Abstract
The STARD4 subfamily of steroidogenic acute regulatory protein (StAR)-related lipid transfer (START) proteins consists of STARD4, STARD5, and STARD6, which share ~ 30 % amino acid identity with each other, ~ 20 % with the StAR/STARD1 and STARD3/metastatic lymph node clone 64 (MLN64) START domains, and ~ 15 % with other START domains. All three have no other protein domains besides START. Since their initial discovery, they were proposed to serve as intracellular sterol transporters. A subsequent decade of research has led to seven high-confidence conclusions: (1) STARD4 expression is regulated by cellular sterol levels via the sterol regulatory element binding protein (SREBP)-2 transcription factor. (2) Endoplasmic reticulum (ER) stress-inducing agents increase STARD5 levels, though the mechanism may be posttranscriptional. (3) STARD4 and STARD5 are cytosolic proteins that may associate loosely with specific subcellular membranes. (4) STARD4 can bind cholesterol and efficiently transfer it between membranes. (5) STARD4 can mediate transfer of cholesterol to the ER resident enzyme acyl-coenzyme A cholesterol acyltransferase (ACAT) for esterification. (6) STARD5 does not have these ACAT effects but has some distinct activities from STARD4. (7) STARD4 null mice lack any clear phenotype. Other conclusions are less certain due to conflicting data but merit further study: ER stress and steroidogenic regulation of STARD4, selective expression of STARD5 in immune and reticuloendothelial cells, nuclear localization of STARD4 and STARD5, binding of bile acids by STARD5, and binding of other sterols besides cholesterol by STARD4 (such as 7α-hydroxycholesterol) and STARD5 (such as 25-hydroxycholesterol). Selective expression of STARD6 in male germ cells strongly suggests a role in fertility, but the functions of STARD4 and STARD5 in normal physiology and disease remain elusive.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- STARD4
- STARD5
- STARD6
- Intracellular cholesterol transport
- SREBP
- Cholesterol esterification
- Endoplasmic reticulum
- Oxysterols
- Bile acids
Introduction: A Personal Account of Discovery
I was an MD-PhD graduate student at Rockefeller University whose primary project was floundering, so my secondary project was a fishing expedition. Our overarching goal was to address the question of how the body responds to dietary cholesterol , such that some individuals are sensitive and increase plasma cholesterol while others are insensitive. Genes in the liver that change with dietary cholesterol seemed like a good place to start, so we fed mice a standard diet (0.02 % cholesterol) versus a high cholesterol diet (0.5 %) for three weeks. At the time around the year 2000, microarrays to measure messenger ribonucleic acid (mRNA) gene expression were the latest in technology. The wide availability of commercial arrays was a year or so away, so we did not have access to the familiar oligonucleotide probe-based systems like Affymetrix GeneChips, which are now being replaced by RNA-sequencing. Instead, we used complementary deoxyribonucleic acid (cDNA) arrays in which entire cDNA clones (each hundreds of base pairs) were spotted onto slides. To obtain and analyze these microarrays, my excellent mentor, Jan Breslow, established a collaboration with Raju Kucherlapati’s laboratory, then at Albert Einstein College of Medicine. I remember taking the subway from Manhattan to the Bronx to see the wondrous custom-built robotic system that spotted approximately 9,000 cDNA clones from 96-well plates onto glass slides. About half of these cDNAs were annotated with gene names, while the rest were unidentified expressed sequence tags (ESTs).
In our initial microarray hybridizations, we noted marked variation across experiments, as the statistical tools to analyze microarray data were also in their infancy. After multiple biological and technical replicates and stringent cutoffs, only six transcripts were consistently downregulated more than twofold by dietary cholesterol, and disappointingly none were convincingly upregulated. Identifying the genes corresponding to six regulated transcripts was not trivial given that the annotation of the mouse genome was far from complete. Five of the six genes turned out to encode enzymes involved in cholesterol or fatty acid synthesis. This was an excellent proof of principle for our experimental system, since these pathways were expected to be downregulated by cholesterol, negative feedback mediated by the sterol regulatory element binding protein (SREBP) transcription factors.
The sixth transcript was a mystery. EST AA239481 was cloned from mouse liver, not annotated with any gene name, and the available 460 bases of sequence in the database did not contain any obvious protein-coding region or homology to known genes. We obtained the EST clone and sequenced its entire insert of 1,114 base pairs, but did not find an open reading frame. Since the cDNA was cloned with oligo-dT for the 3′ polyadenylation site, I suspected that this sequence was in the 3′ untranslated region (UTR) of the gene. I was self-taught at using the relatively primitive bioinformatic resources available, and I remember spending a late evening in the lab at my computer staring at this sequence. The breakthrough happened when I performed a basic local alignment search tool (BLAST) search and found overlap with another uncharacterized EST, extending the cDNA sequence by several hundred nucleotides in the presumed 5′ direction but still not revealing the coding sequence. However, this new EST overlapped with another, and then another, so by walking from one EST to the next across about 3 kilobases I finally found protein coding sequence. Ultimately, alignment of EST and genomic sequence revealed an unannotated six exon gene encoding a 224 amino acid protein, with the final exon including more than 4 kilobases of 3′ UTR including the initial EST.
Evening had turned into night and I had completely lost track of time immersed in this DNA sequence analysis, but the biggest surprise came when I searched for known proteins with homology to this novel one. The first hit was something called “StAR (Steroidogenic Acute Regulatory Protein).” I had never heard of it, but I immediately performed literature searches and was thrilled to find an intracellular cholesterol transport protein, essential for delivering cholesterol to mitochondria for steroid hormone synthesis [1]. The StAR-related lipid transfer (START) domain had been described [2] and the crystal structure of the related metastatic lymph node clone 64 (MLN64) START domain was published [3], with the beautiful hydrophobic cavity to bind lipid. My mentor Jan Breslow was instrumental in the molecular cloning of many apolipoprotein genes [4], which are essential for transport of otherwise insoluble cholesterol and other lipids in the blood. Cholesterol of course has the same problem of insolubility within the cell, and here we had found a putative cholesterol transport protein whose expression was regulated by dietary cholesterol! After a sleepless night with excitement, I rushed into Jan’s office the next morning to share this result. Jan has a famously calm and even demeanor, but I could see his delight as we allowed ourselves to speculate on the potential functions and implications of a novel sterol transporter.
We decided to name this protein CRSP, pronounced “crisp,” for “Cholesterol-Regulated START Protein.” With my new experience searching DNA sequence databases, I found and assembled the whole family of 15 mammalian START domain-containing proteins into a phylogenetic tree [5]. Among characterized START proteins, CRSP was most similar to the known cholesterol-binding proteins StAR and MLN64, but there were two other novel START domain proteins even more like CRSP. I creatively called them “CRSP-like1” and “CRSP-like2,” and we immediately set out cloning and characterizing these three genes. It quickly became my primary PhD thesis project, displacing studies of the bile acid biosynthetic enzyme cholesterol 7 alpha-hydroxylase (Cyp7A1) [6]. I remember reaching out to other researchers in the START field to obtain reagents and advice, such as Walter Miller, Jerry Strauss, and Doug Stocco. I was amazed how helpful and collaborative they were (in retrospect, with more experience in the competitive world of academic biomedical research, I am only more amazed). In fact, several of them had independently noted the cDNAs for one or more of the three novel genes, but none of them rushed to publish first or compete with us.
When we had assembled enough data for our first publication describing this subfamily of three novel START domain proteins [7], the journal required us to submit to the Human Genome Organisation (HUGO) gene nomenclature committee. My name CRSP was shot down, and instead HUGO decided to rename the whole START domain superfamily with the START domain (Stard/STARD#) nomenclature reflected in the official mouse and human gene symbols today. I objected to these generic names since they fail to describe known physiology, regulation, or function of the proteins, and many of the family members have other protein domains besides a START domain. My objections were to no avail, and CRSP, CRSPL1, and CRSPL2 proteins became STARD4, STARD5, and STARD6, whereas previously named proteins StAR, Phosphatidylcholine transfer protein (PCTP), and MLN64 became STARD1, STARD2, and STARD3, respectively. Please don’t blame me for the nomenclature, because I do not like it either!
I finished my PhD work on the STARD4 subfamily, with Fred Maxfield from across the street at Weill Cornell Medical College serving on my thesis committee and advising us on the cell biology of intracellular cholesterol transport . I was never satisfied that we failed to describe a true physiological function for STARD4 and STARD5 in cholesterol metabolism. Two years after my cDNA microarrays, a new and very talented MD-PhD student in the Breslow lab, my good friend Kara Maxwell, performed the same cholesterol feeding experiment yet used Affymetrix microarrays. Of course she found STARD4 again, but she also discovered the proprotein convertase subtilisin/kexin type 9 (Pcsk9) protein [8], which turned out to have a key role in regulating levels of low-density lipoprotein (LDL) “bad” cholesterol and the associated risk of atherosclerosis. Pcsk9 is now an exciting drug target [9], whereas—despite the diligent efforts of very thorough investigators—STARD4 languishes in relative obscurity. I returned to medical school, then clinical training in internal medicine and endocrinology. This choice of subspecialty was guided by my research interest in lipid metabolism, diabetes, and obesity, but perhaps also a little by the role of STARD1/StAR in steroid hormone synthesis. During my clinical fellowship, my attending physician, Carrie Burns noted a patient with congenital adrenal hyperplasia and unusual biochemical results we interpreted as partial StAR deficiency, which had not been described. Sequencing of his StAR gene indeed revealed compound heterozygosity for two mutations, which we shared with Walter Miller’s laboratory for characterization. Along with several other patients, this led to the description of nonclassic/atypical lipoid congenital adrenal hyperplasia due to StAR mutants with partial activity [10]. I had hoped that human disease phenotypes might likewise someday be associated with STARD4 or STARD5, but this has not yet come to pass. My postdoctoral research has taken me away from my beloved START proteins, but I have followed from afar with great interest, and I review the progress here.
Stard4/STARD4 Gene Regulation
As described in detail above, the mouse StarD4 gene was discovered due to its regulation by cholesterol [7]. Cellular cholesterol exerts negative feedback on genes like STARD4 via the SREBP transcription factors, whose mechanism has been elegantly described over the past several decades by the laboratory of Michael Brown and Joseph Goldstein (Fig. 7.1a, reviewed in ref. [11]) Briefly, SREBPs are synthesized as inactive precursors spanning the membrane of the endoplasmic reticulum (ER) . When cellular sterols are adequate, SREBP remains in the ER associated with sterol sensing proteins SREBP cleavage-activating protein (SCAP) and Insulin-induced gene (INSIG). Low sterol results in a conformational change in SCAP, releasing INSIG such that SCAP escorts SREBP in vesicles from the ER to the Golgi, where successive proteolytic cleavages of the SREBP transmembrane region by site 1 and site 2 proteases release the soluble N-terminal transcription factor domain. This mature SREBP then translocates to the nucleus and binds to the sterol response elements (SREs) in gene promoters to activate target genes.
Regulation of SREBP and ER stress transcription factors. a SREBPs are activated upon cholesterol depletion. When cellular cholesterol or certain oxysterols are abundant, SREBPs remain in the ER as inactive precursors associated with SCAP and Insig. When sterols are scarce, SCAP releases Insig allowing SREBP-SCAP trafficking to the Golgi apparatus, where the site 1 and site 2 proteases (S1P and S2P) cleave SREBP. This releases the N-terminal transcription factor nSREBP, which translocates to the nucleus and binds promoter SREs. nSREBPs cooperate with other factors like NF-Y to activate transcription of target genes involved in the synthesis and uptake of cholesterol and fatty acids. b ER stress signals are transduced to the nucleus via three parallel pathways, activating the ATF4, Xbp1, and ATF6 transcription factors. The RNA-dependent protein kinase-like ER kinase (PERK) pathway transiently inhibits global translation but activates translation of ATF4. IRE1 mediates non-traditional splicing of the Xbp1 mRNA to increase its synthesis, while ATF6 is activated by proteolysis in the Golgi similar to SREBP processing. The ER chaperone BiP negatively regulates all three pathways, which may converge in the nucleus with complex interactions on target gene promoters with ER stress response elements (ERSEs). SREBP sterol regulatory element binding protein, ER endoplasmic reticulum, SCAP SREBP cleavage-activating protein, ATF activating transcription factor
Many lines of evidence support the idea that STARD4 is a direct target of SREBP transcription factors, particularly SREBP-2 that predominantly regulates cholesterol metabolism, as opposed to SREBP-1 that predominantly regulates fatty acid metabolism . First, STARD4 is consistently co-regulated by sterols with other classic targets of SREBP-2, such as enzymes in the cholesterol biosynthesis pathway like the rate-limiting enzyme 3-hydroxy-3-methyl-glutaryl (HMG) CoA reductase (HMGCR). In mouse liver, upon 3 weeks of high cholesterol feeding, twofold decreases in STARD4 mRNA are observed on microarrays and validated by quantitative polymerase chain reaction (PCR), coordinately regulated with known SREBP-2 targets [7, 8]. Regulation is even greater in cultured mouse 3T3-L1 cells: sterol depletion with lovastatin to activate SREBP increased STARD4 expression, while addition of 25-hydroxycholesterol to repress SREBP decreased STARD4 expression, with a difference of 14-fold between the conditions [7]. The same sterol regulation of STARD4 mRNA was confirmed in 3T3-L1 fibroblasts and human THP-1 macrophages, and extended to the protein level by STARD4 Western blots, even correlating with the amount of mature SREBP-2 (Fig. 7.2) [12]. STARD4 mRNA is also regulated with HMGCR in other cells culture models where SREBP is affected: cholesterol-loaded macrophages [13], THP-1 cells differentiated into macrophages [12], mouse embryonic fibroblasts deficient in a sterol dehydrogenase involved in cholesterol synthesis [14], and HepG2 cells cultured in lipoprotein-depleted serum [15]. Notably, hepatic STARD4 mRNA and protein levels are induced threefold by statin treatment of mice [16]. Second, transgenic and knockout mice with altered SREBP activities show the expected changes in STARD4 levels: mice transgenic for constitutively active nuclear SREBP-1a or SREBP-2 have higher STARD4 expression, whereas SCAP knockout mice with loss of SREBP activity have decreased STARD4 expression [17]. Furthermore, in the same nuclear SREBP transgenic mice, STARD4 is more highly regulated by SREBP-2 than SREBP-1 [13, 17], supporting a role in sterol metabolism rather than fatty acid metabolism . Third, the promoter for STARD4 in mice and humans has several potential SREs, and a proximal promoter fragment confers sterol regulation in luciferase reporter assays. Sterol regulation of mouse and human STARD4 promoter reporters was abrogated when one potential SRE (called SRE-B) was mutated, or when either of the two nearby CCAAT box elements were mutated (presumably affecting binding of NF-Y, which cooperates with SREBPs) [13]. Fourth, direct binding of SREBP-2 to the STARD4 promoter was observed by chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) [18]. This ChIP-seq experiment revealed 1,800 binding sites in mouse liver treated with lovastatin plus ezetimibe to maximize SREBP-2 processing, and the majority of these sites had an SRE consensus motif of AA(G/A)ATGGC. In retrospect, the functional STARD4 promoter SRE-B motif defined in reporter assays [13] has a sequence of AGGATGGA in mouse or TAAATGGA in human, each with only two disagreements (underlined) from the ChIP-seq consensus motif. Given this preponderance of evidence, there can be little doubt that STARD4 is a direct transcriptional target of SREBP-2. There are only two inconsistent cases in the literature, and both can be explained by the duration of time: (1) sterol depletion and overload of U2OS cells had the expected effects on SREBP-2 processing but failed to affect STARD4 protein levels as detected by Western blot [19] and (2) a 0.5 % cholesterol feeding experiment for one week failed to affect hepatic STARD4 protein levels [16]. In both cases, the treatment time, 2 h for U2OS cells and 1 week for cholesterol feeding, was likely too short to see changes in protein level .
STARD4 expression is regulated by intracellular sterol levels via SREBP-2. a In 3T3-L1 fibroblasts, treatment with sterols represses SREBP-2 processing to its mature form, while treatment with the statin drug mevinolin to deplete sterols activates SREBP-2 processing (Western blot in bottom panel, the nuclear protein Lamin B1 serves as a loading control). In these conditions, mRNA levels of STARD4 and HMG CoA reductase are coordinately regulated (quantitative RT-PCR in upper panel). b STARD4 protein levels show the same pattern of regulation by Western blot (upper panel, cytosolic beta-actin serves as a loading control) with quantification (lower panel). Adapted from Rodriguez-Agudo et al. [12]. SREBP sterol regulatory element binding protein, mRNA messenger ribonucleic acid, RT-PCR reverse transcription polymerase chain reaction
STARD4 expression can be induced during steroidogenesis like StAR/STARD1. In MA-10 mouse Leydig tumor cells treated with cyclic adenosine monophosphate (cAMP), StAR mRNA is induced ~ 50-fold while STARD4 mRNA is induced only ~ 3-fold, and the similar small induction of HMGCR suggests that steroidogenesis may deplete cellular cholesterol and activate SREBP-2 [13]. A more recent experiment in the same MA-10 cell line showed ~ 20-fold cAMP induction of STARD4 protein by Western blot, and these authors propose a role for STARD4 in steroidogenesis [20]. Until further studies are performed, it remains unclear whether STARD4 is specifically regulated by steroidogenic stimuli.
One report indicates that STARD4 is regulated in the early phase of the ER stress response (Fig. 7.1c) [21]. Tunicamycin treatment of HeLa cells followed by cDNA subtraction identified STARD4 along with four well-known ER stress-induced genes; binding immunoglobulin protein (BiP), GRP94, CHOP, and Herp. STARD4 induction peaked at ~ 2.5-fold by 4–6 h of treatment, then came down by 12–24 h. This time course, as well as the different species and cell type, may explain why tunicamycin failed to induce STARD4 after 18–20 h in mouse NIH-3T3 cells [13]. A luciferase reporter driven by the human STARD4 promoter was activated by tunicamycin and other ER stressors thapsigargin, dithiothreitol (DTT), and brefeldin A, as well as by overexpression of the ER stress induced transcription factor activating transcription factor 6 (ATF6) [21]. Notably, overexpression of a dominant negative ATF6 prevented STARD4 reporter activation by thapsigargin. Site-directed mutagenesis of three potential ER-stress response elements (ESREs CCAAT-N9-CCACG) individually in reporters suggested that the second element was most responsible for ER-stress response. Compared to the other human STARD4 luciferase reporters [13], this functional ESRE-like element lies between potential SRE-B and SRE-C. Other SREBP-2 target genes were not studied in this system, and it is notable that in mouse 3T3-L1 cells the SREBP-2 target, HMGCR, shows a similar time course of induction only in early ER stress [22]. It has also been observed that STARD4 expression increases during the differentiation of THP-1 cells from monocytes to macrophages, even at days 5–6 when mature SREBP-2 decreases, but there are increased levels of mature ATF6 [12]. Given that SREBPs and ATF6 are processed by the same proteases after ER to Golgi translocation [23] and may bind to the same DNA elements and coordinately regulate target genes [24]—and that STARD5 is induced by ER stress (see below)—there is potential cross-talk between cholesterol metabolism and ER stress such that regulation of STARD4 by ER stress via ATF6 deserves further study.
Other regulatory effects on STARD4 expression have been reported. STARD4 mRNA is induced in granulosa cells of women with diminished ovarian reserve, an effect also observed for STARD1 [25], though the mechanism for this is uncertain. One report proposes that STARD4 is a target of the p53 transcription factor, based on a p53-binding region identified downstream of the gene by an early ChIP-seq method and p53-dependent gene regulation (though STARD4 expression was down rather than up) [26]. While some authors have hypothesized that this may relate to adipocyte biology [27], further reports connecting p53 and STARD4 are lacking. Taking a similar approach with the abundant genome-wide data now available in the Encyclopedia of DNA Elements (ENCODE) project [28], there are dozens of transcription factors in different cell types that can potentially bind to the STARD4 proximal promoter region in addition to the SREBPs as expected. Furthermore, STARD4 has at least three uncharacterized enhancer regions located in the 50 kb upstream, all of which have factor binding, DNaseI hypersensitivity, and histone marks .
Stard5/STARD5 Gene Regulation
STARD5 does not show regulation by the SREBP-related manipulations that affect STARD4 [13], yet it is induced by the ER stress response. Robust (up to 10-fold) increases in STARD5 mRNA have consistently been shown upon treatment with ER stressors such as tunicamycin, thapsigargin, brefeldin A, and dithiothreitol in mouse NIH-3T3 fibroblasts (Fig. 7.3) [13], human HK-2 kidney tubular cells [29], and mouse 3T3-L1 preadipocytes [22]. In the last study, a dose dependent protein induction by thapsigargin also was shown (using a commercial antibody from Santa Cruz, see below). The three main pathways of the ER stress response are mediated by the transcription factors ATF4, nuclear ATF6, and spliced Xbp1 (Fig. 7.1c), and only expression of spliced Xbp1—but not active ATF6 or ATF4—activated STARD5 expression in 3T3-L1 cells [22]. The mechanism for activation of STARD5 by ER stress and Xbp1 apparently lies outside the proximal promoter, as luciferase reporters driven by ~ 2000 bp upstream of human STARD5 or ~ 400 bp upstream of mouse STARD5 failed to show ER stress regulation [22, 13]. Instead, markedly more stable STARD5 mRNA was observed upon thapsigargin treatment of 3T3L1 cells in the presence of actinomycin D to inhibit new transcription , suggesting a novel posttranscriptional mechanism to increase STARD5 mRNA levels [22]. Thus, even the STARD5 induction by spliced Xbp1 may be indirect via mRNA stability, though increased transcription remains possible and would require a nuclear run-on type assay to test . Some physiologic stimuli that induce ER stress also increased STARD5 expression: cholesterol-loading of mouse macrophages [13] and OVE26 diabetic mouse kidney [29]. The exact role of STARD5 in the response to ER stress is a matter of speculation, as it is possible that lipids stressors as well as unfolded proteins play a role in ER stress. The ER membrane is notably poor in cholesterol, which constitutes only ~ 5 % of its lipid molecules compared to ~ 30 % in the plasma membrane (reviewed in ref. [19]).
STARD5 mRNA levels are increased in ER stress. In NIH-3T3 fibroblasts, treatment with drugs known to induce ER stress (tunicamycin, thapsigargin, brefeldin A, and DTT) all increases mRNA levels of STARD5 as well as the positive control ER stress response gene BiP. (a) and (c) show quantitative RT-PCR, while (b) shows a Northern blot with both STARD5 mRNAs upregulated. This research was originally published in reference 13, and subsequent studies in reference 22 also showed upregulation of STARD5 protein by Western blot, and that the mRNA induction is likely posttranscriptional via increased mRNA stability. Adapted from Soccio et al. [13] © the American Society for Biochemistry and Molecular Biology. mRNA messenger ribonucleic acid, ER endoplasmic reticulum
STARD5 was regulated by the cytokine interleukin (IL)-1β in rat Sertoli cells, though the mRNA was induced and the protein decreased [30]. In contrast, STARD4 expression was not regulated by IL-1β, even though precursor and mature SREBP-1 levels decreased (SREBP-2 was not reported). There are no other reports of inflammatory cytokines regulating STARD5, and the significance of this finding is unknown .
STARD4 Expression Pattern and Subcellular Localization
When overexpressed in HeLa cells, GFP-tagged STARD4 gives a diffuse cytosolic and nuclear localization, in contrast to the vesicular pattern observed for full-length STARD1 (mitochondria) and MLN64 (endosomes) [31], and the same result is seen by immunofluorescence [32]. However, when overexpressed in human keratinocytes [33] or U2OS osteosarcoma cells [19], STARD4 staining is not seen in the nucleus and its cytosolic localization is more punctate, with a more intense perinuclear region.
The most detailed study of STARD4 localization used a polyclonal antibody generated in the laboratory of Dr W. M. Pandak, Virginia Commonwealth University [12]. This antibody was well validated, recognizing in Westerns a sterol-regulated band of the predicted size in human THP-1 macrophages, and a slightly larger band in mouse liver than human—consistent with the predicted 224 amino acid mouse protein versus 205 in humans. In human liver, STARD4 immunostaining was observed in hepatocytes, and it was even more intense in Kupffer cells but absent in endothelial cells . This was confirmed by Western Blots showing expression in both hepatocytes and nonparenchymal cells. In mouse 3T3-L1 cells, basal STARD4 immunostaining was weak and homogenous with some punctate regions, whereas sterol-depletion to increase expression resulted in stronger staining with a peri-nuclear reticular pattern, colocalizing with the ER marker calnexin. Fractionation of these cells confirmed cytoplasm and membrane association, without any detectable protein in nuclei or mitochondria. In THP-1 macrophages, STARD4 similarly co-localized with the ER marker calnexin, but also notably around ER-derived vesicles with bodipy-stained neutral lipids where acyl-coenzyme A cholesterol acyltransferase (ACAT)1 co-localized extensively (Fig. 7.4). Therefore, STARD4 appears to be a widely expressed cytosolic protein that may associate with ER membranes.
STARD4 localization in 3T3-L1 cells and THP-1 macrophages. a A mouse 3T3-L1 fibroblast visualized by phase contrast microscopy (left) and stained with STARD4 (green), calnexin (ER maker, red), and DAPI (nucleus, blue). STARD4 is present in the cytosol colocalizing extensively with calnexin. b STARD4 Western blots were performed on 3T3-L1 cells fractionated into cytosol (marked by IκBα), membranes (marked by Calnexin), nuclei (marked by Lamin B1), and mitochondria (marked by aconitase 2), showing a predominantly cytosolic distribution with some membrane association. c A human THP-1 macrophage, in which phase contrast microscopy (left panel) show the nucleus and lipid droplets. Staining was performed for STARD4 (green), ACAT1 (red), and nucleus 4’,6-diamidino-2-phenylindol (DAPI, blue). STARD4 appears present in the nucleus and the cytosol, where it is enriched surrounding lipid droplets and co-localizing with ACAT1. Adapted from Rodriguez-Agudo et al. [12]. ER endoplasmic reticulum
STARD5 Expression Pattern and Subcellular Localization
RNA expression of STARD5 is detected across multiple mouse tissues, with highest levels in liver and kidney [7]. A polyclonal antibody was generated in the Pandak lab against recombinant human STARD5 and detects a ~ 28kD band in human liver lysates, with subcellular fractionation showing this band in the cytosolic fraction but not the mitochondria or microsomes [34]. However, separation of liver cell types surprisingly showed STARD5 was not present in the isolated hepatocytes, but rather in the nonparencymal fraction [35]. STARD5 immunostaining of liver identifies cells lining sinusoids that stain for CD68, or Kupffer cells. Consistent with this, STARD5 protein was not detected using this antibody in primary human hepatocytes, HepG2 cells, or HUVEC endothelial cells, but it is found in immune cell lines derived from monocyte/macrophages, promyelocytic cells, mast cells, and basophils. Protein expression by Western Blot was also not detected in brain tissue or cell lines from fibroblasts, osteosarcoma, or astrocytes. For all of these Western blot analyses, the corresponding mRNA expression is not published, so it remains uncertain whether mRNA and protein expression correlate. For instance, multiple tissue northern blots show minimal STARD5 mRNA in spleen [7, 36] where many immune cells reside. Furthermore, a different polyclonal antibody against STARD5 generated in the Breslow lab (The Rockefeller University) recognizes a band in HepG2 hepatocellular carcinoma cells [15]. Unfortunately, no publications compare these two antibodies side-by-side, or validate either antibody by showing ER stress regulation or loss of signal in STARD5 knockdown. Further studies will be necessary to definitively address the cell type selective expression of STARD5 protein.
In THP-1 macrophages, STARD5 immunofluorescence with the Pandak laboratory antibody was located in the cytosol with a focal intensity near the nucleus co-localizing with the GM130 Golgi marker and high levels of free cholesterol by filipin staining [35]. Consistent with Golgi localization, STARD5 staining dispersed along with GM130 when the Golgi was disrupted with nocodazole, and it did not colocalize with a marker of the endocytic recycling compartment (ERC). However, fractionation and Western blotting for STARD5 showed only cytosolic localization and none in Golgi or ER, suggesting the observed Golgi association may be loose [35].
The Breslow laboratory antibody was used to study STARD5 expression and localization in kidney, recognizing a ~ 22kD band in mouse bone marrow derived macrophages and kidney [29]. Immunohistochemistry of mouse kidney shows STARD5 localization to proximal tubules in the cortico-medullary region and transitional epithelium lining the renal pelvis, but not in glomeruli. Immunoelectron microscopy showed STARD5 in cytosol and the apical membrane brush border at the base of microvilli where endocytosis occurs. STARD5 also associated with ER but not Golgi or mitochondria in renal tubule cells. In human HK-2 kidney tubule cells, STARD5 colocalized with the ER marker Grp78, both before and after treatment with the ER stressor tunicamycin, which relocated both markers from a diffuse punctate pattern to perinuclear and peripheral pattern. Also, STARD5 did not co-localize with an endosomal marker. The authors of this report suggest that differences in subcellular localization in renal tubule (ER and apical membrane) and macrophages (Golgi, see above) may reflect association with cholesterol-rich membranes rather than with a specific compartment [29]. As noted above and discussed further below, different antibodies could also be a factor.
Nuclear localization was not observed using the Pandak laboratory antibody for endogenous STARD5 in THP-1 macrophages, or primary hepatocytes overexpressing recombinant adenoviral STARD5 [35]. Likewise, using the Breslow laboratory antibody, STARD5 was located in the cytosol but not the nucleus of rat Sertoli cells [30]. However, in mouse 3T3-L1 cells assayed by immunofluorescence and subcellular fractionation using a commercial Santa Cruz antibody, STARD5 was located primarily in the nucleus, though remarkably, it redistributed to the cytosol and perinuclear locations after ER stress with thapsigargin [22]. This regulated subcellular localization of STARD5 is extremely interesting if validated in other systems.
A Note on STARD4 and STARD5 Antibodies
As detailed above, multiple polyclonal antibodies generated in different laboratories have been used to study STARD4 and STARD5, often with conflicting results, particularly for STARD5. Fortunately, there are now commercial antibodies available from Santa Cruz that seem to recognize proteins with the expected regulatory patterns. A STARD5 antibody from Santa Cruz gives a band in 3T3-L1 cells with the expected regulation by ER stress [22], and indeed this antibody showed the regulated nuclear localization above but was not used in prior localization or expression studies. A STARD4 antibody from Santa Cruz (sc-66663) shows loss of the STARD4 band by Western Blot in livers of knockout mice, and decrease in heterozygous mice [16]. The standard use of such well-validated commercial antibodies for STARD4 and STARD5 in the future may help clarify ambiguous and conflicting results about cellular and subcellular localization.
Structural Studies of STARD4 and STARD5
X-ray crystal structures have been published for STARD4 [37] and STARD5 [38], as have nuclear magnetic reasonance (NMR) solution structure models [39, 40]. STARD4 and STARD5 are very similar to each other and to other START domains: globular with a helix-grip fold, such that the curved β-sheet and C-terminal α-helix enclose a hydrophobic cavity large enough to accommodate a single lipid molecule. The four crystal structures of presumed cholesterol-binding START domains (StAR, MLN64, STARD4, and STARD5) all lack any lipid in the cavity, even when efforts were made to include cholesterol in the crystallization solutions. Models for reversible cholesterol binding have been proposed (reviewed in ref [41] and elsewhere in this volume). Comparative structural analysis has identified cavity residues that may mediate ligand specificity [38], and volumetric modelling of the hydrophobic cavities correlates with lipid binding [42]. While structure-function correlations have been performed extensively for STARD1, particularly mutations that cause lipoid congenital adrenal hyperplasia, similar studies have yet to be performed for STARD4 family members. It is obviously of great interest to determine which lipids occupy the binding cavities of STARD4 and STARD5.
STARD4 Lipid Binding and Transfer
STARD4 and STARD5 were initially predicted to bind cholesterol or related sterols based on sequence and structural similarity to STARD1 and STARD3 (MLN64) [7]. Consistent with this, STARD4 and STARD5 both show binding to fluorescently labelled NBD-cholesterol similar to that for STARD1 and STARD3 [43]. Given the volume of the lipid-binding cavity and the size of the nitrobenzoxadiazole (NBD) fluorophore, it is surprising that these START domains specifically bind to NBD-cholesterol at all, but lack of binding by STARD7 served as a negative control. Table 7.1 summarizes all the lipid-binding data reported to date, which is described below.
A direct-binding assay showed STARD4 binding to radiolabelled cholesterol but not 25-hydroxycholesterol or 27-hydroxycholesterol, and no competition by other unlabelled oxysterols [44]. Lipid protein overlay (LPO) assays were also performed, in which sterols were spotted on nitrocellulose membranes, then recombinant GST-STARD4 added and washed prior to detection with anti-Glutathione S-transferase (GST) antibody. By this assay, STARD4 was able to bind cholesterol strongly and 7α-hydroxycholesterol weakly (both were competed away by prebinding GST-STARD4 with cholesterol), but not other sterols. Circular dichroism (CD) spectroscopy also showed cholesterol causes dose-dependent changes in STARD4 far ultraviolet (UV) spectra, consistent with a conformational change upon binding [44].
In vitro lipid transfer assays test for the ability of a purified protein to facilitate movement of lipid from a donor to an acceptor membrane. Like StAR/STARD1, STARD4 was able to increase by 2.6-fold the transfer of [14C]cholesterol from small unilamellar liposomes (50 nm small, unilamellar vesicles, SUVs) to isolated mitochondria [20]. Notably, STARD4 failed to transport normal or peroxidated phosphitidylcholine. STARD4 was even more effective at increasing the transfer of 7α-hydroperoxycholesterol, which the authors go on to show causes oxidative damage to mitochondria and loss of membrane potential, suggesting a potential deleterious effect of START domain mediated lipid transfer of oxidated lipids [20].
Another elegant transfer assay used donor liposome containing the florescent cholesterol analog dihydroergosterol (DHE) and acceptor liposomes with a florescent phopholipid, such that DHE transfer from donor to acceptor results in fluorescence resonance energy transfer (FRET) between the two lipids [19]. In this system, STARD4 was an extremely efficient transporter: it increased the rate of DHE transfer by 5 orders of magnitude versus the spontaneous level, with 1 uM STARD4 as effective as 1000 uM cyclodextrin, and each molecule of STARD4 transferred 7 DHE molecules per minute. STARD4 has a positively charged patch on its surface that was hypothesized to interact with negatively charged lipid head groups. Indeed, the presence of anionic lipids phosphatidylserine and phosphatidylinositol in donor and acceptor membranes increased STARD4 transfer activity 10-fold relative to neutral liposomes. Mutation of lysines in the STARD4 basic patch decreased transfer activity with anionic liposomes by eightfold. Increasing unsaturated fatty acid chains in acceptor (but not donor) liposomal phosphatidylcholine also increased transfer activity 3-fold, and this is relevant because the ER is enriched for unsaturated acyl chains and STARD4 may favor transfer to this membrane (see below) [19]. Overall, STARD4 consistently binds and transfers cholesterol, and perhaps also some 7α-metabolites .
STARD5 Lipid Binding
In vitro binding assays have shown that STARD5 can bind several sterol and non-sterol ligands. One direct binding assay uses recombinant bacterially generated His-tagged human START domains to bind radiolabelled sterols, retaining binding through washes of the nickel-resin and elution. STARD5 was able to bind both [14C]cholesterol and [3H]25-hydroxycholesterol, though cholesterol with higher affinity since in competitive assays it could displace 25-hydroxycholesterol but not vice versa [34]. Cholesterol binding was similar to StAR/STARD1, with saturable 1:1 stoichiometry; however, STARD1 did not bind 25-hydroxycholesterol. Furthermore, STARD5 did not bind [14C]27-hydroxycholesterol, and no competition with [14C]cholesterol was observed using other unlabelled sterols or cholic acid, though this result is difficult to interpret since even 25-hydrocholesterol did not compete. CD spectroscopy showed cholesterol and 25-hydroxycholesterol cause dose-dependent changes in STARD5 far UV spectra, consistent with conformation change upon binding [34].
NMR spectroscopy is another approach taken recently to characterize STARD5 lipid binding (see Chap. 3 of this volume). Similar methods had shown cholesterol binding to STAR and STARD6 , yet the same authors showed that STARD5 did not bind cholesterol or 25-hydroxycholesterol, but rather the bile acids cholic acid and chenodeoxycholic acid [45, 46]. The NMR structure of STARD5 was identical to the X-ray crystal structure, allowing identification of residues in contact with bile acids using the “SAR by NMR” (structure activity relationship) method. Remarkably, the residues all lined the internal lipid binding cavity of STARD5, demonstrating for the first time that a lipid could occupy this cavity. This also presumably rules out long-range allosteric effects or specific binding outside the cavity [46]. In these NMR studies, there was also little perturbation of the overall structure by lipid binding, in contrast to the change in CD spectra above. These authors went on to report the relative affinity of STARD5 for multiple common bile acids , showing STARD5 has the highest affinity—in the physiological range—for unconjugated secondary bile acids without 7α-OH groups (deoxycholic and lithocholic acid) [47]. Overall for STARD5, there is a clear discrepancy between biochemical studies showing cholesterol (and 25-hydroxycholesterol) binding and biophysical NMR studies showing bile acid binding but not cholesterol—even though other START domains of STARD1 and STARD6 bind cholesterol as demonstrated using NMR technology .
STARD4 and STARD5 Activities
There are several pathways in intracellular cholesterol transport that are thought to be non-vesicular and mediated by intracellular transport proteins, including transport to mitochondria mediated by StAR/STARD1 in steroidogenic tissues (reviewed in ref. [48]). Table 7.2 summarizes START domain activities in steroidogenesis and other functional assays, described below. In one of the most common steroidogenesis assays, non-steroidogenic COS-1 cells are transfected with the mitochondrial P450 side change cleavage enzyme system and 3β-hydroxysteroid dehydrogenase. These two enzymes convert cholesterol to pregnenolone then progesterone, which is drastically increased by StAR-like activity of delivering cholesterol to mitochondria. In this cell culture transfection assay, STARD4 and STARD5 increased progesterone production two–threefold, less than the five–sevenfold for STARD1 and STARD3/MLN64 START domains (Fig. 7.5a) [13]. When these same START domains were localized to the cytosolic side of the outer mitochondrial membrane by fusion with the Tom20 protein (as in ref. [49]), then all four had equal steroidogenic activity (Fig. 7.5b, unpublished data from Soccio and Breslow). However, when isolated mitochondria were used for similar steroidogenesis assays, high activity was observed for STARD1, STARD3, and STARD6 , whereas STARD4 had minimal activity and STARD5 had none [43]. When overexpressed by adenovirus in primary mouse hepatocytes, both STARD1 and STARD4 could increase the rate of bile acid synthesis, but STARD5 lacked this activity [44]. Since these cells only use the alternative/acidic pathway of bile acid synthesis initiated by the mitochondrial Cyp27 enzyme, this result is also interpreted as cholesterol delivery to mitochondria . Together, these data suggest that STARD4 and STARD5 may be capable of delivering cholesterol to mitochondria under some conditions, but with less activity than STARD1—even N-62 STAR lacking the mitochondrial import signal.
Some activities common to STARD4, STARD5, STARD1/STAR, and STARD3/MLN64. a A co-transfection steroidogenesis assay was performed in COS-1 cells (see text for details), and the four cholesterol-binding START domains (but not STARD2/PCTP) were able to stimulate progesterone production. STARD1 and STARD3/MLN64 consistently showed higher activity than STARD4 and STARD5. b The same steroidogensis assay was performed with Tom20 fusion proteins localizing the START domains to the cytosolic face of outer mitochondrial membrane. In this case, the four cholesterol-binding START domains had similar activities, with the anti-FLAG tag Western blot in the lower panel showing similar expression levels. c COS-1 cells were transfected with expression plasmids and luciferase reporters for LXR activity, with a pharmacologic LXR agonist serving as the positive control. The four cholesterol-binding START domains (but not PCTP) were able to activate the LXR reporter to various degrees in the absence of agonist. d HEK-293 cells were transfected with an SRE-luciferase reporters to measure endogenous SREBP activity, which is repressed by 25-hydroxycholesterol (25HC) as a positive control. The four cholesterol-binding START domains (but not PCTP) were able to repress SREBP activity. The research in (a) and (c) was adapted from Soccio [13]. © the American Society for Biochemistry and Molecular Biology. (b) and (d) are unpublished data from Soccio and Breslow. MLN64 metastatic lymph node clone 64, START steroidogenic acute regulatory protein (StAR)-related lipid transfer, SREBP sterol regulatory element binding protein
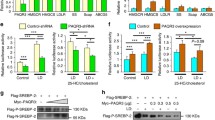
STARD4 and STARD5 may have roles in cholesterol synthesis and esterification, both of which involve the ER, and thus affect cellular levels of free and esterified cholesterol (see Chap. 8 of this volume for further detail). STARD4 overexpression in primary mouse hepatocytes increases neutral lipid staining by Oil Red O and cholesterol ester production from [14C]cholesterol, effects not observed for STARD1 or STARD5 [44]. Remarkably, even isolated mouse liver microsomes showed increased ACAT activity (incorportation of [14C]oleoyl-CoA into cholesterol ester) in the presence of STARD4 but not STARD5 [12]. Notably, overexpressed STARD4 did not affect the production of cholesterol or cholesterol esters from [14C]acetate, showing that STARD4 does not increase cholesterol biosynthesis or the esterification of newly synthesized cholesterol [44]. Together, these data indicate that STARD4 increases esterification of preformed but not newly synthesized cholesterol—suggesting the existence of distinct pools of subcellular cholesterol.
STARD4 knockdown was reported in HepG2 human hepatoma cells by stable short hairpin RNA (shRNA) and resulted in an ~ 50 % decrease in the RNA and protein levels under conditions of high expression in lipoprotein-depleted serum [15]. Consistent with STARD4 delivery of cholesterol to ACAT, STARD4 knockdown cells had ~ 40 % decreased cholesterol ester levels and ~ 60 % decreased ACAT activity (measured by incorporation of [14C]oleate into cholesterol esters). While free cholesterol levels overall were unchanged, filipin staining of free cholesterol showed more cholesterol at the plasma membrane in STARD4 knockdown cells, both in cholesterol-depleted and -replete conditions. Along with apparently more cholesterol in the plasma membrane, there was ~ 70 % less free cholesterol in ER membrane fractions from STARD4 knockdown cells. Furthermore, cholesterol movement to the ERC was studied using DHE and fluorescence recovery after photobleaching (FRAP). In control cells in lipoprotein depleted serum, adding cholesterol resulted in more rapid DHE recovery (t1/2 from 118 to 94 s), while this effect was abrogated in STARD4 knockdown cells (t1/2 not significantly changed from 110 to 117 s). Transferrin recycling was unaffected, indicating that STARD4 selectively affects non-vesicular sterol trafficking to the ERC. STARD5 protein levels were not affected by STARD4 knockdown, but under cholesterol-depleted conditions STARD4 knockdown cells had increased cell surface LDL receptor and decreased NPC1 protein (an endosomal transmembrane cholesterol transporter), suggesting compensatory responses [15].
Elegant experiments involving STARD4 knockdown and overexpression were also reported in U2OS human osteosarcoma cells [19]. Table 7.3 summarizes the consistently opposite effects reported for STARD4 knockdown and overexpression. STARD4 overexpression had many notable effects: ACAT-dependent re-localization of DHE from the plasma membrane to neutral lipid droplets (i.e. esterification), increasing cellular cholesterol esters twofold, and increasing the rate and extent of DHE FRAP in the ERC. In cholesterol-depleted cells upon reloading with cholesterol , SCAP relocates from Golgi to ER resulting in less SREBP activation, and STARD4 accelerated relocation of SCAP—presumably by increasing ER cholesterol. Notably, microinjection of these cells with the nonselective lipid exchanger methylcyclodextrin (MCD) had all of the same effects delivering cholesterol to the ER and ERC. Conversely, small interfering RNA (siRNA) knockdown of STARD4 in U2OS cells increased filipin staining without changing its expression pattern and increased free cholesterol levels by ~ 50 %, and these effects were rescued by expression of an siRNA-resistant STARD4 or by microinjection of MCD. This result can be interpreted based on ER cholesterol sensing by SCAP: lack of STARD4 results in less ER-free cholesterol, such that SREBP2 establishes a higher “set point” for cellular cholesterol. Furthermore, in the setting of cholesterol loading, STARD4 siRNA resulted in defective cholesterol esterification (measured by cholesterol ester content and transfer of DHE to lipid droplets), again rescued by MCD microinjection. Therefore, while STARD4 can apparently mediate transfer of cholesterol to the ER for esterification by ACAT or sensing by SCAP, the non-targeted carrier MCD can have the same effects.
The effects of STARD5 overexpression are different from STARD4. In the same mouse primary hepatocyte system where STARD4 affected cholesterol esterification, overexpression of STARD5 (but not STARD1 or STARD4) increased cellular free cholesterol 12-fold as assessed by filipin staining, similar to what was already observed in primary rat hepatocytes [34, 44]. A correlation between increased STARD5 and higher renal free cholesterol was also observed in the diabetic OVE26 mice [29], and the same positive correlation is seen in human proximal tubule cell lines [50]. However, STARD5 overexpression did not change the distribution of exogenously added DHE in U2OS cells, a system in which STARD4 caused DHE to localize to lipid droplets [19]. In THP-1 human macrophages, overexpression of STARD5 had drastic effects increasing mRNA expression of SREBP-2 and liver X receptor (LXR)α, whereas MLN64/STARD3 and STARD4 did not have these effects [51]. Therefore, while not affecting ACAT, STARD5 appears to affect cellular free cholesterol levels.
The SREBP and LXR transcription factors respond to cellular sterols, so perturbations in cellular sterol metabolism may affect their activities. Consistent with this, overexpression of STARD4 or STARD5 (as well as the START domains of STARD1 and STARD3) could activate luciferase reporters driven by the nuclear receptor LXR (Fig. 7.5c), and this result is interpreted as sterol transport to generate or translocate an LXR ligand oxysterol [13]. Likewise, overexpressed STARD4 and STARD5—again like the STARD1 and STARD3 START domains—could repress SRE driven-reporters, suggesting sterol transport affecting sterol-sensing by SCAP and thus SREBP activity (Fig. 7.5d, unpublished data from Soccio and Breslow). The effect of STARD4 on SREBP-2 processing was tested directly, and both overexpression and knockdown of STARD4 could affect SREBP-2 cleavage in complex ways [19]. Since SREBP-2 activates STARD4, and STARD4 can deliver cholesterol to the ER, then STARD4 negatively feeds back on SREBP activity. Statin drugs rely on SREBP-induced LDL receptor expression to lower serum cholesterol, so statin-induced STARD4 expression (shown in ref. 16) may attenuate this effect such that STARD4 inhibition would increase statin efficacy. If this model is correct, then it is remarkably analogous to Pcsk9, which is induced by SREBP2 but triggers degradation of the LDL receptor, such that Pcsk9 inhibition is desirable to lower serum cholesterol [9]. STARD4 and Pcsk9 may thus both serve as negative feedback “brakes” on the SREBP-2 and LDL receptor pathway (Fig. 7.6).
Like Pcsk9, STARD4 may negatively feedback on the SREBP-2/LDL receptor pathway. a Low levels of ER cholesterol result in proteolytic activation of the SREBP-2 transcription factor. b Mature SREBP-2 activates transcription of genes involved in cholesterol synthesis (HMGCR and other biosynthetic enzymes) and uptake (the LDL receptor), thus restoring cholesterol levels. c SREBP-2 activates Pcsk9, which functions to inactivate the LDL receptor and thus blunt the effect of SREBP-2 on cholesterol uptake. d SREBP-2 also increases expression of STARD4, which can deliver cholesterol to the ER and thus decrease SREBP-2 activity. Widely used statin drugs rely on SREBP-2 mediated LDL receptor activation to lower serum LDL levels, and inhibition of Pcsk9 is known to potentiate this effect. Likewise, inhibition of STARD4 could increase the effects of statins on SREBP-2 and LDL receptor. Pcsk9 proprotein convertase subtilisin/kexin type 9, SREBP sterol regulatory element binding protein, LDL low-density lipoprotein, ER endoplasmic reticulum, HMGCR HMG CoA (3-hydroxy-3-methylglutaryl coenzyme A) reductase
One small study with overexpression of STARD4 in human keratinocytes by transient transfection gave conflicting results, such as decreased incorporation of [14C]acetate into free cholesterol and cholesterol esters, suggesting less cholesterol synthesis [33]. While no changes in “total lipid mass” were noted, it is unclear whether cholesterol esters were measured (the data above suggest STARD4 mediates increased esterification of preexisting cholesterol). There were also changes in gene expression for SREBP-2 (increased, without changes in target genes like HMGCR) and ABCA1 and ABCG4 lipid transporters (though these went in opposite directions despite both being LXR targets induced by oxysterols). These keratinocyte results are difficult to interpret, but support the idea that STARD4 can redistribute cellular sterols to affect lipid metabolic pathways.
In contrast to potential negative effects on statin efficacy, START protein-mediated lipid transfer may have beneficial effects on atherosclerotic disease (see Chap. 5 of this volume). Several studies have ectopically expressed STARD1 in non-steroidogenic cells and tissues that do not normally express high levels, such as hepatocytes, macrophages, and endothelial cells . STARD1 expression in THP-1 macrophages could reduce lipid accumulation and inflammation , likely by increasing LXR signaling and expression of target genes [52]. Similar effects on LXR target genes were seen in endothelial cells [53]. Likewise, viral infection to overexpress STARD1 predominantly in the liver of ApoE null mice resulted in lowering of serum cholesterol, as well as reduction in hepatic steatosis and—most notably—aortic atherosclerotic lesions (Fig. 7.7) [54]. It is likely that effects of STARD1 in these ectopic experimental contexts may mimic the physiology of other related START proteins like STARD4 and STARD5 that are normally expressed in liver and macrophages. Overall, given their potential effects on subcellular cholesterol transport, SREBP processing, and LXR activation, loss of STARD4 or STARD5 protein activity may have complex phenotypes—and in the absence of pharmaceutical inhibitors, knockout mice are an excellent way to study these.
Viral expression of STARD1/STAR decreases atherosclerosis. a ApoE null mice develop aortic atherosclerotic lesions, which stain with Sudan IV dye for neutral lipid in en face preparations. Mice infected with an adenovirus expressing STAR had markedly smaller lesions compared to control mice that were either uninfected (NS) or infected with a control virus (EGFP). b Quantification of lesion area confirms the protective effect of STAR. Notably, in this system STAR is predominantly overexpressed in the liver where it is not normally found, while STARD4 and STARD5 are normally expressed there. Adapted from Ning Y, Xu L, Ren S, Pandak WM, Chen S, Yin L. STAR overexpression decreases serum and tissue lipids in apolipoprotein E-deficient mice. Lipids. 2009 Jun;44(6):511–9. PMID: 19373502
STARD4 Subfamily Knockout Mice
Given that STARD4 can clearly affect cellular cholesterol metabolism, it was hypothesized that the knockout mice might have phenotypes reflecting altered cholesterol transport. However, mice with a whole body null allele for STARD4 were born at normal Mendelian ratios, developed normally, and were apparently healthy with normal male and female fertility and normal litter sizes [16]. Pathological examination (gross and histological) as well as serum chemistries and blood counts failed to show any differences. On chow diet, the STARD4 knockout mice had lower body weights by approximately 2 g (males from weeks 5–12, though females differed significantly only at week 12), but they were also shorter in length such that the body mass index was unchanged. Food intake during weeks 6–8 did not differ significantly . When male mice were placed on a high fat diet for 12 weeks (weeks 8–20), there was no longer any significant weight difference. Dual-energy X-ray absorptiometry (DEXA) scans for body composition at week 8 (prior to high fat diet) and week 20 (after high fat diet) showed no difference in lean, fat, and bone mass. Notably, cholesterol levels in knockout mice did not differ in plasma (total, high-density lipoprotein (HDL), non-HDL, free, esterified) or liver (total, free, and esterified), nor did plasma or liver triglycerides. This lack of difference was seen in both sexes on chow diet, upon statin treatment, or upon cholesterol feeding (with the exception of some small decrease in serum total, LDL, and esterified cholesterol in female mice on high cholesterol diet). Fasting glucose and glucose tolerance were also unchanged. A subtle difference was reported in the bile, which appeared ~ 25 % more dilute in female knockout mice (based on biliary cholesterol , phospholipid, and bile acids , though the last component was not significantly different), but this difference was only seen in female but not male mice, and measurement was only reported on the chow diet, not the other diets tested. Notably, microarray profiling of liver gene expression on a cholesterol-free diet showed no significant differentially regulated genes in the knockout mice . Quantitative reverse transcription (RT)-PCR analysis of candidate genes showed some small differences that were not consistent across diets (i.e., NPC1 was decreased in knockout mice on a 0.0 % cholesterol diet, but not with lovastatin or 0.5 % cholesterol, while STARD5 was slightly increased in knockout mice on statin and 0.5 % cholesterol but not on 0.0 % cholesterol). In conclusion, any of the reported differences between wild-type and STARD4 knockout mice are subtle and inconsistent across conditions (gender, diet, etc.), suggesting that they could merely arise from chance variation when so many parameters were tested in so many combinations. This lack of any overall apparent phenotype suggests that STARD4 functions can be replaced by other redundant cellular proteins [16].
Despite the overall lack of a phenotype in STARD4 null mice, these animals nonetheless merit further study. For instance, stressing the mice with obesity and metabolic syndrome (such as high fat diet-fed or ob/ob mice, in which serum and hepatic lipids are markedly increased) or atherosclerosis (such as ApoE or LDL receptor null mice, in which macrophage cholesterol handling affects lesion progression) may unmask phenotypes. To address potential redundancy among START family members, combined knockout of STARD4 could be generated with STARD1, STARD3, or STARD5. Likewise, studies of isolated STARD4 null primary macrophages or hepatocytes may reveal effects on cholesterol uptake, esterification, or efflux that are compensated in the whole animal. Tissue specific knockouts could also be generated by using the existing LoxP-flanked STARD4 allele with different Cre recombinase drivers, and acute hepatic knockout by infection with adenoviral Cre could reveal effects prior to compensatory mechanisms . Furthermore, studies of mice lacking STARD5 and STARD6 should be undertaken, particularly as these gene-targeted mice are now available via the International Knockout Mouse Consortium [55]. One obvious question is whether male mice lacking STARD6 are fertile, as some defect in sperm maturation or function may exist (see below). Likewise, an effect on bile acid homeostasis in STARD5 null mice would support the notion that STARD5 binds these cholesterol-derived molecules .
What About STARD6?
There have been many fewer studies of STARD6 compared to the other subfamily members STARD4 and STARD5. STARD6 expression by Northern Blot was observed only in the testis [7], and subsequent in situ hybridization studies in rats further localized the mRNA to male germ cells, particularly the maturing round and elongated spermatids [56]. An anti-STARD6 antibody confirmed testis-specific expression by Western Blot, and immunostaining in round and elongated spermatids [56]. This agrees with unpublished observations that STARD6 mRNA is neither detected in MA-10 or freshly isolated Leydig cells nor in the testis of Kit W/Wv mice [57] deficient in germ cells (Fig. 7.8, unpublished data from Soccio and Breslow). There is one conflicting report of STARD6 immunostaining of interstitial Leydig cells of hypothyroid but not control rats [58], but no mRNA or Western analysis was performed to confirm this unexpected localization in Leydig cells. STARD6 protein transfers cholesterol to mitochondria as efficiently as STARD1 [43], and notably the STARD6 gene falls in a mouse quantitative trail locus that affects activity of sperm mitochondrial diaphorase enzyme activity [59]. The localization of STARD6 in mature sperm has not been reported, and this will be informative as mitochondria localize to the midpiece. Cholesterol and its precursor sterols have several essential roles in sperm maturation and function (reviewed in ref. 7 and 56), and STARD6 knockout mice may have impaired fertility due to altered cholesterol metabolism in germ cells.
STARD6 mRNA is absent in germ cell deficient testis from KitW/Wv mice. a Northern blots show abundant STARD6 and StAR mRNA in testis, but none in liver Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) is an RNA loading control). In the testis of KitW/Wv mice which lack germ cells, STARD1 mRNA from Leydig cells remains present, while STARD6 mRNA is absent. The testis of littermate controls with germ cells show mRNA for both STARD6 and STARD1. b Quantitative RT-PCR analysis of STAR and STARD6 expression in testis of the same mice. Unpublished data from Soccio and Breslow. StAR steroidogenic acute regulatory protein, mRNA messenger ribonucleic acid, RT-PCR reverse transcription polymerase chain reaction
STARD6 expression in brain has reported by immunohistochemistry, in nuclei but not cytosol, of neurons and glial cells throughout the central and peripheral nervous system [60]. These authors used the same antibody described above, and could not explain the discrepancy that it failed to detect a band by Western blot in brain [56], nor that STARD6 mRNA is undetectable in brain [7]. Nonetheless, this group has gone on to publish a series of reports about STARD6 immunolocalization in various brain regions. STARD6 immunostaining in rat hippocampus was observed in the nucleus and cytoplasm and increased transiently after induction of seizures [61] or exitotoxicity [62], an effect actually confirmed by Western blots on hippocampal protein, and the localization was distinct from that of STARD1 immunolocalization [63]. STARD1 and STARD6 immunostaining were also reported in Purkinje cells of the cerebellum, potentially responding differentially to hypothyroidism [64]. A potential role in neurosteroid production is proposed, but this remains entirely speculative.
One report implicates STARD6 in non-small cell lung cancer cells response to the chemotherapeutic agent paclitaxel [65]. While this raises the interesting issue of tumors ectopically express spermatogenic proteins that may be functional in neoplasia, there are no other reports of STARD6 expression and function in tumors.
Regulatory elements mediating the male germ cell specific expression of STARD6 have not been identified. Notably, the STARD6 mRNA suggests many levels of regulation. 5′ RACE from testis cDNA identified multiple alternative first exons (three in mouse, two in human) as well as multiple initiation sites in these exons (unpublished data from Soccio and Breslow), showing alternative initiation and splicing. Also notable is the long multiexon 5′ UTR, which includes many potential ATG start codons which cannot encode the START protein, as a TAG stop codon immediately precedes the ATG for that open reading frame [7, 56]. Such upstream ATGs are rare in eukaryotic mRNAs and are thought to mediate translational regulation (reviewed in ref 56).
C. elegans Have One STARD4 Subfamily Member
While mammals have the three STARD4 subfamily members, the nematode worm C. elegans has only one STARD4 subfamily homolog, named K02D3.2, among its six START domain encoding genes [5]. (In comparison, the fruit fly Drosophila melanogaster has four START domain-encoding genes, and none of these is most similar to the mammalian STARD4 subfamily.) The K02D3.2 gene is ~ 23 % identical to mammalian STARD5, and only ~ 17 % to STARD4 and STARD6 . No publications have specifically studied this gene, though two large screens have given suggestive results. First, siRNA knockdown of K02D3.2 resulted in decreased fat storage in the gut lining cells [66]. Second, a genome wide map of protein-protein interactions by yeast two-hydrid showed K02D3.2 binding to T01D1.6 (abu-11) [67], which is activated when the ER stress response is blocked [68]. The expression pattern of K02D3.2 is not published, though a GFP reporter driven by ~ 2.7 kb of its promoter was expressed in seam cells of the embryos and larvae, though it was not affected by cholesterol depletion or tunicamycin treatment (unpublished data, Soccio and Breslow in collaboration with Elliot Perens and Shai Shaham at the Rockefeller University). Seam cells actively secrete the cuticle covering the worm, so they may have high physiological levels of ER stress due to active protein secretion, but this is entirely speculative. Finally, K02D3.2 itself has not been implicated in the ER stress response in nematodes. Further studies specifically addressing K02D3.2 will be necessary to determine the role of this STARD4 subfamily protein in nematodes.
Summary
Despite a decade of work and dozens of publications, there are many unanswered questions regarding the STARD4 subfamily of START proteins. STARD6 expression in male germ cells clearly indicates a role in sperm and male fertility, while the consistent activation of STARD4 by SREBPs and STARD5 by ER stress suggests other potential functions. The preponderance of evidence generally indicates that all three proteins can bind and transfer cholesterol itself, while the results are inconsistent for related molecules (precursors, oxysterols, and bile acids) . Given important roles for these other molecules in metabolism, ligand binding by these START proteins certainly merits further study, particularly comparative efforts looking at panels of START domains in the same assay system. STARD4 can clearly mediate intracellular cholesterol movement, particularly to ER-resident ACAT for esterification, though many of the cellular effects can be mimicked by non-selective transport by cyclodextrin. The STARD4 null mice have unchanged levels of cholesterol and cholesterol ester in liver and plasma, suggesting that other redundant sterol transfer mechanisms may exist. While not affecting esterification, STARD5 can have other drastic effects on cellular cholesterol metabolism that require further characterization, such as large increases in free cholesterol staining with filipin. STARD4 and STARD5 appear to be widely expressed cytosolic proteins, though two intriguing aspects of STARD5 localization deserve careful study: the potential selective localization of STARD5 to macrophage and immune cells and the potential translocation from nucleus to cytoplasm upon ER stress.
In conclusion, there has been considerable progress in the understanding of the STARD4 subfamily’s cellular and subcellular localization, regulation, lipid binding and transfer, and effects of knockdown and overexpression. This progress has yet to reveal their functions in normal and disease physiology, but many of the reagents and animal models to probe these questions now exist. There is a trend towards more publications naming STARD4 and/or STARD5 in their titles, as more than half of these have appeared since 2010 (10 of 19 total, with the other 9 from 2002–2009). The next decade of research on the STARD4 subfamily will hopefully show even more accelerated progress.
Abbreviations
- ACAT:
-
Acyl-coenzyme A Cholesterol Acyltransferase
- ATF6:
-
Activating Transcription Factor 6
- CD:
-
Circular Dichroism
- ChIP-seq:
-
Chromatin Immunoprecipitation Followed by Deep Sequencing
- CHOP:
-
CCAAT/Enhancer-binding Protein Homologous Protein
- DHE:
-
Dihydroergosterol
- ER:
-
Endoplasmic Reticulum
- ERC:
-
Endocytic Recycling Compartment
- EST:
-
Expressed Sequence Tag
- FRAP:
-
Fluorescence Recovery after Photobleaching
- GRP94:
-
Glucose-regulated Protein 94
- Herp:
-
Homocysteine-induced Endoplasmic Reticulum Protein
- HMGCR:
-
HMG CoA (3-hydroxy-3-methylglutaryl coenzyme A) Reductase
- INSIG:
-
Insulin-induced Gene
- LDL:
-
Low-density Lipoprotein
- MCD:
-
Metylcyclodextrin
- NMR:
-
Nuclear Magnetic Resonance
- SCAP:
-
SREBP Cleavage-activating Protein
- SREBP:
-
Sterol Regulatory Element Binding Protein
- StAR:
-
Steroidogenic Acute Regulatory Protein
- STARD4:
-
START Domain Containing 4
- STARD5:
-
START Domain Containing 5
- STARD6:
-
START Domain Containing 6
- START:
-
StAR-related Lipid Transfer
- UTR:
-
Untranslated Region
- Xbp1:
-
X-box Binding Protein 1
References
Lin D, Sugawara T, Strauss JF 3rd, Clark BJ, Stocco DM, Saenger P, Rogol A, Miller WL. Role of steroidogenic acute regulatory protein in adrenal and gonadal steroidogenesis. Science. 1995 March 24;267(5205):1828–31. PMID: 7892608.
Ponting CP, Aravind L. START: a lipid-binding domain in StAR, HD-ZIP and signalling proteins. Trends Biochem Sci. 1999 April;24(4):130–2. PMID: 10322415.
Tsujishita Y, Hurley JH. Structure and lipid transport mechanism of a StAR-related domain. Nat Struct Biol. 2000 May;7(5):408–14. PMID: 10802740.
Breslow JL. Human apolipoprotein molecular biology and genetic variation. Annu Rev Biochem. 1985;54:699–727. PMID: 3896129.
Soccio RE, Breslow JL. StAR-related lipid transfer (START) proteins: mediators of intracellular lipid metabolism. J Biol Chem. 2003 June 20;278(25):22183–6. PMID: 12724317.
Tiemann M, Han Z, Soccio R, Bollineni J, Shefer S, Sehayek E, Breslow JL. Cholesterol feeding of mice expressing cholesterol 7alpha-hydroxylase increases bile acid pool size despite decreased enzyme activity. Proc Natl Acad Sci U S A. 2004 Feb 17;101(7):1846–51. PMID: 14762172.
Soccio RE, Adams RM, Romanowski MJ, Sehayek E, Burley SK, Breslow JL. The cholesterol-regulated StarD4 gene encodes a StAR-related lipid transfer protein with two closely related homologues, StarD5 and StarD6. Proc Natl Acad Sci U S A. 2002 May 14;99(10):6943–8. PMID: 12011452.
Maxwell KN, Soccio RE, Duncan EM, Sehayek E, Breslow JL. Novel putative SREBP and LXR target genes identified by microarray analysis in liver of cholesterol-fed mice. J Lipid Res. 2003 Nov;44(11):2109–19. PMID: 12897189.
Petrides F, Shearston K, Chatelais M, Guilbaud F, Meilhac O, Lambert G. The promises of PCSK9 inhibition. Curr Opin Lipidol. 2013 Aug;24(4):307–12. PMID: 23817198.
Sahakitrungruang T, Soccio RE, Lang-Muritano M, Walker JM, Achermann JC, Miller WL. Clinical, genetic, and functional characterization of four patients carrying partial loss-of-function mutations in the steroidogenic acute regulatory protein (StAR). J Clin Endocrinol Metab. 2010 Jul;95(7):3352–9. PMID: 20444910.
Radhakrishnan A, Goldstein JL, McDonald JG, Brown MS. Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: a delicate balance. Cell Metab. 2008 Dec;8(6):512–21. PMID: 19041766.
Rodriguez-Agudo D, Calderon-Dominguez M, Ren S, Marques D, Redford K, Medina-Torres MA, Hylemon P, Gil G, Pandak WM. Subcellular localization and regulation of StarD4 protein in macrophages and fibroblasts. Biochim Biophys Acta. 2011 Oct;1811(10):597–606. PMID: 21767660.
Soccio RE, Adams RM, Maxwell KN, Breslow JL. Differential gene regulation of StarD4 and StarD5 cholesterol transfer proteins. Activation of StarD4 by sterol regulatory element-binding protein-2 and StarD5 by endoplasmic reticulum stress. J Biol Chem. 2005 May 13;280(19):19410–8. PMID: 15760897.
Cunningham D, Swartzlander D, Liyanarachchi S, Davuluri RV, Herman GE. Changes in gene expression associated with loss of function of the NSDHL sterol dehydrogenase in mouse embryonic fibroblasts. J Lipid Res. 2005 June;46(6):1150–62. PMID: 15805545.
Garbarino J, Pan M, Chin HF, Lund FW, Maxfield FR, Breslow JL. STARD4 knockdown in HepG2 cells disrupts cholesterol trafficking associated with the plasma membrane, ER, and ERC. J Lipid Res. 2012 Dec;53(12):2716–25. PMID: 23033213.
Riegelhaupt JJ, Waase MP, Garbarino J, Cruz DE, Breslow JL. Targeted disruption of steroidogenic acute regulatory protein D4 leads to modest weight reduction and minor alterations in lipid metabolism. J Lipid Res. 2010 May;51(5):1134–43. PMID: 19965609.
Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci U S A. 2003 Oct 14;100(21):12027–32. PMID: 14512514.
Seo YK, Jeon TI, Chong HK, Biesinger J, Xie X, Osborne TF. Genome-wide localization of SREBP-2 in hepatic chromatin predicts a role in autophagy. Cell Metab. 2011 April 6;13(4):367–75. PMID: 21459322.
Mesmin B, Pipalia NH, Lund FW, Ramlall TF, Sokolov A, Eliezer D, Maxfield FR. STARD4 abundance regulates sterol transport and sensing. Mol Biol Cell. 2011 Nov;22(21):4004–15. PMID: 21900492.
Korytowski W, Rodriguez-Agudo D, Pilat A, Girotti AW. StarD4-mediated translocation of 7-hydroperoxycholesterol to isolated mitochondria: deleterious effects and implications for steroidogenesis under oxidative stress conditions. Biochem Biophys Res Commun. 2010 Jan 29;392(1):58–62. PMID: 20059974.
Yamada S, Yamaguchi T, Hosoda A, Iwawaki T, Kohno K. Regulation of human STARD4 gene expression under endoplasmic reticulum stress. Biochem Biophys Res Commun. 2006 May 19;343(4):1079–85. PMID: 16579971.
Rodriguez-Agudo D, Calderon-Dominguez M, Medina MA, Ren S, Gil G, Pandak WM. ER stress increases StarD5 expression by stabilizing its mRNA and leads to relocalization of its protein from the nucleus to the membranes. J Lipid Res. 2012 Dec;53(12):2708–15. PMID: 23053693.
Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, Brown MS, Goldstein JL. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000 Dec;6(6):1355–64. PMID: 11163209.
Zeng L, Lu M, Mori K, Luo S, Lee AS, Zhu Y, Shyy JY. ATF6 modulates SREBP2-mediated lipogenesis. EMBO J. 2004 Feb 25;23(4):950–8. PMID: 14765107.
Skiadas CC, Duan S, Correll M, Rubio R, Karaca N, Ginsburg ES, Quackenbush J, Racowsky C. Ovarian reserve status in young women is associated with altered gene expression in membrana granulosa cells. Mol Hum Reprod. 2012 July;18(7):362–71. PMID: 22355044.
Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, Liu J, Zhao XD, Chew JL, Lee YL, Kuznetsov VA, Sung WK, Miller LD, Lim B, Liu ET, Yu Q, Ng HH, Ruan Y. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006 Jan 13;124(1):207–19. PMID: 16413492.
Bazuine M, Stenkula KG, Cam M, Arroyo M, Cushman SW. Guardian of corpulence: a hypothesis on p53 signaling in the fat cell. Clin Lipidol. 2009 April 1;4(2):231–43. PMID: 20126301.
ENCODE Project Consortium, Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012 Sept 6;489(7414):57–74. PMID: 22955616.
Chen YC, Meier RK, Zheng S, Khundmiri SJ, Tseng MT, Lederer ED, Epstein PN, Clark BJ. Steroidogenic acute regulatory-related lipid transfer domain protein 5 localization and regulation in renal tubules. Am J Physiol Renal Physiol. 2009 Aug;297(2):F380–8. PMID: 19474188.
Ishikawa T, Hwang K, Lazzarino D, Morris PL. Sertoli cell expression of steroidogenic acute regulatory protein-related lipid transfer 1 and 5 domain-containing proteins and sterol regulatory element binding protein-1 are interleukin-1beta regulated by activation of c-Jun N-terminal kinase and cyclooxygenase-2 and cytokine induction. Endocrinology. 2005 Dec;146(12):5100–11. PMID: 16123165.
Alpy F, Tomasetto C. Give lipids a START: the StAR-related lipid transfer (START) domain in mammals. J Cell Sci. 2005 July 1;118(Pt 13):2791–801. PMID: 15976441.
Alpy F, Tomasetto C. START ships lipids across interorganelle space. Biochimie. 2013 Sept 24. [Epub ahead of print] PMID: 24076129.
Elbadawy HM, Borthwick F, Wright C, Martin PE, Graham A. Cytosolic StAR-related lipid transfer domain 4 (STARD4) protein influences keratinocyte lipid phenotype and differentiation status. Br J Dermatol. 2011 March;164(3):628–32. PMID: 20969562.
Rodriguez-Agudo D, Ren S, Hylemon PB, Redford K, Natarajan R, Del Castillo A, Gil G, Pandak WM. Human StarD5, a cytosolic StAR-related lipid binding protein. J Lipid Res. 2005 Aug;46(8):1615–23. PMID: 15897605.
Rodriguez-Agudo D, Ren S, Hylemon PB, Montanez R, Redford K, Natarajan R, Medina MA, Gil G, Pandak WM. Localization of StarD5 cholesterol binding protein. J Lipid Res. 2006 June;47(6):1168–75. PMID: 16534142.
Strauss JF 3rd, Kishida T, Christenson LK, Fujimoto T, Hiroi H. START domain proteins and the intracellular trafficking of cholesterol in steroidogenic cells. Mol Cell Endocrinol. 2003 April 28;202(1–2):59–65. PMID: 12770731.
Romanowski MJ, Soccio RE, Breslow JL, Burley SK. Crystal structure of the Mus musculus cholesterol-regulated START protein 4 (StarD4) containing a StAR-related lipid transfer domain. Proc Natl Acad Sci U S A. 2002 May 14;99(10):6949–54. PMID: 12011453.
Thorsell AG, Lee WH, Persson C, Siponen MI, Nilsson M, Busam RD, Kotenyova T, Schuler H, Lehtio L. Comparative structural analysis of lipid binding START domains. PLoS One. 2011;6(6):e19521. PMID: 21738568.
Dikiy I, Ramlall TF, Eliezer D. 1H, 13C, and 15N backbone resonance assignments of the L124D mutant of StAR-related lipid transfer domain protein 4 (StARD4). Biomol NMR Assign. 2013 Oct;7(2):245–8. PMID: 22918595.
Lorin A, Latourneau D, Lefebvre A, LeHoux JG, Lavigne P. 1H, 13C, and 15N backbone chemical shift assignments of StAR-related lipid transfer domain protein 5 (STARD5). Biomol NMR Assign. 2013 April;7(1):21–4. PMID: 22392336.
Lavigne P, Najmanivich R, Lehoux JG. Mammalian StAR-related lipid transfer (START) domains with specificity for cholesterol: structural conservation and mechanism of reversible binding. Subcell Biochem. 2010;51:425–37. PMID: 20213553.
Chen BY, Honig B. VASP: a volumetric analysis of surface properties yields insights into protein-ligand binding specificity. PLoS Comput Biol. 2010 Aug 12;6(8):e1000881. PMID: 20814581.
Bose HS, Whittal RM, Ran Y, Bose M, Baker BY, Miller WL. StAR-like activity and molten globule behavior of StARD6, a male germ-line protein. Biochemistry. 2008 Feb 26;47(8):2277–88. PMID: 18211099.
Rodriguez-Agudo D, Ren S, Wong E, Marques D, Redford K, Gil G, Hylemon P, Pandak WM. Intracellular cholesterol transporter StarD4 binds free cholesterol and increases cholesteryl ester formation. J Lipid Res. 2008 July;49(7):1409–19. PMID: 18403318.
Latourneau D, Lorin A, Lefebvre A, Frappier V, Gaudreault F, Najmanovich R, Lavigne P, LeHoux JG. StAR-related lipid transfer domain protein 5 binds primary bile acids. J Lipid Res. 2012 Dec;53(12):2677–89. PMID: 23018617.
Latourneau D, Lorin A, Lefebvre A, Cabana J, Lavigne P, Lehoux JG. Thermodynamic and solution state NMR characterization of the binding of secondary and conjugated bile acids to STARD5. Biochim Biophys Acta. 2013 July 16;1831(11):1589–99. PMID: 23872533.
Latourneau D, Lefebvre A, Lavigne P, LeHoux JG. STARD5 specific ligand binding: comparison with STARD1 and STARD4 subfamilies. Mol Cell Endocrinol. 2013 May 22;371(1–2):20–5. PMID: 23337244.
Soccio RE, Breslow JL. Intracellular cholesterol transport. Arterioscler Thromb Vasc Biol. 2004 July;24(7):1150–60. PMID: 15130918.
Bose HS, Lingappa VR, Miller WL. Rapid regulation of steroidogenesis by mitochondrial protein import. Nature. 2002 May 2;417(6884):87–91. PMID: 11986670.
Clark BJ. The mammalian START domain protein family in lipid transport in health and disease. J Endocrinol. 2012 March;212(3):257–75. PMID: 21965545.
Borthwick F, Allen AM, Taylor JM, Graham A. Overexpression of STARD3 in human monocyte/macrophages induces an anti-atherogenic lipid phenotype. Clin Sci (Lond). 2010 June 22;119(7):265–72. PMID: 20491656.
Ning Y, Bai Q, Lu H, Li X, Pandak WM, Zhao F, Chen S, Ren S, Yin L. Overexpression of mitochondrial cholesterol delivery protein, StAR, decreases intracellular lipids and inflammatory factors secretion in macrophages. Atherosclerosis. 2009 May;204(1):114–20. PMID: 18945429.
Ning YX, Ren SL, Zhao FD, Yin LH. Overexpression of the steroidogenic acute regulatory protein increases the expression of ATP-binding cassette transporters in microvascular endothelial cells (bEnd.3). J Zhejiang Univ Sci B. 2010 May;11(5):350–6. PMID: 20443213.
Ning Y, Xu L, Ren S, Pandak WM, Chen S, Yin L. StAR overexpression decreases serum and tissue lipids in apolipoprotein E-deficient mice. Lipids. 2009 June;44(6):511–9. PMID: 19373502.
Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, Jackson D, Severin J, Biggs P, Fu J, Nefedov M, de Jong PJ, Stewart AF, Bradley A. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011 June 5;474(7351):337–42. PMID: 21677750.
Gomes C, Oh SD, Kim JW, Chun SY, Lee K, Kwon HB, Soh J. Expression of the putative sterol binding protein Stard6 gene is male germ cell specific. Biol Reprod. 2005 March;72(3):651–8. PMID: 15564601.
Ohta H, Tohda A, Nishimune Y. Proliferation and differentiation of spermatogonial stem cells in the w/wv mutant mouse testis. Biol Reprod. 2003 Dec;69(6):1815–21. PMID: 12890724.
Chang IY, Shin SY, Kim JW, Yu JM, Kim JS, Song PI, Yoon SP. The changed immunolocalization of START-domain-containing 6 (StarD6) during the development of testes in rat perinatal hypothyroidism. Acta Histochem. 2007;109(4):315–21. PMID: 17462719.
Golas A, Malek P, Piasecka M, Styrna J. Sperm mitochondria diaphorase activity–a gene mapping study of recombinant inbred strains of mice. Int J Dev Biol. 2010;54(4):667–73. PMID: 20209439.
Chang IY, Kim JH, Hwang G, Song PI, Song RJ, Kim JW, Yoon SP. Immunohistochemical detection of StarD6 in the rat nervous system. Neuroreport. 2007 Oct 8;18(15):1615–9. PMID: 17885612.
Chang IY, Kim JK, Lee SM, Kim JN, Soh J, Kim JW, Yoon SP. The changed immunoreactivity of StarD6 after pilocarpine-induced epilepsy. Neuroreport. 2009 July 1;20(10):963–7. PMID: 19434006.
Chang IY, Kim JH, Cho KW, Yoon SP. Acute responses of DNA repair proteins and StarD6 in rat hippocampus after domoic acid-induced excitotoxicity. Acta Histochem. 2013 April;115(3):234–9. PMID: 22883302.
Chang IY, Jeon YJ, Jung SM, Jang YH, Ahn JB, Park KS, Yoon SP. Does the StarD6 mark the same as the StAR in the nervous system? J Chem Neuroanat. 2010 Nov;40(3):239–42. PMID: 20609383.
Chang IY, Ohn T, Ko GS, Yoon Y, Kim JW, Yoon SP. Immunolocalization of steroidogenic acute regulatory protein-related lipid transfer (START) domain-containing proteins in the developing cerebellum of normal and hypothyroid rats. J Chem Neuroanat. 2012 Jan;43(1):28–33. PMID: 22024186.
Cappell KM, Sinnott R, Taus P, Maxfield K, Scarbrough M, Whitehurst AW. Multiple cancer testis antigens function to support tumor cell mitotic fidelity. Mol Cell Biol. 2012 Oct;32(20):4131–40. PMID: 22869527.
Ashrafi K, Chang FY, Watts JL, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature. 2003 Jan 16;421(6920):268–72. PMID: 12529643.
Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, Goldberg DS, Li N, Martinez M, Rual JF, Lamesch P, Xu L, Tewari M, Wong SL, Zhang LV, Berriz GF, Jacotot L, Vaglio P, Reboul J, Hirozane-Kishikawa T, Li Q, Gabel HW, Elewa A, Baumgartner B, Rose DJ, Yu H, Bosak S, Sequerra R, Fraser A, Mango SE, Saxton WM, Strome S, Van Den Heuvel S, Piano F, Vandenhaute J, Sardet C, Gerstein M, Doucette-Stamm L, Gunsalus KC, Harper JW, Cusick ME, Roth FP, Hill DE, Vidal M. A map of the interactome network of the metazoan C. elegans. Science. 2004 Jan 23;303(5657):540–3. PMID: 14704431.
Urano F, Calfon M, Yoneda T, Yun C, Kiraly M, Clark SG, Ron D. A survival pathway for Caenorhabditis elegans with a blocked unfolded protein response. J Cell Biol. 2002 Aug 19;158(4):639–46. PMID: 12186849.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer Science+Business Media New York
About this chapter
Cite this chapter
Soccio, R. (2014). The STARD4 Subfamily: STARD4 and STARD5 in Cholesterol Metabolism. In: Clark, B., Stocco, D. (eds) Cholesterol Transporters of the START Domain Protein Family in Health and Disease. Springer, New York, NY. https://doi.org/10.1007/978-1-4939-1112-7_7
Download citation
DOI: https://doi.org/10.1007/978-1-4939-1112-7_7
Published:
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-4939-1111-0
Online ISBN: 978-1-4939-1112-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)