Abstract
In this chapter, we present an accurate automated 3D liver segmentation scheme for measuring liver volumes in MR images. Our scheme consisted of five steps. First, an anisotropic diffusion smoothing filter was applied to T1-weighted MR images of the liver in the portal-venous phase to reduce noise while preserving the liver boundaries. An edge enhancer and a nonlinear gray-scale converter were applied to enhance the liver boundary. This boundary-enhanced image was used as a speed function for a 3D fast marching algorithm to generate an initial surface that roughly approximated the liver shape. A 3D geodesic active contour segmentation algorithm refined the initial surface so as to more precisely determine the liver boundary. The liver volume was calculated based on the refined liver surface. The MR liver volumetry based on our automated scheme agreed excellently with “gold-standard” manual volumetry (intra-class correlation coefficient was 0.98) and required substantially less completion time (our processing time of 1 vs. 24 min/case in manual segmentation).
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
Introduction
Medical and surgical advancements have brought the global success of liver transplantation with the increasing survival rates after transplantation in the past decades [1–3]. One of the important assessments contributing to the success of a transplantation procedure is the estimation for total and segmental liver volumes. It is a major factor to predict the safe outcome for both donor and recipient. A minimum of 40 % of the standard liver mass is required by recipient while 30–40 % of the original volume is remained for donor to survive [4]. Hence, an accurate estimation of liver volumes is necessary for planning liver transplantation [5, 6]. Noninvasive measurement methods have been revealed by the advanced imaging technologies such as CT and MRI. Manual tracing of the liver on CT images is a current gold-standard method. Although the manual tracing method can obtain accurate results, it is subjective, tedious, and time-consuming. It takes 20–48 min to obtain the liver volume for one patient [7, 8]. In addition, the relatively large intraobserver and interobserver variations still occur in the manual method. To address this issue, the automated liver segmentation has been developed with image analysis techniques, and it has become an important research topic.
Several approaches to computerized liver segmentation on CT images have been published, including image-processing techniques such as thresholding, histogram analysis, morphological operations, and their combinations [9, 10]. A comparison between the semiautomatic liver volumetry and manual method in the living liver donors was presented by Hermoye et al. [11]. An automated scheme based on the combination of thresholding, feature analysis, and region growing was proposed by Nakayama et al. [8]. In comparison with manual tracing, it achieved a correlation coefficient of 0.883. Okada et al. [12] developed an automated scheme based on a probabilistic atlas and a statistical shape model, its performance was evaluated with eight cases. Selver et al. [13] developed a three-stage automated liver segmentation scheme consisting of preprocessing for excluding neighboring structures, k-means clustering, multilayer perceptron for classification, and postprocessing for removing mis-segmented objects and smoothing liver contours. The scheme was evaluated on 20 cases. An iterative graph-cut active shape model was developed by Chen and Bagci [14]. Their scheme combined the statistical shape information embodied in the active shape model with the globally optimal delineation capacity of the graph-cut method. Suzuki et al. [7, 15] developed a computer-aided liver volumetry scheme by means of geodesic active contour segmentation coupled with level set algorithms. They compared their automated scheme with manual segmentation and commercially available interactive software. Their scheme achieved the performance comparable to manual segmentation, while reducing the time required for volumetry by a factor of approximately 70.
In comparison with CT-based schemes, there are fewer publications for an automated liver segmentation scheme on MR images in spite of no risk for ionizing radiation, probably because it is believed that MR liver volumetry has more variations and more difficult than CT. Karlo et al. [16] compared the CT- and MRI-based volumetry of the resected liver specimens with intraoperative volume and weight measurements to calculate conversion factors. A semiautomated dual-space clustering segmentation method was proposed by Farraher et al. [17]. Their semiautomated method required manual drawing of a small region-of-interest (ROI) on the liver first; and then it iteratively evaluated temporal liver segmentations with the repeated adjustment of parameters to obtain the final liver segmentation result. Rusko and Bekes [18] proposed a partitioned probabilistic model to represent the liver. In this model, the liver was partitioned into multiple regions, and the different intensity statistical models were applied to these regions. The scheme was tested on eight cases. Gloger et al. [19] developed a three-step segmentation method based on a region-growing approach, linear discriminant analysis, and probability maps. Their method was evaluated with 20 normal cases and 10 fat cases. It achieved a true-positive volume error (TPVE) of 8.3 % with an average execution time of 11.2 min for each normal case, and a TPVE of 11.8 % with an average execution time of 15.4 min for each fat case.
Although the above studies showed a promise, there is still room for developing the computerized liver segmentation in MRI to make it a routine clinical use. In this chapter, we present an automated liver segmentation scheme in MRI based on geodesic active contour model and fast marching algorithm. The performance of our scheme was evaluated on 23 cases, and the comparison between the computerized volumetry and gold-standard manual volumetry was performed.
Materials and Methods
Liver MRI Datasets
In this study, 23 patients were scanned in the supine position with a 1.5T MRI scanners (Signa HDx/HDxt, GE Medical Systems, Milwaukee, WI; and Achieva, Philips Medical Systems, Cleveland, OH) at the University of Chicago Medical Center. Intravenous gadolinium contrast agent (8–20 mL; mean: 15.3 ± 4.2) was administrated. The post-contrast MRIs were obtained by using the T1-weighted liver acquisition with volume acceleration (LAVA) or T1-weighted high-resolution isotropic volume examination (THRIVE) sequence. The flip angle of 10° was used in context with TR and TE ranged from 3.48 to 3.92 ms and from 1.64 to 1.84 ms, respectively. The scanning parameters included collimation of 5 mm (for the GE system) or 4 mm (for the Philips system) and reconstruction intervals of 2.5 mm (for the GE system) or 2 mm (for the Philips system). Each MR slice had the matrix size of 256 × 256 pixels with an in-plane pixel size ranged from 1.17 to 1.72 mm. The 23 cases in our database had liver diseases.
The manual contours were traced carefully by a broad-certificated abdominal radiologist on each slice containing the liver. The number of slices in each case ranged from 88 to 120 (average: 97.9). The liver volume was calculated by multiplying the areas of the manually traced regions in each slice by the reconstruction interval. Note that the collimation was different from the reconstruction interval and that consecutive slices overlapped. The total liver volume of each case was obtained from the summation of volumes in all slices. We also recorded the time required for the completion of the manual contour tracing. The performance of our computerized liver extraction scheme was evaluated by using manual liver volumes as the “gold standard.”
Computer-Based Measurement Scheme for MR Liver Volumes
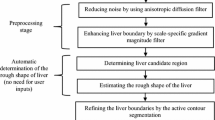
A computerized scheme employing level set algorithms coupled with geodesic active contour segmentation was proposed by our group for CT liver extraction. In this chapter, we present a scheme for the automated liver segmentation on MR images based on the knowledge and techniques acquired in the development of our CT liver extraction scheme. Our MR liver extraction scheme applied to the portal-venous-phase images in T1-weighted (T1w) sequences consists of five steps, as shown in Fig. 6.1. First, a 3D MR volume I(x,y,z) consisting of portal-venous-phase images must be processed to reduce noise and enhance liver structures. This was accomplished by using an anisotropic diffusion algorithm (which is also called nonuniform or variable conductance diffusion proposed by Perona and Malik [20]). The algorithm based on the modified curvature diffusion equation is given by
where c(∙) is a fuzzy cutoff function that reduces conductance at areas of large ∣∇I∣. It can be any of a number of functions. The literature suggested
to be effective. Note that this term introduces a free parameter κ, the diffusion coefficient, which controls the sensitivity of edge contrast. The anisotropic diffusion algorithm smoothes noise in the image while preserving the major liver structures such as major vessels and the liver boundaries. The noise-reduced image was then passed through a Gaussian gradient magnitude filter to enhance the boundaries. This filter is given by
and
where * denotes a convolution operator, σ is the standard deviation of the Gaussian filter controlling the scale of the edges to be enhanced. It was set to 0.5 in our scheme. The enhanced image was used to produce the edge potential image from the gradient magnitude image by using a sigmoid function defined by
where α and β are parameters specifying the range and center, respectively, of intensity to be enhanced. They were set to −2.5 and 8.0 in our scheme. The normalized output image of the sigmoid gray-scale converter was used as a speed function for level set segmentation and fast marching algorithms.
In the following step, the shape of the liver was estimated roughly by a fast marching algorithm [21, 22]. This algorithm was initially proposed as a fast numerical solution of the Eikonal equation:
where F is a speed function and T is an arrival time function. The algorithm requires five to eight initial seed points. From the initial location (T = 0), the algorithm propagates the information in one way from the smaller values of T to larger values based on the first order scheme. This algorithm consists of two main processes. First, all grid points generated from the entire region were categorized into three categories: seed points corresponding to the initial location were categorized into Known; the neighbors of Known points were categorized into Trial with the computed arrival time; and all other points were categorized into Far that the arrival time was set to infinity. An iterative process served points in the Trial and Far list. The Trial point p with the smallest T value was chosen and moved to the Known. The arrival time of neighbors of p was recomputed based on the first order scheme, and the Far points that are neighbors of p were moved to the Trial. This iterative process was terminated when the maximum number of iterations was reached. The salient point of this algorithm is to use a heap data structure that can locate points with the smallest T value rapidly. The output of the fast marching algorithm is a time-crossing map indicating the time traveling to each point. It forms a rough shape of the liver in MR images.
A 3D geodesic active contour algorithm [23] was employed to refine the initial surface determined by the time-crossing map in order to determine the liver boundaries more precisely. This algorithm is based on the relation between active contours and the computation of geodesic or minimal distance curves, which allows boundary detection with large variations of gradients, including gaps. Let ψ(p, t) be a level set function with the initial surface corresponding to ψ(p, t) = 0 (Fig. 6.2). This level set function is then evolved to fit the form of liver following the partial differential equation:
where A(∙) is an advection vector function, F(∙) is a propagation (or expansion) function, and Z(∙) is a spatial modifier function for the mean curvature κ. The scalar constants α, β, and γ allow trading off among three terms: advection, propagation, and curvature. The algorithm requires an initial zero level set containing an initial surface that roughly approximates the liver boundaries. The initial surface was propagated with speed and direction (outwards, inwards) controlled by the propagation function. The spatial modifier term controls the smoothness of the surface where regions of high curvature are smoothed out. The level set evolution was terminated when the convergence criterion or the maximum number of iterations was reached. The convergence criterion was defined in terms of the root mean squared (RMS) change in the level set function. The evolution was considered to be converged if the RMS change is below a predefined threshold. The liver regions extracted by the geodesic active contour algorithm were used to calculate the liver volume. The intermediate results of our scheme for an example case are illustrated in Fig. 6.3. The original MR image in Fig. 6.3a was passed into the anisotropic diffusion filter to reduce noise while preserving the major liver structures such as the portal vein and liver boundary, as shown in Fig. 6.3b. The noise-reduced image was then passed through a Gaussian gradient magnitude filter to enhance the boundaries, as shown in Fig. 6.3c. The edge potential image generated from the enhanced image using the sigmoid gray-scale converter was applied to the fast marching algorithm to generate the initial contour, as shown in Fig. 6.3d. The liver was extracted more precisely by using the geodesic active contour algorithm, as shown in Fig. 6.3e. Corresponding between computer-based liver segmentation (red contour) and “gold-standard” manual liver segmentation (blue contour) is shown in Fig. 6.3f. Liver volume was computed using the extracted regions.
Examples of the resulting images at each step in our automated volumetry scheme. (a) Original axial MR image of the liver. (b) 3D anisotropic diffusion noise reduction. (c) 3D gradient magnitude filter. (d) 3D fast marching algorithm. Time-crossing map indicates the traveling time to each voxel. The majority of vessels inside the liver are excluded at this stage. (e) 3D geodesic active contour segmentation. (f) Corresponding computer-based liver segmentation (red contour) and “gold-standard” manual liver segmentation (blue contour)
Evaluation Criteria
The liver volumes obtained by using our computerized scheme were compared to the “gold-standard” manual volumes determined by the radiologist. The definitions used in evaluation of a computerized liver segmentation compared to the gold-standard manual liver segmentation are shown in Fig. 6.4. True-positive (TP) segmentation was defined as an overlapping region (gray color) between the computerized liver segmentation (indicated by a red contour), C, and a gold-standard manual segmentation (indicated by a blue contour), G; i.e., TP = G ∩ C. False-positive (FP) segmentation (red region) was defined by FP = C − TP. False-negative (FN) segmentation (blue region) was defined by FN = G − TP. True-negative (TN) segmentation was defined by TN = I − G ∪ C, where I is the entire image. We define accuracy, specificity, and sensitivity of the segmentation as
The Dice measurement representing the fraction of the overlapping volume and the volume of two segmentation methods is given by
We also determine the percentage volume error (E) for each computerized volume (V c) and the gold-standard manual volume (V m) as
The association between the computerized volumetry and the manual volumetry was measured by the Pearson product–moment correlation coefficient (r). The significance of correlation coefficient was evaluated by using the Student t test. An agreement between two measurements was assessed by using the intraclass correlation coefficient (ICC) [24, 25]. The two-way random single measure model, ICC(2,1), was used because we assumed that the cases were chosen randomly from population and each case was measured by two volumetric methods. The ICC(2,1) was defined by the following equation:
where n is the number of cases, k is the number of raters (i.e., volumetric methods), BMS is the between-cases mean square, EMS is the error mean square, and RMS is the between-raters mean square. The statistical significance was obtained by the analysis of variance. The post-hoc power analysis using the Walter–Eliasziw–Donner model [26] for ICC-based reliability studies was performed to determine the statistical power in this study. As done in [7], we assumed the type I error (α) of 0.05 and type II error (β) of 0.20 in this analysis. An additional agreement analysis for two measurements was performed by the Bland–Altman method [27] based on the mean difference (bias) and the standard deviation of difference (SD). The limits of agreement, which are given by bias ± 1.96 × SD, were used to consider the degree of agreement.
Results
The comparison on the liver volume between the two measurements is shown in Tables 6.1 and 6.2. The mean gold-standard manual volume was 1,710 cc with a standard deviation of 401 cc (range: 1,013–2,529 cc), while the mean volume of our computerized scheme was 1,697 cc with a standard deviation of 400 cc (range: 1,120–2,418 cc). The mean absolute difference and the percentage volume error (E) were 56 cc and 3.6 %, respectively.
The overall mean of the Dice coefficients was calculated as 93.6 ± 1.7 %, the accuracy was 99.4 ± 0.14 %, the sensitivity was 93.4 ± 3.3 %, and the specificity was 99.7 ± 0.12 %. The relationship between the computerized volumetry and the manual volumetry is shown in Fig. 6.5. The Pearson correlation coefficient was 0.98 at a level that was not statistically significant (p = 23.65). Table 6.3 presents the results from the ICC analysis. Two volumetric methods achieved an excellent agreement with an ICC of 0.98 and no statistically significant difference (p = 0.42). The statistical power in the study was evaluated by using the post-hoc power analysis based on the Walter–Eliasziw–Donner model [26]. The lowest ICC between the computer-based volumetry and the manual volumetry that we should have been able to detect with 23 cases was 0.95, and this study had the power to detect a bias of 0.03 in ICC. The Bland–Altman plot for assessing agreement is also presented in Fig. 6.6. Here the mean difference was −13.2. The limits of agreement with the 95 % confidence interval were −163 to 137 cc which were small enough to show a good agreement between two volumetric methods.
Figure 6.7 illustrates the computerized liver segmentation and manual liver segmentation for a case with a high accuracy (99.7 %). The computerized segmentation agreed almost perfectly with the gold-standard manual segmentation for slices through the superior portion of the liver, as shown in Fig. 6.7b, d. Two other cases with more typical results which have the accuracies close to the average accuracy are presented in Fig. 6.8. Overall, the computerized method was able to extract the livers very accurately. However, there were occasionally over- and under-extractions in the extracted livers. Major FP and FN extraction sources are illustrated in Fig. 6.9. The major FN sources included a lesion attaching to the liver boundary, a low-contrast liver boundary, and inhomogeneous density due to focal fatty and noise. The major FP sources included the heart, kidney, vena cava, and stomach, which abut the liver. They were also from artifact due to the partial volume effect. Other under- and over-extraction sources were convex and concave boundary parts with high curvatures.
Comparisons of computerized liver extraction with “gold-standard” manual liver extraction for the case with a high accuracy (99.7 %). (a) Original axial MR image from the case. (b) Computerized liver extraction (red contour) and “gold-standard” manual liver extraction (blue contour). (c) Original axial MR image (different slice) from the same case. (d) Computerized liver extraction (red contour) and “gold-standard” manual liver extraction (blue contour)
Comparisons of the computerized liver extraction with “gold-standard” manual liver extraction for two cases with accuracies (99.5 % for the upper case; 99.2 % for lower case) close to the average accuracy (99.4 %). (a) Original axial MR image from one of the cases. (b) Corresponding computerized liver extraction (red contour) and “gold-standard” manual liver extraction (blue contour). (c) Original axial MR image from the other case. (d) Corresponding computerized liver extraction (red contour) and “gold-standard” manual liver extraction (blue contour)
Illustrations of major FP and FN sources. (a, c), Original axial MR images. (b, d) computerized liver extraction (red contour) and “gold-standard” manual liver extraction (blue contour). (b) There is an FP due to the heart (a), an FN due to vein (b), and an FN due to a lesion on the liver boundary (c). (d) There is an FP due to the duodenum (d), and an FN due to a low-intensity region (e)
The average processing time of our scheme for liver segmentation was 1.03 ± 0.13 min/case (range: 0.9–1.5 min/case) on a PC (CPU: Intel, Xeon, 2.66 GHz), whereas that for manual method was 24.0 ± 4.4 min/case (range: 18–30 min/case). The difference was statistically significant (p < 0.001).
Discussions
Liver volumetry is performed for hepatectomy to treat patients with liver tumors. Because the liver volume is reduced after hepatectomy, it must be ensured that the remaining liver volume is sufficient to maintain the liver function. In the case of complicated treatment such as chronic liver disease, a larger remaining liver volume is required [28]. Many researchers have tried to estimate the liver volume accurately based on CT images, such as the one using virtual hepatectomy [29]. However, fewer researchers have reported liver volumetry on MRI, probably because it is believed that MR liver volumetry has more variations, and manual MR liver volumetry is more difficult than CT. Furthermore, manual liver volumetry is very time-consuming and not cost-effective. Therefore, it is crucial to investigate the potential of a computerized volumetry for liver MR images. We believe that computerized MR liver volumetry is potentially very useful.
Although our computerized liver volumetry had an excellent agreement with the gold-standard manual liver volumetry (the ICC was 0.98), there were still occasional FNs and FPs which were mainly caused by the similar density of other organs abutting to the liver. The liver segmentation accuracy was also interfered by the partial volume effects and the liver intensity variation among different studies/patients, as the intensity depends on acquisition timing and contrast material characteristics.
Although the volumes obtained by using our computerized method had a strong correlation with those by the gold-standard manual tracing method (Pearson’s product–moment correlation coefficient was 0.98), it does not still reach the minimal variation in CT volumetry between expert radiologists that was reported as 0.997 [30] (Although we could not find a study reporting the variation in MR volumetry, we expect it would be larger than the one in CT). One can increase the overall accuracy by correcting FP and FN extractions manually. This can be accomplished rapidly with routine manipulations. The substantial amount of time saved by using the computerized method may justify the small error rate (average percentage volume error of 3.6 %) compared to the manual tracing method, which the average processing time was 24 min/case.
Direct comparisons of our method with existing methods in literature are not easy because different databases and quality measurements were used. Freiman et al. [31] achieved volume errors of 5.36 and 2.36 % in CT volumetry of their database and a publicly available database (i.e., SLIVER07), respectively. Florin et al. [32] obtained a volume error of 10.72 % in CT volumetry. For evaluation of liver MRI segmentation, Gloger et al. [19] obtained volume errors of 8.3 % for normal livers and 11.8 % for fat livers with runtime of 11.2 and 15.4 min, respectively. Besides volume errors, some researchers used the shape alignment measurement to evaluate the segmentation performance. A robust measurement based on the shape alignment is the modified Hausdorff distance (MHD) which overcomes the noise and outlier sensitivity of the original Hausdorff distance, defined by
where X and Y are two sets of boundary positions of the liver extracted by a manual method and a computerized method, respectively. Our scheme achieved an average MHD of 12.8 ± 2.24 mm for livers with diseases, whereas an average original Hausdorff distance reported in [19] was 20.35 ± 8.66 mm for fat livers. Note that an MHD was not provided in [19].
The 95 % limits of agreement between our computerized volumes and the gold-standard manual volumes were −163.3 and 136.9 cc. These limits are smaller than the results reported by Nakayama et al. [8]: the limits between automated and manual volumes were −230.3 and 327.9 cc; and those between automated and measured liver volumes were −309.3 and 412 cc. In addition, these 95 % limits of agreement are smaller than those in our previous work on CT images [7] which were −211 and 278 cc for agreement between the automated and manual CT volumes. Note that the above comparisons were not direct comparisons due to different databases.
One of the limitations in this study is that the evaluation is performed with the gold-standard manual volumes determined by a single expert radiologist. Ideally, the gold-standard volumes are determined by multiple radiologists who are experts in liver diagnosis. However, this ideal evaluation would not be available at all institutions because not many institutions have a number of such radiologists who are sufficiently experienced in the liver diagnosis. Many publications reported the evaluation using the gold-standard manual volumes. However, none of them used the gold-standard volumes estimated from multiple radiologists. This may result from the above reason. Furthermore, it was shown that the correlation between two radiologists’s manual volumes was 0.997 [30], which may infer that the interobserver variation is small and the difference among manual volumes determined by multiple radiologists and a single radiologist is not significant. We used the manual volumes determined from an experienced radiologist as the gold standard. We thought that the manual volumes from multiple inexperienced radiologists or mixture of inexperienced radiologists and experienced radiologists may be less reliable, compared to volumes determined by an experienced radiologist who traces liver boundaries very carefully.
Conclusions
In this chapter, we developed an automatic scheme for the liver volumetry in MR images by employing the fast marching algorithm combined with the geodesic active contour segmentation. MRI liver volumes obtained by using our scheme agreed excellently with those determined by the current “gold-standard” manual tracing method. With our computerized volumetry, the time required for volumetry was reduced significantly from 24 min per case to a min per case. Therefore, our computerized scheme would be useful for radiologists in liver volumetric analysis on MR images.
References
Thuluvath PJ, Guidinger MK, Fung JJ, Johnson LB, Rayhill SC, Pelletier SJ (2010) Liver transplantation in the United States, 1999–2008. Am J Transplant 10:1003–1019
Biggins SW (2012) Futility and rationing in liver retransplantation: when and how can we say no? J Hepatol 56(6):1404
Monbaliu D, Pirenne J, Talbot D (2011) Liver transplantation using donation after cardiac death donors. J Hepatol 56:474–485
Lo CM, Fan ST, Liu CL et al (1997) Adult-to-adult living donor liver transplantation using extended right lobe grafts. Ann Surg 226:261–270
Radtke A, Sotiropoulos GC, Nadalin S et al (2007) Preoperative volume prediction in adult living donor liver transplantation: how much can we rely on it? Am J Transplant 7:672–679
Kamel IR, Kruskal JB, Warmbrand G, Goldberg SN, Pomfret EA, Raptopoulos V (2001) Accuracy of volumetric measurements after virtual right hepatectomy in potential donors undergoing living adult liver transplantation. AJR Am J Roentgenol 176:483–487
Suzuki K, Epstein ML, Kohlbrenner R et al (2011) Quantitative radiology: automated CT liver volumetry compared with interactive volumetry and manual volumetry. AJR Am J Roentgenol 197:W706–W712
Nakayama Y, Li Q, Katsuragawa S et al (2006) Automated hepatic volumetry for living related liver transplantation at multisection CT. Radiology 240:743–748
Gao L, Heath DG, Kuszyk BS, Fishman EK (1996) Automatic liver segmentation technique for three-dimensional visualization of CT data. Radiology 201:359–364
Bae KT, Giger ML, Chen CT, Kahn CE Jr (1993) Automatic segmentation of liver structure in CT images. Med Phys 20:71–78
Hermoye L, Laamari-Azjal I, Cao Z et al (2005) Liver segmentation in living liver transplant donors: comparison of semiautomatic and manual methods. Radiology 234:171–178
Okada T, Shimada R, Hori M et al (2008) Automated segmentation of the liver from 3D CT images using probabilistic atlas and multilevel statistical shape model. Acad Radiol 15:1390–1403
Selver MA, Kocaoglu A, Demir GK, Dogan H, Dicle O, Guzelis C (2008) Patient oriented and robust automatic liver segmentation for pre-evaluation of liver transplantation. Comput Biol Med 38:765–784
Chen X, Bagci U (2011) 3D automatic anatomy segmentation based on iterative graph-cut-ASM. Med Phys 38:4610–4622
Suzuki K, Kohlbrenner R, Epstein ML, Obajuluwa AM, Xu J, Hori M (2010) Computer-aided measurement of liver volumes in CT by means of geodesic active contour segmentation coupled with level-set algorithms. Med Phys 37:2159–2166
Karlo C, Reiner CS, Stolzmann P et al (2010) CT- and MRI-based volumetry of resected liver specimen: comparison to intraoperative volume and weight measurements and calculation of conversion factors. Eur J Radiol 75:e107–e111
Farraher SW, Jara H, Chang KJ, Hou A, Soto JA (2005) Liver and spleen volumetry with quantitative MR imaging and dual-space clustering segmentation. Radiology 237:322–328
Rusko L, Bekes G (2011) Liver segmentation for contrast-enhanced MR images using partitioned probabilistic model. Int J Comput Assist Radiol Surg 6:13–20
Gloger O, Kuhn J, Stanski A, Volzke H, Puls R (2010) A fully automatic three-step liver segmentation method on LDA-based probability maps for multiple contrast MR images. Magn Reson Imaging 28:882–897
Perona P, Malik J (1990) Scale-space and edge-detection using anisotropic diffusion. IEEE Trans Pattern Anal Mach Intell 12:629–639
Sethian JA (1999) Level set methods and fast marching methods, 2nd edn. Cambridge University Press, New York
Sethian JA (1996) A fast marching level set method for monotonically advancing fronts. Proc Natl Acad Sci U S A 93:1591–1595
Caselles V, Kimmel R, Sapiro G (1997) Geodesic active contours. Int J Comput Vis 22:61–79
Portney LG, Watkins MP (1993) Foundations of clinical research: applications to practice, 2nd edn. Appleton & Lange, Norwalk
Shrout PE, Fleiss JL (1979) Intraclass correlations: uses in assessing rater reliability. Psychol Bull 86:420–428
Walter SD, Eliasziw M, Donner A (1998) Sample size and optimal designs for reliability studies. Stat Med 17:101–110
Bland JM, Altman DG (1986) Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1:307–310
Okamoto E, Kyo A, Yamanaka N, Tanaka N, Kuwata K (1984) Prediction of the safe limits of hepatectomy by combined volumetric and functional measurements in patients with impaired hepatic function. Surgery 95:586–592
Yamanaka J, Saito S, Fujimoto J (2007) Impact of preoperative planning using virtual segmental volumetry on liver resection for hepatocellular carcinoma. World J Surg 31:1249–1255
Sandrasegaran K, Kwo PW, DiGirolamo D, Stockberger SM Jr, Cummings OW, Kopecky KK (1999) Measurement of liver volume using spiral CT and the curved line and cubic spline algorithms: reproducibility and interobserver variation. Abdom Imaging 24:61–65
Freiman M, Eliassaf O, Taieb Y, Joskowicz L, Azraq Y, Sosna J (2008) An iterative Bayesian approach for nearly automatic liver segmentation: algorithm and validation. Int J Comput Assist Radiol Surg 3:439–446
Florin C, Paragios N, Funka-Lea G, Williams J (2007) Liver segmentation using sparse 3D prior models with optimal data support. Inf Process Med Imaging 20:38–49
Acknowledgments
The authors are grateful to members in the Suzuki Laboratory in the Department of Radiology at the University of Chicago for their valuable comments. This work was partly supported by the NIH S10 RR021039, P30 CA14599, and Vietnam Education Foundation.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer Science+Business Media New York
About this chapter
Cite this chapter
Huynh, H.T., Karademir, I., Oto, A., Suzuki, K. (2014). Liver Volumetry in MRI by Using Fast Marching Algorithm Coupled with 3D Geodesic Active Contour Segmentation. In: Suzuki, K. (eds) Computational Intelligence in Biomedical Imaging. Springer, New York, NY. https://doi.org/10.1007/978-1-4614-7245-2_6
Download citation
DOI: https://doi.org/10.1007/978-1-4614-7245-2_6
Published:
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-4614-7244-5
Online ISBN: 978-1-4614-7245-2
eBook Packages: EngineeringEngineering (R0)