, Klejnot J, Lin C (2010) The cryptochrome blue light receptors. Arabidopsis Book 8:e0135
Zhang H, He H, Wang X, Wang X, Yang X, Li L, Deng XW (2011) Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J 65:346–358
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer Science+Business Media New York
About this chapter
Cite this chapter
Jiang, L., Li, S. (2015). Signaling Cross Talk Under the Control of Plant Photoreceptors. In: Björn, L. (eds) Photobiology. Springer, New York, NY. https://doi.org/10.1007/978-1-4939-1468-5_14
Download citation
DOI: https://doi.org/10.1007/978-1-4939-1468-5_14
Published:
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-4939-1467-8
Online ISBN: 978-1-4939-1468-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)