Abstract
The proteomics studies made on the post-mortem changes in meat clearly illustrate its large potential in meat research. Proteomics has especially proved to be a powerful tool to investigate post-mortem protein degradation in meat because it provides valuable information about the complex mechanisms behind post-mortem proteolysis. However, it is important to emphasize that the proteome of a muscle cell or a meat product is extremely complex and that none of the proteomic methods are capable of profiling a complete proteome. Moreover, all methods in proteomics are very complicated and time consuming and only a limited number of samples can be analyzed in each experiment. Hence, it is often necessary to perform several different proteomics studies to reveal a specific mechanism.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Skeletal muscle is the most abundant tissue in animals; on average, 30–40% of the live weight or 40–60% of the carcass of domestic animals consists of muscular tissue (Lawrie and Ledward 2006). In living animals, contractile fibers of skeletal muscle tissues constitute the cellular units that support coordinated excitation–contraction–relaxation cycles for movements and postural control (Gordon et al. 2000). Nutritionally, muscle is a great source of essential amino acids, and to a lesser extent, of other important nutrients such as minerals, vitamins, and fatty acids, so that muscle could provide the majority of the nutrients required for human health. From the food science perspective, due to the high abundance and nutritional value, skeletal muscle is one of the most important food resources for consumers.
Variation in meat quality traits is a well-known problem in the meat industry. Although extensively studied, the underlying mechanisms contributing to different meat quality traits are not fully understood which is essential in order to improve meat quality. Hence, new tools and strategies must be applied in meat research. Proteomics is one of these tools and has been shown to be a powerful tool to investigate post-mortem protein changes.
In proteomics it is possible to separate and quantify hundreds of proteins in one analysis. A major advantage of proteomics is that it is possible to carry out a detailed characterization of the single proteins and identify cleavage sites and protein modifications such as protein oxidation and phosphorylation. The use of proteomics in meat research has a large potential and will lead to a much more detailed characterization of the post-mortem protein changes. Proteomics can also be applied in other aspects of meat research and become a valuable tool in the investigation of protein changes during meat processing, effects of meat packaging, animal growth, and genetic variation. Hence, proteomics can provide valuable information on the mechanisms influencing the different quality traits to gain a better understanding of these mechanisms. This information can be used to optimize meat production and improve meat quality. This chapter focuses on the challenges regarding muscle proteomic study, the technologies used in proteomics, and the recent results obtained with proteomics in meat research.
2 The Challenges Regarding Proteomic Analysis of Skeletal Muscle
Skeletal muscle proteomics aims at the global identification, characterization of the entire protein complement in muscle under normal or abnormal development, or different treatments. Over the past decade, the rapid developments in mass spectrometry and bioinformatics tools have driven the remarkable progress of proteomic science. However, proteomics study in skeletal muscle is still a very challenging task because of the wide-ranging biochemical heterogeneity of the muscle proteins.
Muscle proteome is notorious for its high dynamicity and diversity (Ohlendieck 2010). The amount of protein expression varies greatly in muscle. Myosin heavy chain, actin, titin, and nebulin account for the majority of muscle proteins whereas the other proteins are only present in relatively low amounts, in a large dynamic range on the order of 106 magnitude. The wide and dynamic expression range of proteins makes it impossible to separate and detect all protein species with current techniques because the extremely intensive signals from high abundant proteins often overlap or cover the signals from other low abundant proteins during separation and MS identification. In addition, skeletal muscle is heterogeneous in composition. Aside from the multiple fiber types, such as slow oxidative (type I), moderately fast oxidative glycolytic (type IIa), and fast glycolytic (type IIb) fibers, muscle also contains connective tissues, capillaries, and nerve cells.
The complex tasks of muscles are performed by numerous muscle proteins with specialized functions, structures, and interactions. A considerable amount of muscle proteins are integral membrane proteins and high molecular mass complexes. It is a notable feature of contractile fibers that some of the largest protein species are highly expressed in skeletal muscle, such as nebulin of 600–800 kDa and titin with a molecular mass exceeding 1,200 kDa (Ottenheijm and Granzier 2010a, b). Supramolecular membrane assemblies are also highly present in muscle, such as the abundant actomyosin machinery with its regulatory troponin–tropomyosin system, the ryanodine receptor Ca2+ release channel of the triad junctions, the dystrophin–glycoprotein complex of the sarcolemma, and the respiratory chain of muscle mitochondria (Ohlendieck 2011). The large number of membrane-associated proteins, the high molecular mass of many muscle components, and extensive post-translational modifications in various muscle proteins and their assembly in highly complex supramolecular structures make it extremely difficult to carry out conventional biochemical studies of muscle. All these analytical facts have to be considered when one performs a skeletal muscle proteomic study. Standard proteomic methods such as gel electrophoresis and liquid chromatography are not capable of separating all muscle proteins in one run. Some sample fractionation and subcellular proteomic strategies can be employed to analyze membrane proteins and proteins from subcellular organelles (Ohlendieck 2011).
3 Approaches for Proteomic Study of Muscle Food
For muscle food, mass spectrometry-based large-scale proteomic studies have already provided a plethora of new information by comparison of different meat quality, processing condition, and genetic backgrounds in most farm animals (Bendixen 2005; Bendixen et al. 2011; Hollung et al. 2007). Methodologically, proteomics routinely employ diverse separation techniques including gel-based approaches (one and two-dimensional gel electrophoresis) and gel-free approaches (different chromatographic methods), in combination with advanced mass spectrometric methods for the identification of peptides and proteins of interest (Baggerman et al. 2005; Falk et al. 2007). The proteomic data are interpreted by diverse bioinformatics tools and can be verified by using immunoblotting, activity assays, or other methods.
3.1 Gel-Based Proteomic Approaches
Gel-based proteomic (one-dimensional gel electrophoresis (1DE), two-dimensional polyacrylamide gel electrophoresisis (2DE)) comprise the most classic and versatile methods for global protein separation and quantification (Baggerman et al. 2005). They can screen the protein expression at large scale with lower cost as compared with gel-free proteomics. Despite the apparent advantages of 2DE for separation of complex protein mixtures, the technique suffers from a number of major drawbacks. The proteins identified from 2-D gel are mainly high abundant proteins; the low abundant proteins are not easily detected by gel-based approaches. Some pre-fractionation methods can be used to overcome this drawback to some extent. Proteins with high (>150 kDa) and low (<10 kDa) molecular weight and proteins with extreme isoelectric points, in particular basic proteins, are usually not detected in standard 2DE. Another drawback concerns the hydrophobic proteins that are not soluble in the buffers used for sample loading or precipitate during the electrophoresis process (Lescuyer et al. 2004). These limitations of the 2DE make it very difficult to investigate many of the structural proteins in muscle as discussed before. 2DE also has some technical limitations. Briefly, the process is time consuming, labor intensive, and requires significant technical expertise to generate reproducible gels (Rabilloud 2002).
3.2 Gel-Free Proteomic Approaches
The limitations of gel-based approaches have promoted the development of alternative gel-free proteomic approaches. In these approaches, complex protein mixtures are digested in solution. The resulting peptide mixture is fractionated by one or several steps of chromatographic methods and analyzed in a data-dependent manner by LC-MS/MS for separation, quantification, and identification of the proteins (Aebersold and Mann 2003; Yates 2004). However, no single chromatographic method is capable of separating the complex proteome of a cell or tissue. Hence, it is necessary to use a combination of different separation steps such as ion exchange, size exclusion, and reverse-phase chromatography. The gel-free methods have the advantage over 2DE in that they allow examination of high or low abundance proteins in the same analysis and are unbiased with respect to molecular weight, isoelectric point, and hydrophobicity of the proteins. Furthermore, all steps may be automated for high-throughput analysis.
The development in LC-MS/MS methods in proteomics is enormous and many new and smarter methods are constantly being introduced. However, the methods all have some common limitations. For instance, the use of LC-MS/MS in the high-throughput gel-free approaches results in an enormous dataflow and thousands of spectra are generated during the MS analysis; a very strong bioinformatics platform is required to analyze the raw data. Datasets from different LC-MS/MS runs can be combined, but there is a high likelihood that different sets of peptides will be identified in each experiment, so how to interpret the preliminary results is also a big challenge. Another limitation is that only a few samples can be analyzed in each LC-MS/MS run; this makes comparison of multiple samples difficult to undertake. Lastly, in order to get satisfactory results, the requirement for advanced equipment and experienced personal is also very high and expensive.
4 Application of Proteomics in Early Post-Mortem Meat
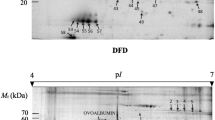
The quality of raw pig meat is influenced by changes in the muscle/meat proteome caused by different factors such as animal growth, age, rate of glycolysis, and post-mortem protein degradation (Hopkins and Thompson 2002; Rosenvold and Andersen 2003). Meat scientists have performed a substantial amount of research on their factors, which has led to considerable quality and compositional improvements. However, the underlying biochemical and physicochemical mechanisms behind the influence of these factors on meat are to some extent still not fully understood. The recent application of proteomics in the field of meat science has provided some interesting and promising results. Early post-mortem protein changes in muscle have been investigated with proteomics in several studies (D’Alessandro et al. 2011; Jia et al. 2007; Lametsch et al. 2003; Promeyrat et al. 2011). The studies revealed that a large part of the proteome changes post mortem. The post-mortem changes in pig LM during the first 48 h after slaughter were monitored with 2DE and more than 600 protein spots were separated and quantified; more than 100 proteins were found to change post mortem as illustrated in Fig. 6.1 (Lametsch and Bendixen 2001). The mechanism behind these post-mortem changes are to some extent still unclear. However, the main cause of post-mortem protein changes is probably protein degradation, as many of the identified changes are protein fragments that increase in spot intensity post mortem (Lametsch et al. 2002). It has been suggested that protein stability is changed post mortem leading to precipitation and aggregation (Bjarnadottir et al. 2010). But changes in protein modification such as phosphorylation or oxidation that change the isoelectric point of the proteins or release from protein complexes have also been found to contribute to post-mortem protein changes (Lametsch et al. 2011; Lund et al. 2007). Even protein expression may to some extent cause some changes post mortem, however, it is unlikely that protein expression causes major changes once the muscle has entered the state of rigor mortis because protein expression is an energy-requiring process and the energy is nearly depleted after reaching rigor mortis (Henckel et al. 2002).
Postmortem protein changes. The spot intensity at 4, 8, 24 and 48 h postmortem of more than 600 individual spots are plotted against the spot intensity at slaughter. If the spot intensity does not change postmortem it is plotted on the diagonal. If the spot intensity increases it is plotted above the diagonal and vice versa
5 Future Perspectives
The proteomics studies made on the post-mortem changes in meat clearly illustrate their large potential in meat research. Proteomics have especially proven to be a powerful tool to investigate post-mortem protein degradation in meat because they provide valuable information about the complex mechanisms behind post-mortem proteolysis. Proteomics can provide information on the cleavage sites and post-mortem degradation pattern of the proteins. Furthermore the resulting protein fragments may be used as biomarkers to measure the activity of specific proteolytic enzymes. Such biomarkers could be applied in breeding or to optimize animal slaughter and meat processing to gain more tender meat. Proteomics can also be applied to study post-mortem metabolism to provide further knowledge of undesirable meat characteristics such as PSE. Proteomics will also be an effective research tool to investigate the relation between meat quality and post-mortem protein modifications such as protein oxidation. However, it is important to emphasize that the proteome of a muscle cell or a meat product is extremely complex and that none of the proteomic methods is able to profile a complete proteome. Moreover, all methods in proteomics are very complicated and time consuming and only a limited number of samples can be analyzed in each experiment. Hence, it is often necessary to perform several different proteomics studies to reveal a specific mechanism.
References
Aebersold R, Mann M (2003) Mass spectrometry-based proteomics. Nature 422:198–207
Baggerman G, Vierstraete E, De Loof A, Schoofs L (2005) Gel-based versus gel-free proteomics: a review. Comb Chem High Throughput Screen 8:669–677
Bendixen E (2005) The use of proteomics in meat science. Meat Sci 71:138–149
Bendixen E, Danielsen M, Hollung K, Gianazza E, Miller I (2011) Farm animal proteomics – a review. J Proteomics 74:282–293
Bjarnadottir SG, Hollung K, Faergestad EM, Veiseth-Kent E (2010) Proteome changes in bovine longissimus thoracis muscle during the first 48 h postmortem: shifts in energy status and myofibrillar stability. J Agric Food Chem 58:7408–7414
D’Alessandro A, Marrocco C, Zolla V, D’Andrea M, Zolla L (2011) Meat quality of the longissimus lumborum muscle of Casertana and large white pigs: metabolomics and proteomics intertwined. J Proteomics 75:610–627
Falk R, Ramstrom M, Stahl S, Hober S (2007) Approaches for systematic proteome exploration. Biomol Eng 24:155–168
Gordon AM, Homsher E, Regnier M (2000) Regulation of contraction in striated muscle. Physiol Rev 80:853–924
Henckel P, Karlsson A, Jensen MT, Oksbjerg N, Petersen JS (2002) Metabolic conditions in porcine longissimus muscle immediately pre-slaughter and its influence on peri- and post mortem energy metabolism. Meat Sci 62:145–155
Hollung K, Veiseth E, Jia XH, Faergestad EM, Hildrum KI (2007) Application of proteomics to understand the molecular mechanisms behind meat quality. Meat Sci 77:97–104
Hopkins DL, Thompson JM (2002) Factors contributing to proteolysis and disruption of myofibrillar proteins and the impact on tenderisation in beef and sheep meat. Aus J Agric Res 53:149–166
Jia XH, Ekman M, Grove H, Faergestad EM, Aass L, Hildrum KI, Hollung K (2007) Proteome changes in bovine longissimus thoracis muscle during the early postmortem storage period. J Proteome Res 6:2720–2731
Lametsch R, Bendixen E (2001) Proteome analysis applied to meat science: characterizing post mortem changes in porcine muscle. J Agric Food Chem 49:4531–4537
Lametsch R, Roepstorff P, Bendixen E (2002) Identification of protein degradation during post-mortem storage of pig meat. J Agric Food Chem 50:5508–5512
Lametsch R, Karlsson A, Rosenvold K, Andersen HJ, Roepstorff P, Bendixen E (2003) Postmortem proteome changes of porcine muscle related to tenderness. J Agric Food Chem 51:6992–6997
Lametsch R, Larsen MR, Essen-Gustavsson B, Jensen-Waern M, Lundstrom K, Lindahl G (2011) Postmortem changes in pork muscle protein phosphorylation in relation to the RN genotype. J Agric Food Chem 59:11608–11615
Lawrie RA, Ledward DA (2006) Lawrie’s meat science, 7th edn. Woodhead, Cambridge
Lescuyer P, Hochstrasser DF, Sanchez JC (2004) Comprehensive proteome analysis by chromatographic protein prefractionation. Electrophoresis 25:1125–1135
Lund MN, Lametsch R, Hviid MS, Jensen ON, Skibsted LH (2007) High-oxygen packaging atmosphere influences protein oxidation and tenderness of porcine longissimus dorsi during chill storage. Meat Sci 77:295–303
Ohlendieck K (2010) Proteomics of skeletal muscle glycolysis. Biochim Biophys Acta 1804:2089–2101
Ohlendieck K (2011) Skeletal muscle proteomics: current approaches, technical challenges and emerging techniques. Skelet Muscle 1:6
Ottenheijm CAC, Granzier H (2010a) New insights into the structural roles of nebulin in skeletal muscle. J Biomed Biotech 968139:1–6
Ottenheijm CAC, Granzier H (2010b) Role of titin in skeletal muscle function and disease. Muscle Biophys Mol Cells 682:105–122
Promeyrat A, Sayd T, Laville E, Chambon C, Lebret B, Gatellier P (2011) Early post-mortem sarcoplasmic proteome of porcine muscle related to protein oxidation. Food Chem 127:1097–1104
Rabilloud T (2002) Two-dimensional gel electrophoresis in proteomics: old, old fashioned, but it still climbs up the mountains. Proteomics 2:3–10
Rosenvold K, Andersen HJ (2003) Factors of significance, for pork quality – a review. Meat Sci 64:219–237
Yates JR (2004) Mass spectral analysis in proteomics. Annu Rev Biophys Biomol Struct 33:297–316
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer Science+Business Media New York
About this chapter
Cite this chapter
Huang, H., Lametsch, R. (2013). Challenges and Applications of Proteomics for Analysis of Changes in Early Postmortem Meat. In: Toldrá, F., Nollet, L. (eds) Proteomics in Foods. Food Microbiology and Food Safety, vol 2. Springer, Boston, MA. https://doi.org/10.1007/978-1-4614-5626-1_6
Download citation
DOI: https://doi.org/10.1007/978-1-4614-5626-1_6
Published:
Publisher Name: Springer, Boston, MA
Print ISBN: 978-1-4614-5625-4
Online ISBN: 978-1-4614-5626-1
eBook Packages: Chemistry and Materials ScienceChemistry and Material Science (R0)