Abstract
In land plants, at least five groups of photoreceptors perceive information about light conditions and diurnal rhythm, as well as about ambient temperature, presence of pathogens or competing neighbors, direction of the gravity vector and other factors. The photoreceptor toolkit allows plants to integrate environmental information and “make decisions” necessary for survival and successful reproduction, i.e., whether to enter or exit dormancy, accelerate or stop growth, promote or delay flowering; choose the direction of growth, induce or suppress the formation of side shoots, as well as regulate the synthesis of volatile substances affecting the growth of neighbors or pathogens. These photoreceptors include phytochromes, cryptochromes, phototropins, the ZTL (ZEITLUPE) family proteins, and the ultraviolet-B receptor UVR8. In spite of the diversity of plants photoreceptors, their functionality follows several “common rules.” Transformation of the information on light quality and quantity into metabolic and morphogenetic responses occurs via controlled degradation of transcription factors mediated by interactions of the active form of a photoreceptor and the СОР1-SPA1 E3-ubiquitine ligase complex in the nucleus. Apart from interacting with СОР1, the active forms of photoreceptors in the nucleus can directly bind to transcription factors and trigger their degradation. Phytochromes belong to the largest (the molecular mass of a monomer is ca. 125 kD) and most sophisticated plant photoreceptors. In addition to the abovementioned mechanisms, they also regulate alternative splicing and the selection of alternative promoters for thousands of plant genes. The interaction of phytochrome with jasmonate signaling is of special interest, as phytochromes regulate the jasmonate-mediated cessation of growth in response to stress. This review focuses on data revealing the potential for the application of novel information on plant photoreceptors for the generation of crop varieties capable of high performance under stress conditions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
A thorough understanding of the mechanisms regulating photosynthesis and sink-source relations in plants is of primary importance for research aimed to increase crop productivity [1]. Adolf T. Mokronosov, famous for his research of crop photosynthesis and the development of sink–source relations during plant ontogenesis, pointed at the importance of photoperiodic regulation both for photosynthetic performance and the establishment of the architecture of plant body ultimately influencing the distribution of photoassimilates between plant organs [1]. In plants, at least five groups of photoreceptors perceive information on light conditions and photoperiod, as well as on ambient temperature, on the presence of pathogens or competing neighbors, gravitational forces and other factors [2–4]. These are: phytochromes perceiving red- (R) and far-red- (FR) light; cryptochromes, phototropines and the proteins of the ZEITLUPE family perceiving UV-A radiation, blue (B) and green (G) light; and the receptor for ultraviolet B-radiation (UV-B), the UVR8 protein [5–11]. It is likely that plants contain further, yet undiscovered, photoreceptors, including a type specific for G [12]. Altogether, a complex combination of photoreceptors enables plants to integrate environmental information and ‘make decisions’ necessary for survival and successful reproduction: enter or exit a dormant state, accelerate or stop growth, promote or retard the transition from vegetative growth to flowering, determine the optimal direction for growth, start the formation of tillers, and regulate the synthesis of volatiles able to influence the propagation of pathogens or the growth of competing neighboring plants, and the transition to programmed cell death [13–18]. Such a wide spectrum of physiological responses is enabled by interactions of photoreceptor-based signaling pathways with phytohormone systems and with signaling pathways based on the production of reactive oxygen species (ROS) in the photosynthetic apparatus and in mitochondria [19–21]. Apart from this, recent studies have shown that photoreceptors can lead to switches in genetic programs via the regulation of alternative splicing as well as through selection of alternative promoters for thousands of plant genes [22, 23]. Here, a short summary of current knowledge on plant photoreceptors will be presented, with a focus on the interactions of phytochrome-based signaling with jasmonate signaling with the potential to be used for the breeding of crop varieties able to ensure high yield under stress conditions.
CLASSES OF PLANT PHOTORECEPTORS
Thus far, at least 14 photoreceptors have been identified in the model plant Arabidopsis thaliana. These include five phytopchromes (PhyA-E), three cryptochromes (СRY1-3), two phototropins (PHOT1, PHOT2), three proteins of the ZEITLUPE family (ZTL, FKF1, LKP2) and a UV-B receptor, UVR8. These classes of photoreceptors are present in bacteria, fungi, algae and land plants but not in all subgroups of these organisms [24].
The Red-Light Receptors: Phytochromes
Phytochromes are the key regulators of plants growth and development. With monomers of 125 kD, they represent the largest and most sophisticated plant photoreceptors. The N-terminal part of phytochrome performs light sensing. It consists of three conserved domains, PAS, GAF and PHY; the GAF domain binds the chromophore, phytochromobiline, via a thioester bond with a cysteine residue [25]. The С-terminal part forms a serine-threonine kinase which can perform autophosphorylation required to form a dimer, the active form of a phytochrome. Phytochromes are synthesized in the cytoplasm in the dark phase as inactive proteins. Absorption of a light quantum by phytochromobiline leads to minor conformational shifts (in the order of less than one angstrom) of parts of the chromophore molecule which, due to the particular structure of the photoreceptor protein, lead to major conformational changes in the phytochrome, its activation and mobilization to the nucleus and ultimately to a switch in the activity of genetic programs in the cell [25]. Upon absorption of a light quantum, the С15=С16 double bond within the phytochromobiline molecule undergoes isomerization leading to the rotation of the D-ring plane by 180° [25]; an alternative mechanism is the rotation of the A-ring plane by 90° around the double bond С4=С5 [26]. Phytochromes form unique three-dimentional structures: the ‘knot’ (formed by a ‘looping’ of polypeptide chain around itself) necessary for stabilization of hydrophobic interactions within the protein, and the ‘hairpin’ formed by antiparalleled beta-sheets. Upon illumination with R, the liberation and rotation of the ‘hairpin’ lead to the formation of a single alpha-helix in the place of two beta-sheets within the PHY domain; upon illumination with FR, the phytochrome structure returns to the initial inactive state [25]. The changes of the conformation of the phytochrome also lead to the activation of its kinase activity.
The absorption maximum of the chromophore in inactive phytochromes lies in the red part of the light spectrum (660 nm). After illumination with R, the activated phytochrome acquires the second absorption maximum of the chromophore in FR region (730 nm); these two forms of phytochromes are designated as Pr and Pfr, respectively. Illumination with FR promotes the transformation of the active Pfr to the inactive Pr. Due to a gradual thermal relaxation of phytochromobiline, the reversion of Pfr to Pr will also occur in the dark. The absorption of both Pfr and Pr is negligible in the region of 450–550 nm, and low in the UV-A region [9]. However, in the cells containing very high amounts of phytochrome, the absorption by the inactive Pr form of even very low amounts of light from regions of the solar spectrum other than R, including FR, becomes physiologically significant [27]. This situation takes place in etiolated Arabidopsis seedlings accumulating very high amounts of PhyA, leading to a ratio of PhyA, PhyB, PhyC, PhyD and PhyE of 85 : 10 : 2 : 1.5 : 1.5 [28]. In this case, even very low intensities of FR or any other light will, with high probability, enable the transition of at least several PhyA molecules into their active form, and these molecules will suffice for the induction of the photomorphogenetic program. This explains the special role PhyA plays in de-etiolation, and also means that in etiolated seedlings, photomorphogenesis, once activated, cannot be ‘switched off’ by irradiation with FR [27]. The photomorphogenetic program in seedlings activated by PhyA includes the cessation of hypocotyl growth in dicots (and of mesocotyl growth in monocots, respectively), the growth of coleoptile in monocots, unfolding of cotyledons, greening of seedlings and the completion of the development of the photosynthetic apparatus in plastids. The opposite program, scotomorphogenesis, includes the activation of hypocotyl growth in dicots and of mesocotyl growth in monocots, the inhibition of the expression of many genes encoding components of the photosynthetic apparatus, the enzymes of chlorophyll biosynthesis etc. [27].
PhyA is a photolabile phytochrome which in high light undergoes photodestruction. PhyA can be activated by less than 1 μmol/m2 light quanta of any wavelength within the 300–780 nm region. Upon activation, it mediates the complex of reactions designated as ‘very low fluence response’ (VLFR). However, PhyA can be activated also by high light intensities (1000 µmol/m2 and more quanta) in response to an increase in FR, or in response to a decrease of light quantity [29]. This type of responses is summarized as ‘high irradiance response’ (HIR). Owing to the HIR, seedlings can complete deetiolation even under a dense canopy [27, 30].
In the dark, PhyA is located in the cytosol, as the apoprotein lacks a nuclear localization signal. Here, PhyA can regulate gravitropism [31]; the cytoplasmic phytochromes might directly interact with plasma membrane proteins [32]. Many publications report phytochrome-mediated plant responses occurring within 10 min after illumination with R or FR, i.e. not enough time for changes in gene expression patterns [33]. An example of such a response is the migration of the chloroplasts within the cells of the alga Mougeotia sp. initiated by the illumination of cells with polarized R [34]. In vascular plants, an example would be the rapid (within a few seconds) depolarization of plasma membrane in barley root tips in response to R, abolished by FR (the so-called Tanada effect [35]), or the folding of leaves of some Fabaceae at the onset of the dark phase, etc. (reviewed in [33]). These reactions might be mediated by interactions of cytosolic phytochromes with other photoreceptors, namely with plasma membrane-anchored phototropines (see below). Such interactions were demonstrated in cells of Physcomitrella patens [36] but not yet in vascular plants; they might be mediated by the cytosolic PKS (phytochrome kinase substrate) proteins [37]. At last, phytochromes in the cytosol can regulate the translation of mRNAs as was demonstrated for transcripts encoding protochlorophyllide oxidoreductase A [38].
Upon perception of light of any wavelength, PhyA rapidly translocates into the nucleus. This translocation is mediated by FHY and FHL proteins which contain a nuclear localization signal [27]. They bind the phosphorylated form of PhyA, which then undergoes dephosphorylation in the nucleus and uncouples from FHY and FHL. In the nucleus, PhyA directly interacts with the COP1-SPA1 (Constitutive Photomorphogenic 1/Suppressor of PhyA 1) Е3-ubiquitin ligase complex, and with the transcriptional regulators PIF1 and PIF3 (see below).
In contrast to PhyA, PhyB and all other phytochromes in Arabidopsis are stable in the light. In seedlings grown at constant illumination, PhyB becomes the dominating phytochrome, as indicated by the ratio of PhyA, PhyB, PhyC, PhyD and PhyE (5 : 40 : 15 : 15 : 25, as shown by immunoblotting [28]). However, under these conditions, the levels of PhyB are still much lower than PhyA levels in etiolated seedlings; therefore, neither very low R nor high FR nor any other wavelength can activate a physiologically significant number of PhyB molecules. Altogether, only R of an intensity in the range of 1 to 1000 µmol/m2 can lead to a specific activation of the PhyB and thereby of the complex of PhyB-regulated reactions called ‘low fluence response’ (LFR). This explains the inhibition of the PhyB-activated photomorphogenetic programs by FR [27]. As PhyB contains a nuclear localization signal, its default localization is in the nucleus; there, it interacts with СОР1-SPA1 and with transcription factors such as PIF4, PIF5, PIF7, and also performs other functions (see below). In the light, PhyB triggers photomorphogenesis which includes the completion of the development of the photosynthetic apparatus, intensification of the width growth of leaf laminae, compactification of rosettes and sprouts, and the onset of tillering [13]. In mature plants, PhyB also inhibits the opposite program, scotomorphogenesis, also called a shade avoidance syndrome (SAS). In mature plants, inactivation of PhyB by a decrease in the R : FR ratio in the incident light leads to the elongation of petioles and internodes as well as to leaf hyponasty and pale green coloration of leaves due to incomplete development of the photosynthetic apparatus [13]. Under high light conditions, it is the dysfunction of PhyB, not of PhyA. that results in a pale-green coloration of the leaves [30, 39]. In a rice mutant lacking PhyB, expression of the genes encoding the Mg-chelatase subunit ChlH and an activator of this enzyme, GUN4, was inhibited, leading to the establishment of a pale-green phenotype and the onset of SAS under direct light [40].
PhyB triggers formation of stomata [41]. The demonstration that the PhyB-regulated signals mediating the differentiation of stomata in leaf epidermis move cell-to-cell, i.e., act non-cell-autonomously, was the first evidence for the systemic action of PhyB [42]. In leaves, the PhyB gene is expressed in the guard cell precursors, in guard cells and in the epidermis. The number of stomata per leaf area unit was lowered in the Arabidopsis mutant lacking PhyB but restored upon complementation with the phyB cDNA, notably, not only under control of a guard cell-specific promoter, but also of any promoter active in cells other than stomata, including the phloem-specific promoter of the sucrose transporter AtSUC2 [42]. Additionally, the non-cell autonomous action of PhyB was confirmed by complementation of negative gravitropism in hypocotyls of the phyb knockout mutant by a PhyB construct expressed in the epidermis of hypocotyls [43].
In summary, PhyA represents a key ‘switch’ between scoto- and photomorphogenetic programs in etiolated seedlings as well as in mature plants grown in the shade, while PhyB in responsible for this function in green seedlings and in mature plants grown under high light [27]. This applies to both dicots and monocots. There are, however, some differences in the organization of the phytochrome system between dicots and monocots. In contrast with Arabidopsis, rice, wheat and barley contain only three phytochromes PhyA, PhyB and PhyC. While PhyB represses flowering in dicots, in all cereals studied it promotes flowering in combination with PhyC [44, 45]. Thus far, no other functional differences between dicot and monocot phytochromes have been revealed.
Many phya and phyb mutant alleles have been characterized where specific phytochrome functions are ‘switched off’ owing to mutations leading to amino acid exchange in different domains of the apoprotein; for instance, PhyA-302 plants lack the HIR response but retain the VLFR. Very recently, plant phytochromes were shown to function as thermoreceptors [2]. A mutant form of PhyB where Tyr 276 is exchanged with His is constitutively active and does not revert to the inactive form in the dark; experiments with the corresponding plants showed that the function of phytochromes as thermosensors is related to the rate of Pfr to Pr reversion in the dark [2]. The higher the temperature during the night, the faster is the reversion of wild-type PhyB into the inactive form. Apart from this, the higher the temperature, the weaker is the binding of PhyB to the promoters of PhyB-regulated genes. This mechanism differs from the mechanism of temperature sensing in cyanobacteria: there, membrane integral two-component sensor histidine kinases Hik33, similar to phytochromes, function as sensors of both light and temperature conditions, but, in contrast with phytochromes, perceive the temperature-induced changes in membrane fluidity [46] while temperature sensing of phytochromes has been attributed to a temperature-responsive half-life of Pfr [2]. In Arabidopsis, five phytochromes regulate different plant functions in response to temperature: in warm conditions, seed germination is controlled mainly by PhyB, but in cold conditions by PhyE; at the same time, after a prolonged cold period seeds need PhyA for germination [47]. Inhibition of flowering is regulated by PhyE in cold conditions and by PhyB in warm conditions [47].
Phytochromes are major regulators of transcriptional activity of the plant genome. Both PhyA and PhyB can directly bind to the promoters of the regulated genes [2, 48]. In seedlings transferred from darkness to light, phytochromes regulate the expression of several thousands of genes and alternative splicing of transcripts of over one thousand of genes [22]. Another mechanism is the light-dependent selection of promoters [23]. The occurrence of genes with alternative promoters opens the possibility to produce mRNAs differing in 5'-UTR, which can influence the stability of the mRNA, as well as the amino acid sequence at the N-terminus of the encoded protein, depending on the selection of transcriptional start site. Peptide signals targeting proteins to mitochondria or chloroplasts usually reside at the N-terminus of the protein. PhyB has been shown to mediate promoter selection for transcription of more than two thousands of genes, probably by direct binding to their promoters; this can lead to the synthesis of proteins locating either to the cytosol or to organelles, depending on light conditions [23]. For instance, in A. thaliana growing in the shade, both the chloroplastic as well as a novel cytosolic isoform of the photorespiration enzyme, glycerate kinase, were detected; both isoforms are products of one and the same gene but, depending on PhyB-mediated promoter selection, the N-terminal chloroplast signal peptide is either present or absent. The cytosolic isoform functions in a ‘photorespiratory shunt’ important for the protection from photoinhibition during the transfer of plants from the shade to high light [23].
While the mechanisms of synthesis and degradation of the phytochrome proteins, and the related signaling function of phytochromes, are objects of intensive study, much less is known about the regulation of the activity of the phytochrome-encoding genes. The regulation of PhyA transcription can involve the (de)methylation and (de)acetylation of histones, likely under participation of PhyB [10]. According to other data, in the light PhyB mediates the methylation of the promoter of PhyA and inhibits its expression [27]. In Arabidopsis leaves, the expression of PhyA was inhibited by light, probably via PhyA itself, while the expression levels of PhyB did not change in response to light; in roots, expression of genes encoding both PhyA and PhyB, but in particular PhyB, was enhanced when roots were exposed to light [49]. In the dark, the expression levels of both PhyA and PhyB increased in Arabidopsis leaves, however, these data were obtained using plants expressing promoter-GUS, constructs which precluded the analysis of possible regulation of the stability of phytochrome genes transcripts in vivo based on 3'-UTR sequences [49]. A similar method was used to study PhyA expression patterns in rice [50], where PhyA was found to be active in all tissues in etiolated seedlings but the expression became confined to leaf vascular bundles upon illumination with R but not FR; this restriction was mediated by PhyB [50].
Another mechanism regulating the expression levels of phytochrome genes might be related to the levels of 5-aminolevulinic acid, a common biosynthetic precursor of chlorophylls, heme and phytochromobiline. As phytochromobiline synthesis occurs in plastids, this type of regulation would provide a ‘feedback’ mechanism to coordinate the biosynthesis of phytochrome apoproteins and chromophores, respectively. Recently, joint regulation of light-responsive genes by phytochromes and by retrograde signals was demonstrated [21]. Phytochromes play a major role in the regulation of chlorophyll biosynthesis in seedlings [51].
Phytochrome-encoding genes have been found in the genomes of some fungi and in brown algae (including genomes of brown algal viruses), but are absent from the genomes of haptophytes, red algae and green algae [24]. Phytochromes of charophytes and land plants have a common origin. In liverworts, hornworts and heterosporous lycophytes, phytochromes are encoded by a single gene, while in mosses, homosporous lycophytes, ferns and seed plants, phytochrome genes underwent diversifications. Of special interest are neochromes, the photoreceptors found in zygnemous algae, hornworts and ferns, where the ‘phytochrome-like’ photosensoric N-terminus is fused with a ‘phototropin-like’ C-terminus, and the protein binds both chromophores [24, 52].
The Blue Light Receptors: Phototropins, Proteins of the ZEITLUPE Family, Cryptochromes
Phototropins are light-activated protein kinases. They usually localize to the plasma membrane but do not represent integral membrane proteins; they also can be associated with the chloroplast outer envelope [6]. Arabidopsis has two phototropins: photostable PHOT1 and photolable PHOT2. In Arabidopsis, the light-induced activation and autophosphorylation of phototropins promotes their translocation either to the cytosol (PHOT1) or to Golgi membranes (PHOT2). Phototropins carry FMN-binding LOV1 and LOV2 domains at the N-terminus, and a Ser/Thr-kinase domain required for autophosphorylation upon illumination with B at the C-terminus [5]. Multiple phosphorylation at the C-terminus leads to dimerization and activation of phototropins. In the dark, phototropins lack kinase activity, until the absorption of a light quantum by the chromophore within the LOV2 domain enables kinase activation. Apart from phosphorylation at Ser and Thr, B induces structural changes, most notably the partial unfolding of the LOV Jalpha-helix [53]. Within LOV domains, upon absorption of a light quantum of 450 nm wavelength, the oxidized FMN non-covalently bound to the protein forms a covalent bond between a C-atom of FMN and a S-atom of a nearby cysteine residue. This adduct is formed within several microseconds and is unable to B absorption; its formation mediates the transition of the phototropin into the active state. In the dark, the photoreceptor reverts to the inactive state within several tens or hundreds of seconds. An incomplete list of phototropin-regulated functions in plants includes the phototropism of shoots, roots and leaves, migration of chloroplasts in response to light and temperature signals, and the movement of stomata [6, 54]. The regulatory mechanism best studied is the opening of stomata in response to B: phototropins activate the BLUS1 (blue-light signaling 1) kinase specific for guard cells, which in turn activates the plasma membrane Н+-ATPase leading to hyperpolarization of the membrane and inward currents of potassium via inwardly-rectifying channels, swelling of guard cells and stomata opening [55].
As the rate of the so-called ‘dark’ reactions of photosynthesis depends on the temperature, one and the same light level can result in a different ‘excitation pressures’ on the photosystems depending on the ambient temperature. Therefore, under similar light conditions, at low temperatures chloroplasts move to the anticlinal cell walls of mesophyll cells, while at high temperatures they move to the periclinal cell walls [54]. This observation suggested that phototropins in plants can function as thermosensors. Using Marchantia polymorpha, an organism containing only a single copy of the MpPHOT gene, it was demonstrated that the migration of chloroplasts, of the nucleus and of peroxisomes in response to the lowering of the ambient temperature is controlled by B and MpPHOT [54]. Combination of B and low temperature leads to an increase in the autophosphorylation levels of MpPHOT and in the half-life of its activation state. Together, this promotes the migration of the chloroplasts to the anticlinal cell walls, in order to prevent photooxidation [54, 56]. Thus, phytochromes and phototropins are plant thermoreceptors functioning under different light conditions and on different time scales (the life time for the activated form of the photoreceptor is several tens of minutes for photochromes but only tens of seconds for phototropins) [54].
The ZEITLUPE family proteins ZTL, FKF1 and LKP1 are Е3-ubiquitin ligases which are activated by illumination with B and facilitate the ubiquitination and proteasome degradation of their specific substrates [6, 57]. The N-termini of these proteins contain a single LOV domain with the FMN chromophore, followed by an F-box domain responsible for specific binding of the E3-ubiquitin ligase to its substrate, and by a KELCH domain mediating protein-protein interactions. ZTL exerts control over circadian rhythms, and FKF1 controls the transition to flowering, while LKP1 is required for both processes. Photoactivation of the LOV domain changes the affinity of these proteins to the regulatory protein GIGANTEA (GI), and also leads to an increase in the levels of E3-ubiquitin ligase activity. The GI-FKF1 complex triggers the degradation of CDFs (Cycling DOF Factors), the repressors of CO (CONSTANS) gene expression. CONSTANS, in turn, activates transcription of the FT gene encoding ‘florigen’, the main activator of flowering in Arabidopsis. The GI-ZTL complex, on the contrary, restricts the capacity of ZTL to ubiquitinylate its substrates, components of circadian oscillator. In the ZEITLUPE proteins, similar to phototropins, the photoactivation of the FMN chromophore promotes formation of a cysteinyl adduct, but the degradation of this adduct is slow in FKF1 (the life time of the adduct can reach several days), while in ZTL it occurs with the same rate as in phototropins [6, 57]. In the dark, ZTL mediates the degradation of Timing of CAB expression 1 (TOC1), the repressor of core circadian genes, while B inhibits it: GI binds to ZTL leading to the accumulation of TOC1. Similarly, GI binds to FKF1 (after its activation by B), but this leads to an increase in the ubiquitin ligase activity of the latter, and, as a result, the long day photoperiod leads to the degradation of CDFs, the repressors of CO and FT genes, via the FKF1-GI complex [6, 57].
Cryptochromes, another class of B photoreceptors, evolved as FAD-dependent DNA photolyases, i.e. as the enzymes repairing breaches in DNA caused by B. In land plants, some cryptochromes, e.g. cryptochromes of the CRY-DASH group (Drosophila, Ara-bidopsis, Synechocystis, Homo), are capable of binding DNA and still retain the DNA photolyase activity [58]; notably, these cryptochromes thus far have not been shown to function as photoreceptors. The PHR (photolyase homology region) domain binds two chromophores: FAD which absorbs light of 320–500 nm, and pterine (5,10-methenyltetrahydrofolate) which functions as an antenna transferring the energy of near UV light (370–390 nm) to FAD [11]. With oxidized FAD, a cryptochrome is in its inactive form; after photoreduction, protonation and formation of a neutral semiquinone FADH, the photoreceptor has achieved its active conformation [59]. Further reduction of the chromophore to FADH2 (e.g. upon absorption of green light quanta) again promotes the transition of the cryptochrome to its inactive form [59]. However, fully reduced FADH2 is required for DNA photolyase activity. Cryptochromes bind AMP and ATP which might stabilize the active semiquinone-containing form of the photoreceptor [6]. Within cryptochromes, there is an intramolecular electron transport chain: electrons are transferred from tryptophan residues to FAD after photoactivation of the latter; an aspartyl residue within the polypeptide chain provides a proton for FAD protonation. The C-termini of cryptochromes (except for CRY-DASH cryptochromes) contain a highly variable signaling CCE (cryptochrome C-terminal extension) domain [11]. Although cryptochromes do not contain kinase domains, their activation requires multiple phosphorylation events at the C-terminus, followed by dimerization. The activation leads to the exposure of the so-called NC-motif at the С-terminus which consists of 80 amino acids. Expression of a cDNA encoding only these 80 amino acid residues can complement the late flowering phenotype in Arabidopsiscry knock-out mutants [11].
In Chlamydomonas rheinhardtii, single cryptochrome perceives both B and R. The Physcomitrella patens genome contains two cryptochrome genes, while the genome of the fern Adianthum capillus-ve-neris contains five. In rice, there are two CRY1 and one CRY2 cryptochrome while wheat contains two CRY1, one CRY2 and one CRY-DASH [11]. There are three cryptochromes in Arabidopsis: a photostable CRY1 activated by high light, a photolabile СRY2 which perceives only very weak light and undergoes degradation in high light, perhaps via the СОР1-SPA1 Е3-ubiquitin ligase complex [60], and CRY3 which in Arabidopsis belongs to the CRY-DASH group. CRY1 and CRY2 localize to the nucleus while CRY3 occurs in chloroplasts and mitochondria. Cryptochrome-based light responses are best studied for Arabidopsis. In natural shade, cryptochromes sense the B : G ratio [61, 62]. It was shown that the SAS can be elicited not only via a decrease in the R : FR ratio but also via an increase of the G proportion in the incident light; the latter mechanism involves cryptochromes [12, 61, 62]. Notably, Zhang et al. [12] have found that some Arabidopsis responses to G do not involve phytochromes or cryptochromes but rather a novel, not yet identified photoreceptor.
Under B illumination, the active cryptochromes in the nucleus interact with two groups of transcription factors, PIFs (phytochrome-interacting factors) and CIBs (cry-interacting bHLH), mediating their degradation via the СОР1-SPA1 ubiquitin ligase complex. Apart from this, СRY bind to СОР1-SPA1 and promote its removal from the nucleus, triggering photomorphogenesis (see below) and the transition to flowering (in combination with other photoreceptors), as the ubiquitinylation and degradation of CONSTANS by СОР1 in the nucleus is required for the onset of flowering. The active form of CRY2 binds the CIB1 transcription factor, leading to the activation of the expression of the ‘florigen’ gene, the FT. This process also involves CIB2 and СIB5. Also, in Arabidopsis, CRY2 negatively regulates the inhibitor of flowering, PhyВ, upon exposure to B or to white light. Thus, CRY2 is the main positive regulator of flowering in Arabidopsis. The activation of photomorphogenesis by cryptochromes involves the same transcription activators HY5, HFR1, and HYH, as the activation by phytochromes (see below). Cryptochromes can upregulate the expression of all nuclear genes encoding photosynthetic proteins, such as САВ and RbcS. CRY1 mediates the greening of roots, i.e. the photomorphogenesis of plastids in root cells, upon illumination with B [63]. When the intensities of B decrease, cryptochromes initiate SAS which, in contrast to SAS induced via phytochromes, is uncoupled from the inhibition of the plant immune responses (see below) [62, 64]; this fact is potentially promising for practical use. Altogether, cryptochromes regulate circadian rhythms, the onset of flowering, B-dependent photomorphogenesis and other functions [6].
In response to illumination with B, Arabidopsis CRY1 migrates into the nucleus and activates anion (Cl–) channels [11]. This presumably leads to the temporary depolarization of the plasma membrane within 30 s after illumination, although the exact mechanism is unknown. The reduced states of the cryptochrome chromophore, semiquinone FADH· and FAD\({\text{H}}_{2}^{ - }\), can mediate the reduction of oxygen and production of reactive oxygen species (ROS) such as superoxide radical and hydrogen peroxide [11].
The Photoreceptor of Ultraviolet B Light: UVR8
Ultraviolet light of B-range (UV-B) light makes up only a small part of the solar spectrum (less than one percent). Nevertheless, plants possess photoreceptors capable of specific perception of UV-B photons. Currently, only one of such photoreceptors is known in Arabidopsis, the UV RESISTANCE LOCUS 8 (UVR8) protein [65]. However, homologous UVR8 genes occur in all genomes of land plants, mosses and algae sequenced thus far [8]. The UVR8 protein in Arabidopsis is a ‘beta-propeller’, a dimer in its inactive state, located in the cytosol. After absorption of UV-B photons, it dissociates into two active monomers which can migrate into the nucleus. A major portion of the active monomeric form is retained in the cytosol, although its role there is not known. The UV-B-absorbing chromophores are tryptophan residues within the polypeptide chain of UVR8. In dimeric UVR8, the tryptophan residues interact with nearby arginine residues forming ‘cation-pi’ bonds; the absorption of UV-B quanta by tryptophans breaks these bonds and triggers the dissociation of the UVR8 dimer into monomers [8]. In the dark, the photoreceptor will undergo re-dimerization and inactivation after several hours; this process is facilitated by the regulatory WD40 proteins RUP1 and 2 (Repressor of UV-B photomorphogenesis) [8].
In the nucleus, UVR8 directly binds the promoters of genes encoding transcriptional activators of photomorphogenesis, such as HY5. Furthermore, in the nucleus, UVR8 interacts with the main repressor of photomorphogenesis, the СОР1 Е3-ubiquitin ligase. However, in contrast with other photorepectors which bind СОР1 to perform ‘the inhibition of the inhibitor’, СОР1 acts as an activator in UV-B mediated signaling [8]. UVR8 can directly interact with СОР1 independently of SPA1 and escape the degradation; moreover, although СОР1 remains in the nucleus, the elicitation of phototropic responses and UV-B dependent photomorphogenesis occurs including de-etiolation, the arrest of hypocotyl growth, activation of flavonoid biosynthesis, regulation of circadian rhythms and an increase in resistance to pathogens and chewing insects [8]. UV-B light was shown to increase plant resistance to pathogens both dependent and independent of jasmonate signaling, probably by means of biosynthesis of flavonoids; moreover, natural levels of UV-B in the incident light are required for plant immunity responses to be established [14]. Thus, further study of photoreception and signal transduction pathways of UV-B is of special interest for potential applications in agriculture, because the UV-B-mediated increase in plant resistance is not related to growth arrest, as it is the case for jasmonate signaling (see below). In spinach and potato, activation of PhyB led to increased resistance of plants to UV-B [66, 67], suggesting that the signal transduction pathways for PhyB and UVR8 overlap.
COMMON MECHANISMS OF PHOTORECEPTOR FUNCTIONING
In spite of the diversity of plants photoreceptors, their functionality follows several ‘common rules’. For phytochromes, cryptochromes as well as phototropins, the active form represents a dimer the formation of which relies on (auto)phosphorylation; the activation is triggered by absorption of shorter-wavelength photon (B or R depending on the photoreceptor type), while inactivation occurs after absorption of longer-wavelength photon (G or FR). For phytochromes, cryptochromes and phototropins, two types of photoreceptors exist: one specifically perceives very low light levels and undergoes photodestruction in high light (for instance, PHOT2, CRY2 and PhyA in Arabidopsis), while the other one specifically perceives high light and is photostable (for instance, PHOT1, CRY1 and PhyB in Arabidopsis).
Transduction of the light signal perceived by photoreceptors can be mediated by second messengers such as Ca2+, cAMP, cGMP, G-proteins and other components [32]. Transformation of the information on light quality and quantity into metabolic and morphogenetic responses occurs via controlled degradation of transcription factors: the active form of a photoreceptor (phytochrome, cryptochrome or UVR8) enters the nucleus and binds the СОР1 E3-ubiquitine ligase, the enzyme which performs ubiquitinylation for the regulators of genetic programs and their degradation via 26S proteasome [60]. In the nucleus, СОР1 inhibits photomorphogenesis, regulates circadian rhythms and flowering, plant imminuty, interactions between various groups of phytohormones, movements of stomata and other functions. Photoreceptors induce the removal of СОР1 from the nucleus, and thereby switch on or off genetic programs [60]. Apart from the СОР1-dependent pathway, the active forms of photoreceptors can directly bind transcriptional repressors in the nucleus, interfering with their action, another mechanism leading to activation of a number of genetic programs in the cell [27]. The ZEITLUPE family proteins do not interact directly with СОР1 but instead represent themselves Е3-ubiquitin ligases which, similar to СОР1, enter the nucleus and mediate the degradation of major regulatory proteins via the proteasome. Thus far, it is unknown whether phototropins can be transcriptional regulators of a number of genes. A recent study in tomato showed the phototropin-dependent regulation of more than hundred genes including those encoding proteins required for chromatine reorganization, regulation of transcription and translation [68]. However, the underlying mechanism remains unknown; it possibly involves phytochrome signaling [69].
The inhibition of СОР1 in the light results in the accumulation of various transcription factors in the nucleus, which activate photomorphogenesis. Of these, the bZIP transcription factors HY5 and HYH, the bHLH transcription factor HFR1 and the MYB-protein LAF1 are central for the regulation of transcriptional activity by light and temperature signals [20]. In the dark, these proteins undergo СОР1-mediated proteasomal degradation. Furthermore, in the dark, these master regulators interact with PIFs (Phytochrome-interacting factors), another system controlling plant responses to changing light conditions.
Phytochrome-interacting factors (PIFs) are bHLH transcription factors. The PIF1, PIF3, PIF4, PIF5, PIF6, PIF7 and PIF8 proteins activate scotomorphogenesis and inhibit photomorphogenesis throughout the life of the plant, mediating light-dependent growth regulation; even in high light, a basal level of PIFs activity is required to maintain growth [70]. PIFs enable growth by multilayered regulation of several classes of phytohormones. They mediate the expression levels of the YUCCA genes which encode the enzymes of auxin biosynthesis [71]; also, PIF3 and PIF4 bind DELLA proteins, the negative inhibitors of gibberellin-responsive genes, while PIF5 activates the accumulation of repressors of gibberellin biosynthesis. Furthermore, DELLA and PIFs directly bind BZR1, the regulator of brassinosteroid-responsive genes [13]. Notably, activation of the biosynthesis of auxin and gibberellins via PIFs occurs in response to a decrease of the R proportion in the incident light, while the PIFs-mediated activation of brassinosteriod signaling occurs in response to a decrease of the B proportion [62]. Another type of interaction between DELLA and PIFs mediates the growth arrest that occurs as part of the plant stress response induced via jasmonate signalling [70]. In the absence of jasmonates, the JAZ repressors bind the transcription factors MYC2/3/4, the activators of jasmonate-inducible genes, inhibiting their functions [72]. JAZ proteins also interact with DELLA, thereby partially decreasing the levels of JAZ available to bind MYC factors, and leading to a further decrease in the levels of DELLA-PIFs complexes; the unbound PIFs thus can function in growth promotion. The biosynthesis of jasmonates induces the degradation of JAZ and thereby increases the binding of DELLA to PIFs, leading to a decrease in the growth rate. This way, the balance between the MYC-dependent activation of jasmonate-responsive genes and PIF-dependent growth in low light is determined to a certain extent by gibberellin biosynthesis and DELLA proteins; gibberellin binding DELLA activates plant growth via PIFs and reduces the expression levels of jasmonate-responsive genes [72].
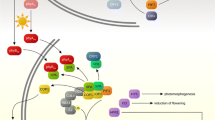
PIF transcription factors contain DNA-binding domains and domains mediating the interactions with phytochromes: the APB (Active PhyB binding) domain, and, in case of PIF1 and PIF3, also the APA (Active PhyA binding) domain. PhyA was shown to interact directly with PIF1 and PIF3, while PhyB interacts with most PIFs. When bound to PIFs, phytochromes promote phosphorylation, ubiquitinylation and proteasome degradation of the latter in the course of several minutes, while also inhibiting the binding of PIFs to the promoters of target genes [13]. Recent studies reveal that PIFs represent master switches of plant development, a regulatory hub receiving signals not only from phytochromes but also from other photoreceptors: cryptochromes and UVR8 [71, 73, 74]. Altogether, very different signals from most photoreceptors in plants are transduced to three major regulatory modules: the repressor of photomorphogenesis, COP1; the activator of photomorphogenesis, HY5; and the systemic integrators, PIFs (Fig. 1).
A simplified scheme of the integration of signals elicited from photoreceptors activated by various regions of the light spectrum in Arabidopsis thaliana. Arrows show activation and inhibition, respectively. The double arrow shows the direct interaction of UVR8 with COP1 and the induced response. The hatched arrow shows interaction between phototropin-based and phytochrome-based signaling. Aux—auxins; GA—gibberellins; Brs—brassinosteroids; JA—jasmonates.
THE ‘SHADE AVOIDANCE SYNDROME’ AND ITS IMPORTANCE FOR CROP PRODUCTION
The shade avoidance syndrome (SAS) is a scotomorphogenetic program activated either by a decrease in the R : FR ratio in the incident light, or by a decrease in the total amounts of R, B, white or UV-B light, and, according to recent data, also by a decrease in the B : G ratio in the incident light [62]. All these changes are typical for light conditions under a dense canopy [4, 13]. This suggests that all photoreceptors participate in the establishment of SAS. Notably, a decrease in the R : FR ratio in the incident light can induce SAS even in high light: this makes plants aware of neighbors, i.e. the future competitors for the light resource, before light becomes limiting, and leads to the preemptive acceleration of growth and the induction of other adaptive reactions of the SAS program. A decrease in the R : FR ratio in the incident light occurs because of the absorption of R and reflection of FR by green plant parts; as a result, plants perceive the FR‑enriched light horizontally reflected by stems of neighboring plants [4]. The decrease in the R : FR ratio promotes the inactivation of PhyB, the main inhibitor of the SAS in high light. In the shade, the main inhibitor of SAS is PhyA, specifically its dephosphorylated, relatively photostable form which appears in the cell in course of the activation of the HIR. The induction of SAS is mediated via СОР1-dependent degradation of HY5 and HFR1, the activators of photomorphogenesis, and also via activation of PIF4- and PIF5-regulated genes.
SAS activation leads to the elongation of petioles, leaf hyponasty, arrest of width growth combined with acceleration of length growth of leaf laminae; elongation of internodes, retardation of chlorophyll biosynthesis and other scotomorphogenic reactions. During the SAS in monocot crops, the transcription factor GRASSY TILLERS increases the dormancy of lateral buds and inhibits tillering; GRASSY TILLERS is activated via another protein, TEOSINTHE BRANCHED, which is inhibited by PhyB [75]. Activation of the SAS inhibits the emission of jasmonate-inducible volatiles, probably because of a decrease in active phytochrome levels in the cells [76]. The establishment of the SAS also involves mechanosensitive channels [4]. Mechanostimulation increases the strength of SAS symptoms in seedlings while decreasing it in mature plants. This can be explained by the fact that for seedlings, mechanostimulation signals the presence of neighbors, while for a mature plant, it indicates that the plant has grown over the grass canopy, where natural mechanostimulation is increased due to air migration and windblasts, and thus has escaped the potential competition from neighboring plants [4].
Jasmonates inducing plant defense against necrotrophic pathogens and chewing insects, and salicylates inducing defense against biotrophic pathogens, represent the backbone of plant immunity [14]. In plants growing in the shade compared to light grown plants, the extent of biotrophic and necrotrophic infections is higher, and feeding larvae are bigger [14]. In fact, SAS induced by a decrease in the R : FR ratio often includes a decrease in plant pathogen resistance due to the weakening of responses to jasmonate signaling [14]. Furthermore, the inactivation of PhyB in the shade leads to a reduction of the biosynthesis of salicylic acid [14, 77]. Earlier, these phenomena have been interpreted as a means to redirect the energy and other plant resources from defense to growth, to outrun the competitors for light availablilty [14].
In crops, the induction of SAS results in a decrease in yield: for instance, in cereals, it leads to a redirection of the resources from leaves and ears to ‘economically not significant’ stems. At the same time, during thousands of years of domestication, along with other important traits, crops were selected for the ability to produce high yield at dense planting, i.e. for weakening of the SAS [78]. Thus, the question of how to inhibit the SAS and enable a further increase in planting density for crops, simultaneously with a change in biomass balance in favor of economically important plant organs, while not inhibiting plant immunity, is a primary issue for breeders and bioengineers. In this context, the possibility to manipulate plant responses to light, including ‘switching off’ the SAS, is of special interest [79]. For this purpose, Wille et al. [80] performed chemical mutagenesis of six varieties of spring wheat followed by multiple screening steps, first with a green light filter and then under conditions of a decreased R : FR ratio, to identify the wheat mutant lines showing no growth acceleration under these conditions. After analysis of more than 1000 lines, five mutants were obtained displaying a high extent of reduction of the SAS: at higher planting density, they showed good growth, early flowering, wider leaves and higher biomass than the parent varieties under similar conditions. In another study aimed at revealing the components of signaling pathways regulating the SAS, the authors for the first time used phenotypic profiling [81], resulting in the identification of 18 new SAS-mediating genes (among them, the gene encoding a guard cell-specific potassium channel KAT1), and the description of three groups of genes controlling three types of shade responses, respectively: the genes controlling hypocotyl elongation, the genes controlling petiole elongation, and the genes controlling the transition to flowering. It should be pointed out that, because of the convenience of using a decreased R : FR ratio to induce the SAS, the most approaches targeted the SAS induced by inactivation of PhyB while studies of cryptochrome-induced or UVR8-induced SAS lag behind, although they also should have the potential for agricultural use [64, 77].
INTERACTIONS BETWEEN PHYTOCHROME-MEDIATED AND JASMONATE-MEDIATED SIGNALING
As discussed above, the induction of SAS can occur either by a decrease of the proportion of the active form of the main SAS inhibitor, PhyB, due to an increase of the FR ratio in the incident light, or of other photoreceptors like CRY1 or UVR8, in response to a decrease of light of the respective wavelength. A decrease in the active forms of photoreceptors in cells leads to the activation of PIFs and of PIF-based scotomorphogenetic reactions including accelerated elongation growth. The mediation of SAS involves almost all phytohormones [82]. In the shade, gibberellin levels rise [82], increasing the degradation rate of the DELLA proteins. In high light, DELLA proteins bind PIFs and interfere with the induction of SAS; however, DELLA proteins also interact with another group of transcription factor repressors, JAZ [72]. JAZ proteins inhibit the transcription factors MYC2/3/4, the activators of jasmonate-responsive genes including genes encoding enzymes involved in the biosynthesis of volatiles and responsible for growth arrest. In Arabidopsis, there are up to 10 JAZ proteins performing similar functions [83].
More and more studies reveal a tight interaction between phytochrome- and jasmonate-based regulatory systems [83–91]. The biosynthesis of the conjugate of jasmonate with isoleucine promotes the degradation of JAZ proteins via the Е3-ubiquitin ligase complex COI1 (CORONATINE-INSENSITIVE 1) [72]; however, recent studies have shown that this degradation requires a presence of PhyA [84]. Furthermore, the JAZ10 protein is required for the perception of the R : FR ratio and the inhibition of PhyB [83]. The active form of the JAZ10.4 protein is a product of alternative splicing [83]; it would be interesting to pinpoint the role of phytochromes in the regulation of splicing of the JAZ10.4 transcripts. In an Arabidopsis mutant, the absence of phytochromes resulted in an increase in the biosynthesis of jasmonate [76], while in a rice mutant, the inhibition of jasmonate biosynthesis changed the ratio of different spectral forms of PhyA [86, 87].
The identification of signals and mechanisms underlying the growth arrest under stress conditions is of special interest for agriculture [91–94]. The leading role in these phenomena belongs to jasmonate-regulated genes, and the biosynthesis of jasmonates is strongly induced under stress [89, 91, 93]. A recent study demonstrated that growth arrest and the onset of stress response reactions are two different genetic programs which function in close cooperation, but nevertheless can be completely separated in mutants of respective key regulators of these programs [89]. Such mutants of A. thaliana displayed high levels of stress resistance and growth vigor under stress conditions, similar to, or even exceeding (probably due to transgressive effects), that of wild-type plants in the absence of stress [89]. In this study, jasmonic acid was the inductor of stress responses and PhyВ was the activator of the growth arrest in response to jasmonate signaling. Notably, manipulations of the levels of phytochrome activity influence not only the growth of shoots, but also of roots: the role of phytochromes (those occurring both in shoot and root cells) in the growth of roots was recently demonstrated [92]. The root phytochromes participate in root gravitropism and in the change of root growth patterns, including the perception of jasmonate signals [95]. The attempts to manipulate the expression levels of phytochrome genes in transgenic plants in order to shift the balance of biomass production to economically important plant organs had begun earlier and continue until now [96].
CONCLUSION
The research on the control of plant growth and development via photoreceptor systems, and their functions under stress conditions, currently experiences a boost. These studies provide a potential basis for breeding of crop varieties capable of high yield production under stress conditions.
REFERENCES
Mokronosov, A.T., Ontogeneticheskii aspekt fotosinteza (Ontogenetic Aspect of Photosynthesis), Moscow: Nauka, 1981.
Jung, J.H., Domijan, M., Klose, C., Biswas, S., Ezer, D., Gao, M., Khattak, A.K., Box, M.S., Charoensawan, V., Cortijo, S., Kumar, M., Grant, A., Locke, J.C., Schäfer, E., Jaeger, K.E., et al., Phytochromes function as thermosensors in Arabidopsis, Science, 2016, vol. 354, pp. 886–889.
Lee, H.J., Ha, J.H., Kim, S.G., Choi, H.K., Kim, Z.H., Han, Y.J., Kim, J.I., Oh, Y., Fragoso, V., Shin, K., Hyeon, T., Choi, H.G., Oh, K.H., Baldwin, I.T., and Park, C.M., Stem-piped light activates phytochrome B to trigger light responses in Arabidopsis thaliana roots, Sci. Signal., 2016, vol. 9: ra106.
Pierik, R. and de Wit, M., Shade avoidance: phytochrome signalling and other aboveground neighbour detection cues, J. Exp. Bot., 2014, vol. 65, pp. 2815–2824.
Briggs, W.R. and Christie, J.M., Phototropins 1 and 2: versatile plant blue-light receptors, Trends Plant Sci., 2002, vol. 7, pp. 204–210.
Christie, J.M., Blackwood, L., Petersen, J., and Sullivan, S., Plant flavoprotein photoreceptors, Plant Cell Physiol., 2015, vol. 56, pp. 401–413.
Schultz, T.F., The ZEITLUPE family of putative photoreceptors, in Handbook of Photosensory Receptors, Ch. 16, Briggs, W.R. and Spudich, J.L., Eds., Weinheim: Wiley, 2005, pp. 337–347. https://doi.org/10.1002/352760510X.ch16
Tilbrook, K., Arongaus, A.B., Binkert, M., Heijde, M., Yin, R., and Ulm, R., The UVR8 UV-B photoreceptor: perception, signaling and response, Arabidopsis Book, 2013, vol. 11: e0164. https://doi.org/10.1199/tab
Sineshchekov, V.A., Fitokhrom A: polimorfizm i polifunktsional’nost' (Phytochrome A: Polymorphism and Polyfunctionality), Moscow: Nauch. Mir, 2013.
Wang, H. and Haiyang, H., Phytochrome signaling: time to tighten up the loose ends, Mol. Plant, 2015, vol. 8, pp. 540–551.
Mishra, S. and Khurana, J.P., Emerging roles and new paradigms in signaling mechanisms of plant cryptochromes, Crit. Rev. Plant Sci., 2017, vol. 36, pp. 89–115.
Zhang, T., Maruhnich, S.A., and Folta, K.M., Green light induces shade avoidance symptoms, Plant Physiol., 2011, vol. 157, pp. 1528–1536.
Casal, J.J., Photoreceptor signaling networks in plant response to shade, Annu. Rev. Plant Biol., 2013, vol. 64, pp. 403–427.
Ballare, C.L., Light regulation of plant defense, Annu. Rev. Plant Biol., 2014, vol. 65, pp. 335–363.
Kuznetsov, E.D., Sechnyak, L.K., Kindruk, N.A., and Slyusarenko, O.K., Rol’ fitokhroma v rasteniyakh (The Role of Phytochrome in Plants), Moscow: Agropromizdat, 1986.
Song, Y.H., Shim, J.S., Kinmonth-Schultz, H.A., and Imaizumi, T., Photoperiodic flowering: time measurement mechanisms in leaves, Annu. Rev. Plant Biol., 2015, vol. 66, pp. 441–464.
Chai, T., Zhou, J., Liu, J., and Xing, D., LSD1 and HY5 antagonistically regulate red light induced-programmed cell death in Arabidopsis, Front. Plant Sci., 2015, vol. 6: 292. https://doi.org/10.3389/fpls.2015.00292
Maurya, J.P. and Bhalerao, R.P., Photoperiod- and temperature-mediated control of growth cessation and dormancy in trees: a molecular perspective, Ann. Bot., 2017, vol. 120, pp. 351–360.
Kreslavski, V.D., Los, D.A., Schmitt, F.J., Zharmukhamedov, S.K., Kuznetsov, Vl.V., and Allakhverdiev, S.I., The impact of the phytochromes on photosynthetic processes, Biochim. Biophys. Acta—Bioenergetics, 2018, vol. 1859, pp. 400–408.
Chen, D., Xu, G., Tang, W., Jing, Y., Ji, Q., Fei, Z., and Lina, R., Antagonistic basic helix-loop-helix/bZIP transcription factors form transcriptional modules that integrate light and reactive oxygen species signaling in Arabidopsis, Plant Cell, 2013, vol. 25, pp. 1657–1673.
Martιn, G., Leivar, P., Ludevid, D., Tepperman, J.M., Quail, P.H., and Monte, E., Phytochrome and retrograde signalling pathways converge to antagonistically regulate a light-induced transcriptional network, Nat. Commun., 2016, vol. 7: 11431.
Shikata, H., Hanada, K., Ushijima, T., Nakashima, M., Suzuki, Y., and Matsushita, T., Phytochrome controls alternative splicing to mediate light responses in A-rabidopsis, Proc. Natl. Acad. Sci. USA, 2014, vol. 111, pp. 18781–18786.
Ushijima, T., Hanada, K., Gotoh, E., Yamori, W., Kodama, Y., Tanaka, H., Kusano, M., Fukushima, A., Tokizawa, M., Yamamoto, Y.Y., Tada, Y., Suzuki, Y., and Matsushita, T., Light controls protein localization through phytochrome-mediated alternative promoter selection, Cell, 2017, vol. 171, pp. 1316–1325.
Li, F.W., Melkonian, M., Rothfels, C.J., Villarreal, J.C., Stevenson, D.W., Graham, S.W., Wong, G.K., Pryer, K.M., and Mathews, S., Phytochrome diversity in green plants and the origin of canonical plant phytochromes, Nat. Commun., 2015, vol. 6: 1038. https://doi.org/10.1038/ncomms8852
Burgie, E.S. and Vierstra, R.D., Phytochromes: an atomic perspective on photoactivation and signaling, Plant Cell, 2014, vol. 26, pp. 4568–4583.
Nagatani, A., Phytochrome: structural basis for its functions, Curr. Opin. Plant Biol., 2010, vol. 13, pp. 565–570.
Casal, J.J., Candia, A.N., and Sellaro, R., Light perception and signalling by phytochrome A, J. Exp. Bot., 2014, vol. 65, pp. 2835–2845.
Sharrock, R.A. and Clack, T., Patterns of expression and normalized levels of the five Arabidopsis phytochromes, Plant Physiol., 2002, vol. 130, pp. 442–456.
Ballaré, C.L., Scopel, A.L., and Sánchez, R.A., Photocontrol of stem elongation in plant neighbourhoods: effects of photon fluence rate under natural conditions of radiation, Plant Cell Environ., 1991, vol. 14, pp. 57–65.
Brouwer, B., Gardeström, P., and Keech, O., In response to partial plant shading, the lack of phytochrome A does not directly induce leaf senescence but alters the fine-tuning of chlorophyll biosynthesis, J. Exp. Bot., 2014, vol. 65, pp. 4037–4049.
Rösler, J., Klein, I., and Zeidler, M., Arabidopsis fhl/fhy1 double mutant reveals a distinct cytoplasmic action of phytochrome A, Proc. Natl. Acad. Sci. USA, 2007, vol. 104, pp. 10737–10742.
Kreslavski, V.D., Carpentier, R., Klimov, V.V., and Allakhverdiev, S.I., Transduction mechanisms of photoreceptor signals in plant cells, J Photochem. Photobiol., C: Photochem. Rev., 2009, vol. 10, pp. 63–80.
Hughes, J., Phytochrome cytoplasmic signaling, Annu. Rev. Plant Biol., 2013, vol. 64, pp. 377–402.
Haupt, W., Regulation der Chloroplastenverteilung in der Zelle durch Lichtintensitaet und Lichtrichtung, Ber. Dtsch. Bot. Ges., 1973, vol. 86, pp. 403–406.
Tanada, T., A rapid photoreversible response of barley root tips in the presence of 3-indoleacetic acid, Proc. Natl. Acad. Sci. USA, 1968, vol. 59, pp. 376–380.
Jaedicke, K., Lichtenthaler, A.L., Meyberg, R., Zeidler, M., and Hughes, J., A phytochrome-phototropin light signaling complex at the plasma membrane, Proc. Natl. Acad. Sci. USA, 2012, vol. 109, pp. 12231–12236.
Fankhauser, C., Yeh, K.C., Lagarias, J.C., Zhang, H., Elich, T.D., and Chory, J., PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis, Science, 1999, vol. 284, pp. 1539–1541.
Paik, I., Yang, S., and Choi, G., Phytochrome regulates translation of mRNA in the cytosol, Proc. Natl. Acad. Sci. USA, 2012, vol. 109, pp. 1335–1340.
Zhao, J., Zhou, J.J., Wang, Y.Y., Gu, J.W., and Xie, X.Z., Positive regulation of phytochrome B on chlorophyll biosynthesis and chloroplast development in rice, Rice Sci., 2013, vol. 20, pp. 243–248.
Inagaki, N., Kinoshita, K., Kagawa, T., Tanaka, A., Ueno, O., Shimada, H., and Takano, M., Phytochrome B mediates the regulation of chlorophyll biosynthesis through transcriptional regulation of ChlH and GUN4 in rice seedlings, PLoS One, 2015, vol. 10: e0135408.
Boccalandro, H.E., Rugnone, M.L., Moreno, J.E., Ploschuk, E.L., Serna, L., Yanovsky, M.J., and Casal, J.J., Phytochrome B enhances photosynthesis at the expense of water-use efficiency in Arabidopsis, Plant Physiol., 2009, vol. 150, pp. 1083–1092.
Casson, S.A. and Hetherington, A.M., Phytochrome B is required for light-mediated systemic control of stomatal development, Curr. Biol., 2014, vol. 24, pp. 1216–1221.
Kim, J., Song, K., Park, E., Kim, K., Bae, G., and Choi, G., Epidermal phytochrome B inhibits hypocotyl negative gravitropism non-cell-autonomously, Plant Cell, 2016, vol. 28, pp. 2770–2785.
Osugi, A., Itoh, H., Ikeda-Kawakatsu, K., Takano, M., and Izawa, T., Molecular dissection of the roles of phytochrome in photoperiodic flowering in rice, Plant Physiol., 2011, vol. 157, pp. 1128–1137.
Pearce, S., Kippes, N., Chen, A., Debernardi, J.M., and Dubcovsky, J., RNA-seq studies using wheat phytochrome B and phytochrome C mutants reveal shared and specific functions in the regulation of flowering and shade-avoidance pathways, BMC Plant Biol., 2016, vol. 16: 1186. https://doi.org/10.1186/s12870-016-0831-3
Sinetova, M.A. and Los, D.A., New insights in cyanobacterial cold stress responses: genes, sensors, and molecular triggers, Biochim. Biophys. Acta, 2016, vol. 1860, pp. 2391–2403.
Franklin, K.A., Light and temperature signal crosstalk in plant development, Curr. Opin. Plant Biol., 2009, vol. 12, pp. 63–68.
Chen, F., Li, B., Li, G., Charron, J.B., Dai, M., Shi, X., and Deng, X.W., Arabidopsis phytochrome A directly targets numerous promoters for individualized modulation of genes in a wide range of pathways, Plant Cell, 2014, vol. 26, pp. 1949–1966.
Somers, D.E. and Quail, P.H., Temporal and spatial expression patterns of PHYA and PHYB genes in Ara-bidopsis, Plant J., 1995, vol. 7, pp. 413–427.
Baba-Kasai, A., Hara, N., and Takano, M., Tissue-specific and light-dependent regulation of phytochrome gene expression in rice, Plant Cell Environ., 2014, vol. 37, pp. 2654–2666.
Belyaeva, O.B., Svetozavisimyi biosintez khlorofilla (The Light-Dependent Chlorophyll Biosynthesis), Moscow: Lab. Znanii, 2015.
Nozue, K., Kanegae, T., Imaizumi, T., Fukuda, S., Okamoto, H., Yeh, K.C., Lagarias, J.C., and Wada, M., A phytochrome from the fern Adiantum with features of the putative photoreceptor NPH1, Proc. Natl. Acad. Sci. USA, 1998, vol. 95, pp. 15826–15830.
Harper, S.M., Neil, L.C., and Gardner, K.H., Structural basis of a phototropin light switch, Science, 2003, vol. 301, pp. 1541–1544.
Fujii, Y., Tanaka, H., Konno, N., Ogasawara, Y., Hamashima, N., Tamura, S., Hasegawa, S., Hayasaki, Y., Okajima, K., and Kodama, Y., Phototropin perceives temperature based on the lifetime of its photoactivated state, Proc. Natl. Acad. Sci. USA, 2017, vol. 114, pp. 9206–9211.
Inoue, S., Takemiya, A., and Shimazaki, K., Phototropin signaling and stomatal opening as a model case, Curr. Opin. Plant Biol., 2010, vol. 13, pp. 587–593.
Casal, J.J. and Ouesta, J.I., Light and temperature cues: multitasking receptors and transcriptional integrators, New Phytol., 2018, vol. 217, pp. 1029–1034.
Zoltowski, B.D. and Imaizumi, T., Structure and function of the ZTL/FKF1/LKP2 group proteins in Arabidopsis, Enzymes, 2014, vol. 35, pp. 213–239.
Huang, Y., Baxter, R., Smith, B.S., Partch, C.L., Colbert, C.L., and Deisenhofer, J., Crystal structure of cryptochrome 3 from Arabidopsis thaliana and its implications for photolyase activity, Proc. Natl. Acad. Sci. USA, 2006, vol. 103, pp. 17701–11706.
Banerjee, R., Schleicher, E., Meier, S., Viana, R.M., Pokorny, R., Ahmad, M., Bittl, R., and Batschauer, A., The signaling state of Arabidopsis cryptochrome 2 contains flavin semiquinone, J. Biol. Chem., 2007, vol. 282, pp. 14916–14922.
Lau, O.S. and Deng, X.W., The photomorphogenic repressors COP1 and DET1: 20 years later, Trends Plant Sci., 2012, vol. 17, pp. 584–593.
Sellaro, R., Crepy, M., Trupkin, S.A., Karayekov, E., Buchovsky, A.S., Rossi, C., and Casal, J.J., Cryptochrome as a sensor of the blue/green ratio of natural radiation in Arabidopsis, Plant Physiol., 2010, vol. 154, pp. 401–409.
Keller, M.M., Jaillais, Y., Pedmale, U.V., Moreno, J.E., Chory, J., and Ballare, C.L., Cryptochrome 1 and phytochrome B control shade-avoidance responses in A-rabidopsis via partially independent hormonal cascades, Plant J., 2011, vol. 67, pp. 195–207.
Usami, T., Mochizuki, N., Kondo, M., Nishimura, M., and Nagatani, A., Cryptochromes and phytochromes synergistically regulate Arabidopsis root greening under blue light, Plant Cell Physiol., 2004, vol. 45, pp. 1798–1808.
Cerrudo, I., Keller, M.M., Cargnel, M.D., Demkura, P.V., Wit, M., Patitucci, M.S., Pierik, R., Pieterse, C.M.J., and Ballare, C.L., Low red/far-red ratios reduce Arabidopsis resistance to Botrytis cinerea and jasmonate responses via a COI1-JAZ10-dependent, salicylic acid-independent mechanism, Plant Physiol., 2012, vol. 158, pp. 2042–2052.
Rizzini, L., Favory, J.J., Cloix, C., Faggionato, D., O’Hara, A., Kaiserli, E., Baumeister, R., Schafer, E., Nagy, F., Jenkins, G.I., and Ulm, R., Perception of UV-B by the Arabidopsis UVR8 protein, Science, 2011, vol. 332, pp. 103–106.
Kreslavski, V.D., Kosobryukhov, A.A., and Shmarev, A.N., Introduction of the Arabidopsis PHYB gene increases resistance of photosynthetic apparatus in transgenic Solanum tuberosum plants to UV-B radiation, Russ. J. Plant Physiol., 2015, vol. 62, pp. 204–209.
Kreslavski, V.D., Khristin, M.S., Shabnova, N.I., and Lyubimov, V.Yu., Preillumination of excised spinach leaves with red light increases resistance of photosynthetic apparatus to UV radiation, Russ. J. Plant Physiol., 2012, vol. 59, pp. 717–723.
Hloušková, P. and Bergougnoux, V., A subtracted cDNA library identifies genes up-regulated during PHOT1-mediated early step of de-etiolation in tomato (Solanum lycopersicum L.), BMC Genomics, 2016, vol. 17: 291.
Demarsy, E., Schepens, I., Okajima, K., Hersch, M., Bergmann, S., Christie, J., Shimazaki, K., Tokutomi, S., and Fankhauser, C., Phytochrome kinase substrate 4 is phosphorylated by the phototropin 1 photoreceptor, EMBO J., 2012, vol. 31, pp. 3457–3467.
Leivar, P. and Monte, E., PIFs: systems integrators in plant development, Plant Cell, 2014, vol. 26, pp. 56–78.
Goyal, A., Karayekov, E., Galvão, V.C., Ren, H., Casal, J.J., and Fankhauser, C., Shade promotes phototropism through phytochrome B-controlled auxin production, Curr. Biol., 2016, vol. 26, pp. 3280–3287.
Kazan, K. and Manners, J.M., The interplay between light and jasmonate signalling during defence and development, J. Exp. Bot., 2011, vol. 62, pp. 4087–4100.
Sun, J., Qi, L., Li, Y., Zhai, Q., and Li, C., PIF4 and PIF5 transcription factors link blue light and auxin to regulate the phototropic response in Arabidopsis, Plant Cell, 2013, vol. 25, pp. 2102–2114.
De Wit, M., Keuskamp, D.H., Bongers, F.J., Hornitschek, P., Gommers, C.M.M., Reinen, E., Martínez-Cerón, C., Fankhauser, C., and Pierik, R., Integration of phytochrome and cryptochrome signals determines plant growth during competition for light, Curr. Biol., 2016, vol. 26, pp. 3320–3326.
Whipple, C.J., Kebrom, T.H., Weber, A.L., Yang, F., Hall, D., Meeley, R., Schmidt, R., Doebley, J., Brutnell, T.P., and Jackson, D.P., grassy tillers1 promotes apical dominance in maize and responds to shade signals in the grasses, Proc. Natl. Acad. Sci. USA, 2011, vol. 108: E506–E512.
Zhai, Q., Li, C.B., Zheng, W., Wu, X., Zhao, J., Zhou, G., Jiang, H., Sun, J., Lou, Y., and Li, C., Phytochrome chromophore deficiency leads to overproduction of jasmonic acid and elevated expression of jasmonate-responsive genes in Arabidopsis, Plant Cell Physiol., 2007, vol. 48, pp. 1061–1071.
De Wit, M., Spoel, S.H., Sanchez-Perez, G.F., Gommers, C.M.M., Pieterse, C.M.J., Voesenek, L.A.C.J., and Pierik, R., Perception of low red: far-red ratio compromises both salicylic acid- and jasmonic acid-dependent pathogen defences in Arabidopsis, Plant J., 2013, vol. 75, pp. 90–103.
Carriedo, L.G., Maloof, J.N., and Brady, S.M., Molecular control of crop shade avoidance, Curr. Opin. Plant Biol., 2016, vol. 30, pp. 151–158.
Warnasooriya, S.N. and Brutnell, T.P., Enhancing the productivity of grasses under high-density planting by engineering light responses: from model systems to feedstocks, J. Exp. Bot., 2014, vol. 65, pp. 2825–2834.
Wille, W., Pipper, C.B., Rosenqvist, E., Andersen, S.B., and Weiner, J., Reducing shade avoidance responses in a cereal crop, AoB Plants, 2017, vol. 9, no. 5: plx039.
Nozue, K., Tat, A.V., Devisetty, U.K., Robinson, M., Mumbach, M.R., Ichihashi, Y., Lekkala, S., and Maloof, J.N., Shade avoidance components and pathways in adult plants revealed by phenotypic profiling, PLoS Genet., 2015, vol. 11: e1004953. https://doi.org/10.1371/journal.pgen.1004953
Yang, C. and Li, L., Hormonal regulation in shade avoidance, Front. Plant Sci., 2017, vol. 8: 1527.
Cerrudo, I., Caliri-Ortiz, M.E., Keller, M.M., Degano, M.E., Demkura, P.V., and Ballare, C.L., Exploring growth-defence trade-offs in Arabidopsis: phytochrome B inactivation requires JAZ10 to suppress plant immunity but not to trigger shade-avoidance responses, Plant Cell Environ., 2017, vol. 40, pp. 635–644.
Robson, F., Okamoto, H., Patrick, E., Harris, S.R., Wasternack, C., Brearley, C., and Turnera, J.G., Jasmonate and phytochrome A signaling in Arabidopsis wound and shade responses are integrated through JAZ1 stability, Plant Cell, 2010, vol. 22, pp. 1143–1160.
Chen, J., Sonobe, K., Ogawa, N., Masuda, S., Nagatani, A., Kobayashi, Y., and Ohta, H., Inhibition of Arabidopsis hypocotyl elongation by jasmonates is enhanced under red light in phytochrome B dependent manner, J. Plant Res., 2013, vol. 126, pp. 161–168.
Riemann, M., Müller, A., Korte, A., Furuya, M., Weiler, E.W., and Nick, P., Impaired induction of the jasmonate pathway in the rice mutant hebiba, Plant Physiol., 2003, vol. 133, pp. 1820–1830.
Dhakarey, R., Peethambaran, P.K., and Riemann, M., Functional analysis of jasmonates in rice through mutant approaches, Plants, 2016, vol. 5, no. 1: 15. https://doi.org/10.3390/plants5010015
Hsieh, H.L. and Okamoto, H., Molecular interaction of jasmonate and phytochrome A signaling, J. Exp. Bot., 2014, vol. 65, pp. 2847–2857.
Campos, M.L., Yoshida, Y., Major, I.T., Ferreira, D.O., Weraduwage, S.M., Froehlich, J.E., Johnson, B.F., Kramer, D.M., Jander, G., Sharkey, T.D., and Howe, G.A., Rewiring of jasmonate and phytochrome B signalling uncouples plant growth-defense tradeoffs, Nat. Commun., 2016, vol. 7: 12570. https://doi.org/10.1038/ncomms12570
Moreno, J.E., Tao, Y., Chory, J., and Ballare, C.L., Ecological modulation of plant defense via phytochrome control of jasmonate sensitivity, Proc. Natl. Acad. Sci. USA, 2009, vol. 106, pp. 4935–4940.
Wasternack, C. and Hause, B., Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development, Ann. Bot., 2013, vol. 111, pp. 1021–1058.
Costigan, S.E., Warnasooriya, S.N., Humphries, B.A., and Montgomery, B.L., Root-localized phytochrome chromophore synthesis is required for photoregulation of root elongation and impacts root sensitivity to jasmonic acid in Arabidopsis, Plant Physiol., 2011, vol. 157, pp. 1138–1150.
Ahmad, P., Rasool, S., Gul, A., Sheikh, S.A., Akram, N.A., Ashraf, M., Kazi, A.M., and Gucel, S., Jasmonates: multifunctional roles in stress tolerance, Front. Plant Sci., 2016, vol. 7: 813.
Ding, H., Lai, J., Wu, Q., Zhang, S., Chen, L., Dai, Y.S., Wang, C., Du, J., Xiao, S., and Yang, C., Jasmonate complements the function of Arabidopsis lipoxygenase3 in salinity stress response, Plant Sci., 2016, vol. 244, pp. 1–7.
Warnasooriya, S.N. and Montgomery, B.L., Spatial-specific regulation of root development by phytochromes in Arabidopsis thaliana, Plant Signal. Behav., 2011, vol. 6, pp. 2047–2050.
Robson, P.R., McCormac, A.C., Irvine, A.S., and Smith, H., Genetic engineering of harvest index in tobacco through overexpression of a phytochrome gene, Nat. Biotechnol., 1996, vol. 14, pp. 995–998.
ACKNOWLEDGMENTS
The author thanks Prof. Katharina Pawlowski (Stockholm University, Sweden) and the anonymous reviewer for critical comments.
Funding
Support by the Russian Science Foundation (project no. 14-16-00120) is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
Additional information
Abbreviations: APA—ACTIVE PhyA BINDING; APB—ACTIVE PhyB BINDING; B—blue light; BZR1—BRASSINAZOLE-RESISTANT 1; CCE—CRYPTOCHROME C-TERMINAL EXTENSION; CDF—CYCLING DOF FACTORS; ChlH—a subunit of the ChlH Mg-chelatase; CIB—CRY-INTERACTING bHLH; CO—CONSTANS; COI1—CORONATINE-INSENSITIVE 1; СОР1—CONSTITUTIVE PHOTOMORPHOGENESIS 1; СRY—cryptochrome; CRY-DASH—DASH (Drosophila, Arabidopsis, Synechocystis, Homo)-class cryptochrome; FHL—FHY1-like protein; FHY—FAR-RED ELONGATED HYPOCOTYL 1; FKF1—FLAVIN-BINDING, KELCH REPEAT, F-BOX 1; FR—far red light; FT—FLOWERING LOCUS T; G—green light; GAF—cGMP-specific phosphodiesterases, adenylyl cyclases, FhlA domain; GI—GIGANTEA; GUN4—GENOMES UNCOUPLED 4; R—red light; HFR1—LONG HYPOCOTYL IN FAR-RED 1; HIR—high irradiance response; HY5—elongated hypocotyl 5; HYH—homolog of long hypocotyl in far-red 1; JAZ—jasmonate-ZIM domain; KELCH—a 50-amino acids-motif in proteins which repetitions form a ‘propeller’ structure; LAF1—long after far-red; LFR—low fluence response; LKP2—LOV Kelch Protein 2; LOV—light-oxygen-voltage-sensing domain; PAS—Per-Arnt-Sim domain; pAtSUC2—promoter of the AtSUC2 gene; Pfr—active phytochrome; PHOT—phototropine; PHR—photolyase homology region; Phy—phytochrome; PIF—phytochrome-interacting factor; Pr—inactive phytochrome; RUP—REPRESSOR OF UV-B PHOTOMORPHOGENESIS; SAS—shade avoidance syndrome; SPA1—SUPPRESSOR OF PHYTOCHROME A; TOC1—TIMING OF CAB1; UV-B—ultraviolet B radiation; UVR8—UV RESISTANCE LOCUS 8; VLFR—very low fluence response; ZTL—ZEITLUPE.
Rights and permissions
About this article
Cite this article
Voitsekhovskaja, O.V. Phytochromes and Other (Photo)Receptors of Information in Plants. Russ J Plant Physiol 66, 351–364 (2019). https://doi.org/10.1134/S1021443719030154
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1021443719030154