Abstract
Intestinal microbiota is now considered as a “new organ” which, over and above their genetic origin, de-orphans the pandemic development of metabolic diseases. The trillions of bacteria and their corresponding million genes which inhabit our gut provide a unique source of molecular hypotheses to explain the wide diversity of metabolic diseases and hence form a basis to reach the important objective of personalized medicine. The gut and more recently the tissue microbiome could be the source of: (1) new biomarkers predicting and classifying metabolic diseases to help the clinician to propose the best therapeutic strategy, and (2) new pharmacological and nutritional strategies to treat the cause rather than the consequence of diabetes and obesity. The field of immunometabolism should be extended to microbio-immunometabolism, thus reconciling the role of the environment, the genetic background, and individual diversity in relation to the onset and development of metabolic diseases.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Diabetes
- Bacterial translocation
- Immune system
- Incretins
- Inflammation
- Immunometabolism
- Intestine
- Metabolic diseases
- Metabolic inflammation
- Metabolic infection
- Microbiome
- Obesity
- Tissue microbiota
11.1 Introduction
Evidence now shows that the pandemic progression of diabetes and obesity, as well as their numerous complications (cardiovascular, hepatic, renal, neuronal), is caused by lifestyle factors including sedentarity and fat-enriched diet which has replaced fiber-enriched diet (Fig. 11.1). A dramatic outcome of this epidemic is the increasing number of cardiovascular events leading to mortality [1–4] – the mechanisms at play need to be delineated in order to define new therapeutic strategies. Over the last decade lethal cardiovascular events associated with diabetes have progressed by 62 % [5, 6]. This is much higher than the risk linked to cholesterol levels or hypertension. The incidence of type 2 diabetes is 4–5 % in Europe, 8–10 % in the USA, and higher in South Asia [7]. These numbers have more than doubled over the last 20 years. Therefore, one can suggest that even if genetic analyses provide the basis for such an epidemic, changes in our genome cannot be solely responsible. One interpretation is that our genome is no longer adapted to environmental factors. Numerous environmental hypotheses have been proposed. First, epigenetic noncoded functions that are independent of genomic factors could have an impact. Second, perhaps more realistically, the impact of changes in feeding habits and social behavior is likely to be an important cause of the growing incidence of metabolic diseases. This phenomenon could be linked to the microbiota, our “second genome” harboring almost ten times more prokaryotic cells than eukaryotic cells in our body [8]. All mammalian organisms are born sterile, without any microbiota which is inherited at birth. During the first hours, days, and weeks of life, microbiota from the mother and the environment colonize the body of the newborn in a specific order [9]. The microbiota colonizes the intestine during the first 3–5 years of life and is species-, age-, and sex-dependent [10]. The initial infant gut microbiota is a simple structure usually dominated by bifidobacteria, and through a series of successions and replacements, it shifts to a more complex adult pattern [11–15]. The microbiota also undergoes substantial changes at the extremes of life, in infants and older people, the ramifications of which are still being explored [16]. Then, throughout life the bacterial ecology tends to vary mostly according to the environment [17]. Each individual has at least 160 shared species and a number of well-balanced host-microbial molecular relationships that define groups of individuals [18, 19]. This second genome is the metagenome. The importance of this metagenome resides in its gene repertoire, 100 times superior to the eukaryotic nuclear genome [18, 19], thus providing a huge genetic diversity susceptible to convey a plethora of functions [18, 19]. Indeed, the tremendous efforts that have been made in bioinformatic analyses have allowed the encoding and the deciphering of all sequences. Humans host different metagenomes from multiple locations such as the skin, lungs, vagina, and the mouth in addition to the intestine [8, 20]. The human gut hosts 100 trillion microorganisms, encompassing up to thousands of species at an average concentration of 1014 per ml and weighing in average 1.5 kg [21]. A major observation is that the metagenomic diversity is extremely large and represents a signature of each individual. Its plasticity is significant and depends on numerous environmental and genetic factors that can evolve over time and could explain the rapid development of metabolic diseases. The adult intestinal microbiota has been shown to be relatively stable over time [22] and is sufficiently similar between individuals to allow identification of a core microbiome comprising 66 dominant operational taxonomic units (OTUs) that correspond to 38 % of sequence reads from 17 individuals [18, 19]. The core microbiota changes to become distinct in elderly subjects from that observed for younger adults with a greater proportion of Bacteroides spp. and typical abundance patterns of Clostridium groups. Interestingly, the onset of metabolic diseases increases with age and is associated with a change in intestinal microbiota as observed during aging [16, 23, 24]. Similarly, it was shown that changes in gut microbiota characterize obesity and diabetes [18, 19]. This suggests that each member can interact in a perfect mutual symbiosis defining a steady microbiota [8, 20, 25]. Hence, a new concept has emerged with an important influence on our understanding of these pathologies. Major advances have been made over the course of the last decade, thanks to the development of high-throughput sequencing of the microbiota and to the use of germ-free mice. These have allowed the demonstration of the causality of the microbiota from the gut [26], from the oral cavity, and more recently from the tissue [27] on the development of metabolic diseases. The following decades will be dedicated to the identification of the molecular crosstalk between the microbiota and the host to understanding mechanisms controlling diabetes and obesity. The intestinal immune system appears to be a major player in the field since at birth bacterial colonization of the intestine represents an outbreak of antigens that can educate the immune system, as well other major functions such as the vascular and the nervous systems. The latter through its connection with the brain could participate in the maturation of the central nervous system over the course of a lifetime [28]. Consequently, beyond the key physiological role of the intestinal microbiota in normal development, numerous pathological issues could be the consequence of an impaired microbiota. In addition to the first described role of the microbiota on the development of intestinal bowel diseases, there is now evidence that a change in the intestinal bacterial ecology could affect metabolic, vascular, liver, heart, oral, and neurodegenerative diseases. More evidence is required to confirm a role in arthritis and skin immunological diseases. The recent increase in the use of bariatric surgery for the treatment of massive obesity and incidental diabetes has generated further evidence reinforcing the role of intestinal microbiota in the control of metabolic diseases.
The pandemic origin of metabolic diseases. The impact of genetics, diet, and other factors on the incidence and the development of metabolic disease pandemic could have a change in gut microbiota as an origin. Gut microbiota would also favor the development of complications of metabolic diseases such as cardiovascular, liver, eye, and skin complications
New clinical approaches for prevention and therapy are now being planned. Functional food and pharmaceutical strategies, based on the targeting of the microbiota to host interactions, can be initiated. In the face of the large diversity of metabolic phenotypes, i.e., a large spectrum of fasted and postprandial glycemia and different fat mass distribution, “intelligent and directed” food complements can be proposed. They should not prevent or treat massively the populations but will focus on subgroups of patients with similar microbiota-related diseases. The relevance of treating the impact of gut microbiota on the diseases requires the development of companion biomarkers. They should first define subgroups of patients with similar microbiota profiles and should be able to follow up the efficacy of the microbiota change in correlation with the metabolic phenotype treated. Hence, they will drive the therapeutic approach toward appropriate subgroups of patients to improve the efficacy of the treatment and reduced secondary effects. Pharmaceutical approaches targeting a mechanism central to the molecular crosstalk between microbiota and the host, such as metabolic inflammation, have currently been developed by means of fecal transplant and immunomodulation. Therefore, it is now time to consider the intestinal microbiota as a new organ controlling metabolism. This organ is characterized by a high level of plasticity so that it can adapt to a change in the host behavior in a reciprocal manner for the control of broad host physiological functions. The understanding of its molecular components will lead to a totally new way of interpreting physiological, clinical, and therapeutic data. Therefore, the limit between nutritional and pharmacological strategies is vanishing. Intelligent food supplements and cause-based pharmacological approaches will be the treatments of tomorrow that still have to be identified.
Pharmaceutical strategies will benefit from the gain of knowledge generated based on the intestinal microbiota to host relationship. The corresponding molecular crosstalk is currently being assessed and will be reviewed below. It includes mechanisms controlling immunomodulation, bile acid conjugation, the intestinal barrier, energy harvesting, and entero-endocrine hormone secretion. The bacterial factors, i.e., the metafactors controlling these eukaryotic functions, are of inflammatory origin such as the lipopolysaccharides, peptidoglycan fragments, flagellin, or derived from fermentation products. Hence, pharmacological strategies should emerge from these concepts and target subgroups of patients with these new drugs acting on the host to microbiota relationship.
An important matter relates to the identification of subgroups of patients sensitive to these new approaches, most likely characterized by a specific dysbiosis and impaired gut and tissue microbiota. Hence, microbiota-based biomarkers precisely identifying these subgroups will help the clinician to treat the patients with the appropriate pharmaceutical strategy.
11.2 From the Main Features of the Pathophysiology of Metabolic Diseases to Microbiota
Metabolic diseases are all characterized by alterations in energy balance which explains, at least in part, the occurrence of obesity. The disease is the consequence of either an increased energy uptake or a reduced energy expenditure demonstrating that different mechanisms are responsible for increased body weight. Furthermore, the accumulation of fat in the body is compartmentalized, i.e., abdominal, subcutaneous. or intratissular such as in the liver, the heart, or the pancreatic islets with different consequences on overall health. The incidence of diabetes is much increased in patients with normal body weight but with abdominal fat accumulation which is considered the most deleterious for health [29]. Similarly, the accumulation of triglycerides within the Langerhans islets impairs insulin secretion [30] that can be restored by leptin treatment [31] activating uncoupling proteins such as UCP2 [32]. At the onset of obesity, the storage of energy is associated with hypertrophy of the adipocytes and their hyperplasia to ensure a sufficient number of cells [33–35]. The signals favoring adipose depot development could be linked to increased adipocyte metabolism leading to a local hypoxia [36] and the recruitment of adipocyte precursors [37, 38]. The maintenance of the adipose depot architecture is ensured by a concomitant proliferation of endothelial precursors [39, 40], increasing the adipose tissue capillaries and hence blood and energy supply [40]. Hyperphagia, supplying large amounts of energy, and hyperinsulinemia, although associated with normal blood glucose profiles, also suggest a neuroendocrine origin of the disease notably, the gut to brain axis which is recruited in response to an oral glucose load [41] or food intake [42, 43]. A key enteroendocrine factor, glucagon-like peptide 1 (GLP-1), triggers the gut–brain axis [42, 44, 45] which is impaired during high-fat-diet-induced metabolic diseases [46]. The brain is also sensitive to hormones from the periphery such as insulin and leptin which no longer efficiently control food intake and energy partitioning. Altogether, impaired gut and peripheral nutrients and hormone sensing systems, as well as signaling effectors toward the adipose depot via the brain, are impaired during obesity. The increase of brain to adipose depot signals then enhances the capacity of the adipose depot to store energy (Fig. 11.1). However, it should be noted that increasing food intake and activating the storage of energy in fat depots cannot be considered as a pathological mechanism since it is a normal behavior of the body following a fasting period to replenish the fat stores. The pathology starts when this process is no longer downregulated. Different levels of deregulation occur involving either early control of food intake and energy distribution or processes that control each step of the obesity process. The intestinal microbiota has been proposed to interfere with hyperphagia and to some extent with energy storage – this will be reviewed below. The emerging role of gut microbiota in the gut to brain axis for the control of neural development, behavior, and food intake will certainly generate new molecular hypotheses regarding the development of obesity [28, 47–49].
Type 2 diabetes arises due to impaired secretion and action of insulin which evolve either simultaneously or independently according to numerous influencing factors (Fig. 11.2). An increased glucose intolerance with glycemic profiles higher than controls is a common feature at the onset of the disease [50, 51]. This is associated with hepatic insulin resistance where insulin does not efficiently reduce hepatic glucose production following a meal [50–52]. Liver insulin resistance is hence involved in glucose intolerance and type 2 diabetes and explains fasting glycemia [53–56]. Simultaneously, insulin-stimulated glucose uptake is reduced which is most likely a consequence of an impaired cellular lipid metabolism [57]. Numerous hypotheses could explain insulin resistance and among those the role played by a low-grade inflammation, called metabolic inflammation since it is chronic and not related to infection or cancer [58]. Briefly, metabolic inflammation is characterized by an increased infiltration of immune cells within organs involved in the glycemic control such as the adipose tissue, the liver, and muscles [59, 60]. Cells from the innate and adaptive immune systems secrete cytokines such as TNFa and IL1-b that impair insulin signaling, thus affecting both glucose and lipid metabolism (Fig. 11.2) [59, 60]. Inflammation is increasingly regarded as a key process underlying metabolic diseases [59–64]. In the adipose tissue of individuals with metabolic diseases, this mechanism includes features characteristic of active local inflammation [63, 65, 66]. The cytokines released impair insulin signaling [67–69], thus leading to cytokine-mediated insulin resistance [70, 71]. Macrophage infiltration of adipose tissue has been described in both mice and humans [64–66, 72–74]. It is suggested that these cells express TNF-alpha and iNOS that are characteristic of M1-type macrophages which are responsible for almost all adipose tissue TNF-alpha expression and significant amounts of iNOS and IL-6 expression [73, 75, 76]. In addition, using functional analyses and microarray technology, it has been demonstrated that adipocyte progenitors and macrophages are characterized by a closed genome and phenotypome [77, 78]. Based on measurements of phagocytic activity and gene profiling analysis of different progenitor cells, we revealed that the origin of inflammation could also be attributed to cells initially present in adipose fat pads such as preadipocytes [77, 78]. Therefore, both infiltrating and resident cells are most likely involved in the processes characterizing adipose tissue inflammation. Similarly, lymphocytes are associated with adipose tissue inflammation [64, 79, 80]. CD8 T cells are present in obese mouse adipose tissue even before the infiltration of the tissue by macrophages. The results also showed that the immunological and genetic depletion of CD8+ T cells lowered macrophage infiltration and adipose tissue inflammation and improved systemic insulin resistance. Conversely, adaptive transfer of CD8+ T cells to CD8-deficient mice aggravated adipose inflammation. Co-culture and other in vitro experiments revealed a cycle of interactions between CD8+ T cells, macrophages, and adipose tissue. Moreover, CD4+ regulatory T lymphocytes (Treg) (CD4+FoxP3+), inhibitory cells of the immune system, decrease in obese adipose tissue [80]. Increases in Treg by antibody treatment (IL-2/anti-IL-2-induced Treg proliferation) improve HFD-induced insulin resistance [80]. Furthermore, transfer of CD4+ T lymphocytes with anti-inflammatory properties decreases HFD-induced glucose intolerance and insulin resistance [79]. The reasons for adipose tissue T lymphocyte and macrophage infiltration are unknown, but it was suggested that the corresponding antigens could be related to intestinal microbiota [6, 27, 81].
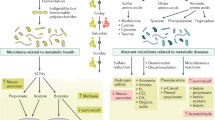
Metabolic functions putatively controlled by gut microbiota. Metafactors, i.e., bacterial compounds such as short-chain fatty acids, acetate, propionate, butyrate (SCFA), lipopolysaccharides (LPS), peptidoglycan (PGN), flagellin, and trimethylamine oxide (TMAO), could be involved in the control of metabolic functions, by means of gene expression and by installing a metabolic inflammation leading to cytokine production which could then impair all metabolic functions
Altogether, little is known about the sequence of events which lead to an increased number of macrophages and lymphocytes in metabolic tissues. The origin of this cascade of events could be related to a change in intestinal microbiota and, as detailed below, to a change in tissue microbiota [27]. This is linked to a translocation of bacteria and bacterial components from the intestine to tissues establishing a tissue microbiota leading to metabolic inflammation. Hence, the role of gut microbiota could be related to its direct role on molecular targets controlling insulin secretion and action, hepatic glucose production, adipose tissue development, and therefore, the incidence of diabetes. These hypotheses will be detailed below.
Although more than 150 genetic loci are associated with the monogenic or multifactorial forms of obesity and type 2 diabetes, their impact on the incidence of the disease is rather low (5–10 % and 2 %, respectively) [82]. In genetically identical twins the incidence of type 2 diabetes is 20–60 % [83, 84]. Even in type 1 diabetes, genetically identical twins develop the disease with an incidence of no more than 50 %. A major impact of the environment has recently been attributed to the role of intestinal microbiota where genes involved in the recognition of bacterial patterns were involved [85, 86]. The innate immunity to microbiota relationship was causally implied. It was shown that the commensal microbial community alters sex hormone levels and regulates autoimmune disease fate in individuals with high genetic risk [87]. On the other hand, environmental factors such as stress, a sedentary lifestyle, and nutritional habit could explain the pandemic progression of metabolic impairment. Humans could be considered as “super-organisms” as a result of their symbiotic association with the gut microbiota [88].
11.3 Lessons from the Gut Microbiota to Metabolic Diseases Relationship
Complex microbial ecosystems occupy the skin, mucosa, and alimentary tract of all mammals, including humans [8]. The species that make up these communities vary between hosts as a result of restricted migration of microorganisms between weak and strong ecological interactions within hosts. Furthermore, diet, genotype, and colonization history also influence this ecology [20]. Hence, a mutual relationship characterizes the host to microbiota crosstalk by which each partner has its own interest and informs the other of the environmental and metabolic situation. The microbiota is now considered as a symbiont that shares with its host the influence of the environment, diet, stress, and the physiological state. Specific communities inhabit the different epithelia according to the physical and biochemical characteristics of each location. With regard to the gastrointestinal tract, it is colonized by a vast community of symbionts and commensals that have important effects on immune function, nutrient processing, and a broad range of other host activities [89].
The precise role of intestinal microbiota on the control of metabolic diseases emerged in 2004 with the discovery that germ-free mice resist high-fat-diet-induced obesity [90]. It was shown that germ-free mice colonized with microbiota harvested from the cecum of a healthy mouse gain 60 % body fat content and became insulin resistant within 14 days despite reduced food intake. The mechanism was due to an increased production of monosaccharides. These molecules are generated from the metabolism of polysaccharides by the cecal microbiota, thus providing an additional source of carbohydrate to the body. However, a rate-limiting factor was that a large part of the microbiota cannot be identified since it is highly difficult to culture. This major problem has been overcome by the use of very-high-throughput sequencing techniques coupled with new bioinformatics approaches. This strategy allowed the identification of the taxons within the microbiota from human and animal intestinal content [91, 92] as well as the overall catalog of bacterial genes [18, 19]. This has allowed the correlation of metabolic diseases to specific bacteria or groups of bacterial genes [93]. An increased Firmicutes-to-Bacteroidetes ratio seems to be a signature of metabolic diseases influencing processes related to energy harvesting, intestinal permeability, bile acid metabolism, brain functions related to metabolism, and immunomodulation.
11.3.1 The Gut Microbiota as a Signature of Metabolic Diseases
An altered gut microbiota has been linked to metabolic diseases including obesity [26, 94], diabetes [19], and cardiovascular diseases [95]. A core microbiome can be found at the gene level, despite large variations in community membership, and that variations from the core are associated with obesity [25, 94, 96–98]. Using a protocol for a metagenome-wide association study (MGWAS) based on deep shotgun sequencing of the gut microbial DNA from 345 Chinese individuals, approximately 60,000 type 2 diabetes-associated markers have been identified and validated, thus establishing the concept of a metagenomic linkage group [19]. This MGWAS analysis showed that patients with type 2 diabetes were characterized by a moderate degree of gut microbial dysbiosis, a decrease in the abundance of some universal butyrate-producing bacteria, and an increase in various opportunistic pathogens, as well as an enrichment of other microbial functions conferring sulfate reduction and oxidative stress resistance. Three enterotypes could be identified from the Chinese samples which were primarily made up of several highly abundant genera, including Bacteroides, Prevotella, Bifidobacterium, and Ruminococcus. However, no significant relationship between enterotype and type 2 diabetes status was found. However, when using the gene reference (KEGG orthologue genes and eggNOG group profiles) rather than the phylogenic profiles, a total of 1,345 biomarkers were identified. Type 2 diabetes-associated biomarkers were mostly involved in membrane transport systems. By contrast, control-enriched markers were frequently involved in cell motility and metabolism of cofactors and vitamins. When studying pathway levels, the gut microbiota of type 2 diabetic patients showed enrichment in membrane transport of sugars, branched-chain amino acid (BCAA) transport, methane metabolism, xenobiotics degradation and metabolism, and sulfate reduction. By contrast, there was a decrease in the level of bacterial chemotaxis, flagellar assembly, butyrate biosynthesis, and metabolism of cofactors and vitamins. Seven of the markers were also related to oxidative stress resistance suggesting that the gut environment of type 2 diabetic patients stimulates bacterial defense mechanisms against oxidative stress [19]. A further specific mathematical model for biomarker identification has been developed and showed compositional and functional alterations in the metagenomes of a specific cohort of 145 women with type 2 diabetes [99]. Type 2 diabetic women were characterized by an increased abundance of four Lactobacillus species and decreases in the abundance of five Clostridium species. The total Lactobacillus species correlated positively with fasting glucose and HbA1c (glycosylated hemoglobin), whereas the Clostridium species correlated negatively with fasting glucose, HbA1c, insulin, C-peptide, and plasma triglycerides. Importantly, impaired glucose tolerance could be identified on the basis of this mathematical modeling. The impact of medication and hyperglycemia on the metagenome was not considered as major confounding factors. Comparisons with a Chinese cohort demonstrated that the biomarkers identifying type 2 diabetes were different from the European population suggesting that the metagenome analysis to predict type 2 diabetes should be specific for the age and geographical location of the populations studied. It should be further noted that the mechanisms at the origin of this observation are numerous and hence will most likely depend upon each individual.
11.3.1.1 Impacts of Diet on Microbiota-Related Metabolic Diseases
Socio-demographic and environmental factors have a great influence on the incidence of metabolic [100] and cardiovascular diseases [101], introducing the possibility of identifying functional metagenomic factors under the control of environmental factors such as stress, food habits, and sedentarity. Diet and nutritional status are among the most important modifiable determinants of human health, and gut bacteria feed on the nutrients absorbed during a meal leading to changes in metabolism, the overall intestinal ecology, and the way bacteria interact with the host. The first analyses of metagenomic sequencing have been performed on obese patients followed up during 1 year of a restricted calorie diet [97, 98, 102]. A clear metagenomic signature was identified in obese patients characterized by a reduction in the relative abundance of the Bacteroidetes-to-Firmicutes ratio which represents more than 80 % of the overall bacterial population in feces [97, 98] (Fig. 11.3). This ratio evolved toward that of lean patients during weight loss showing that the microbiota can evolve according to the environmental factors. It is clear that dietary manipulation, including HF feeding, profoundly alters the profile of the gut microbiota [27, 81, 103–107]. An enrichment in gram-negative to gram-positive bacteria appeared to be associated with the early onset of high-fat-diet-induced diabetes [81]. An elegant validation of the role of diet on human microbiota has been performed in germ-free mice colonized with human microbiota and fed with a fat-enriched diet [104]. A single day of fat-enriched diet was sufficient to change the overall ecological homeostasis within the gut microbiome, the corresponding metabolic pathways, and hence the microbiome gene expression. The influence of high-fat diet on gut microbiota has been validated in other species such as the pig [108, 109] which is highly influenced by pre- and probiotics [110] and sucrose [111]. Defined lipids such as oleic-acid-derived molecules and a combination of n-3 fatty acids markedly increased total bacterial density and restored the proportions of clostridial cluster, Enterobacteriales and Bifidobacterium that were changed during HFD feeding [112] along with the metabolic status. Conversely saturated fat increased the Firmicutes-to-Bacteroidetes ratio [113, 114]. In addition to the tremendous impact of dietary fibers, other molecules such as polyphenols influence gut microbiota and further control metabolism [115, 116]. Type 2 diabetes has also its own metagenomic signature, and type 2 diabetes-associated markers have been associated with the disease and can be useful to classify the different subgroups of type 2 diabetic patients [19]. Among these Akkermansia muciniphila seems to be a good candidate with a recently identified role in the control of the intestinal barrier [107]. A reduction of Akkermansia muciniphila, which is involved in mucus degradation, has also been observed [107]. These bacteria, when used as a probiotic, control body weight gain [107] and its prevalence increased in response to prebiotics [117] accompanying metabolic control through a mechanism that could involve gut peptide secretion [118]. Insulin action also has a metagenomic signature in humans [119]. Furthermore, hypotheses regarding the functional role of gut microbiota can be generated by studying the genes characterizing the microbiome. A decrease in the abundance of some universal butyrate-producing bacteria and an increase in various opportunistic pathogens, as well as an enrichment of other microbial functions conferring sulfate reduction and oxidative stress resistance, were also characterized. A change in intestinal microbiota has also been characterized in non-alcoholic fatty liver diseases (NAFLD) where the incidence is between 16 % and 30 % of the general population [120] and further rises toward 80 % in patients with obesity and type 2 diabetes [121]. Some biomarkers have been identified such as a reduction of Faecalibacterium and Anaerosporobacter and a higher abundance of Parabacteroides, Allisonella, certain Lactobacillus species, and selected members of the phylum Firmicutes (Lachnospiraceae; genera, Dorea, Robinsoniella, and Roseburia) [122]. The causal role of gut microbiota on NAFLD has been shown by microbiota transfer in the mice. The colonization of germ-free mice with the gut microbiota from a high-fat-diet-induced NAFLD mouse induced the disease. Sequencing of the 16S ribosomal RNA revealed differences at the phylum, genera, and species levels [123]. Some mechanisms have been proposed that implicate intestinal permeability, low-grade inflammation and immune balance in the development of hepatosteatosis [124], the modulation of dietary choline and bile acid metabolism, and the production of endogenous ethanol [125] that will be discussed below. Another important feature of the change in intestinal microbiota is that in addition to the change of phylum ratio obesity is associated with phylum-level changes in the microbiota and reduced bacterial diversity [97, 98, 126].
Intestinal functions and microbiota interplay. Upon a change in diet or a specific genetic background, the gut microbiota changes. A modification of the Firmicutes-to-Bacteroidetes ratio leading to a change in polysaccharide fermentation, bile acid metabolism, an accumulation of Proteobacteria (gram negative; releasing LPS, peptidoglycans, and other antigens), a reduced in Faecalibacterium- and Clostridium-producing butyrate, a reduced in Akkermansia muciniphila, and segmented filamentous bacteria (at least in rodents, SFB) would alter intestinal functions. Intestinal permeability, defenses (mucin and defensin production), innate (APC, antigen-presenting cells) and adaptive (T lymphocytes, TL) immunity, and lipid metabolism could lead to bacterial translocation and tissue microbiota dysbiosis, metabolic endotoxemia, and metabolic inflammation of the targeted tissues. Altogether, the tissue biology would be impaired and be considered as a risk factor for the development of cardiometabolic diseases
11.3.1.2 Impact of the Host Genome
In addition to the role of the environment on the shaping of gut microbiota during metabolic diseases, the impact of the host genome cannot be totally ruled out. The concordance of type 2 diabetes within the homozygote population is higher than between heterozygote twins [83, 84, 127] or in response to overfeeding [128, 129] suggesting the important role of the genetic background. The results reveal that the human gut microbiome is shared among family members but that each individual’s gut microbial community varies in the specific bacterial lineages present, with a comparable degree of covariation between adult monozygotic and dizygotic twin pairs [126]. These results demonstrate that a diversity of organismal assemblages can however yield a core microbiome at a gene level and that deviations from this core are associated with different physiological states, for example, obese versus lean. However, the concordance of the metabolic phenotype is not absolute between twins – this could be linked to differences in gut microbiota since the adult monozygotic twins are no more similar to one another in terms of their gut bacterial community structure than are adult dizygotic twins [126, 130]. However, the impact of the host genome on the microbiota seems to depend on the microbiota location considered. In the saliva twins resemble each other more closely than the whole population at all time points but become less similar to each other when they age and no longer cohabit [131]. The sequencing of gut microbiota from the general population and across countries and ethnic origin showed that shared features of the functional maturation of the gut microbiome are identified during the first 3 years of life [24], suggesting an imprinting from the mother which remains but is not absolute. Hence, room is available for the impact of environmental changes throughout life. Numerous studies now report that the influence of the genetic background is due to the impact of the immune system that shapes the microbial community [132–135]. Hence, the immunogenetic traits of an individual appear to be major regulators of gut microbiota. Mutations in the receptors to bacterial determinants such as TLRs which are pathogen-associated molecular pattern recognition receptors that recognize highly conserved microbial molecules (PAMPs) notably TLR2 [136], TLR4 [137], and TLR5 [132] and NLRs notably NOD2 [138–140] shape the microbiota although some controversies do exist [141].
11.3.1.3 Impact of Birth
Importantly, the host immune system to microbiota relationship continues to be educated throughout life since the immune system matures along with the microbiota at birth. The gastrointestinal tract of a normal fetus is sterile. During the birth process and rapidly thereafter, microbes from the mother and surrounding environment colonize the gastrointestinal tract of the infant until a dense, complex microbiota develops [9]. It is now clear that the composition and temporal patterns of the microbial communities vary widely from baby to baby [142]. The distinct features of each baby’s microbial community are recognizable for intervals of weeks to months which show that each baby’s microbiota, until the first year of life, follows an idiosyncratic law. However, it then converges toward a profile characteristic of the adult gastrointestinal tract. Therefore, during the first year of life, while the microbial ecology is not yet set up, the inheritance of gut microbiota could also be influenced during pregnancy and at delivery according to the influence of the mother’s microbiota. Furthermore, the mode of delivery influences the infant’s microbiota since it was shown that vaginally delivered infants acquire bacterial communities resembling their mother’s vaginal microbiota, dominated by Lactobacillus, Prevotella, or Sneathia spp., whereas C-section infants harbored bacterial communities similar to those found on the skin surface, dominated by Staphylococcus, Corynebacterium, and Propionibacterium spp. [11, 143]. However, the delivery mode did not influence the prevalence of obesity in children [144]. Conversely, antibiotic treatment during the first 6 months of life increased the risk of overweight in children with normal-weight mothers but reduced the risk of obesity in children with overweight mothers. Hence, at birth and during the first year of life, the early colonization of the gut by microbiota can influence the incidence of metabolic diseases [93]. Alterations in the microbiota composition of mothers may be transferred to infants and lead to an increased risk of weight gain. During pregnancy, gut microbiota changes dramatically between the first and third trimesters, with a vast expansion of diversity among mothers, an overall increase in Proteobacteria and Actinobacteria, and reduced richness [145]. This change in gut microbiota during pregnancy was functional since colonization of germ-free mice with the microbiota from the third trimester induced greater adiposity and insulin resistance when compared to mice colonized with the microbiota from the first trimester [145]. This original observation could suggest that the change in gut microbiota during pregnancy programs infant body weight. It has also been suggested that the microbiota from obese mothers could influence the prevalence of obesity in children. This was further supported by the fact that the infants’ fecal microbial composition was related to the weight and weight gain of their mothers during pregnancy [146]. The fecal Bacteroides and Staphylococcus concentrations were significantly higher in infants of overweight mothers, whereas the prevalences of Akkermansia muciniphila, Staphylococcus, and Clostridium difficile groups were lower in infants of normal-weight mothers and of mothers with normal weight gain during pregnancy. This was confirmed in that the concentration of the gram-negative family Enterobacteriaceae was significantly higher in obese/overweight children and the levels of Desulfovibrio and Akkermansia muciniphila-like bacteria were significantly lower when compared to lean controls [147]. Chapter 8 “Metabonomics in neonatal and pediatric research: Studying and modulating gut functional ecology for optimal growth and development” provides a comprehensive overview of the state of the art of metabonomics and gut microbiota studies in neonatal and pediatric research.
11.3.2 Molecular Crosstalk Between Gut Microbiota and the Host for the Control of Metabolic Diseases
11.3.2.1 The Lipopolysaccharides Hypothesis and Metabolic Inflammation
Whereas extensive analyses demonstrate the important impact of gut microbiota on host biology, the mechanisms of the crosstalk between the host and the microbiota remain to be delineated. As mentioned above, metabolic inflammation is a leading mechanism responsible for the impairment of glycemia and body weight regulation. One hypothesis involves bacterial factors from the gut, such as lipopolysaccharides (LPS). Lipopolysaccharides are components of the wall from gram-negative bacteria [1]. They are potent endotoxins, involved in the acute-phase response to bacterial infection, inducing a cytokine-mediated systemic inflammatory response that can cause shock and severe multiple organ failure [2, 3]. These bacterial antigens bind to their receptors TLR4 and CD14 on numerous cells types notably those of the immune system, i.e., macrophages and dendritic cells. Adipocytes also express TLR4 and can bind LPS and could be involved directly in the activation of intracellular inflammatory pathways [148, 149].
11.3.2.1.1 Mechanisms of High-Fat-Diet-Induced Blood Bacterial Molecular Patterns
LPS accumulates in blood and contributes to inflammation and insulin intolerance [6, 81]. A 1-month high-fat feeding in humans increased endotoxemia by 71 % [150], suggesting that therapeutic agents that reduce intestinal LPS permeability could control metabolic endotoxemia and hence systemic inflammation in patients with metabolic syndrome. Similarly, in type 1 diabetic patients, metabolic endotoxemia was associated with dyslipidemia, insulin resistance, obesity, and chronic inflammation [151]. The mechanism is associated with an increase of the gram-negative to gram-positive ratio within the intestinal microbiota [81]. LPS need to be transported across the intestinal epithelial barrier to reach the blood. A mechanism involving the synthesis and production of chylomicron is required [152]. The uptake of LPS is observed within hours of lipid absorption in human [153] and could hence be considered as a blood nutritional signal informing the body of a change in feeding behavior and in microbiota ecology. The accumulation of LPS in the blood is also linked to an increased intestinal permeability where tight junctions become leaky due to a reduction of the expression of specific proteins such as zonula occludens [81, 106] and the putative role of endocannabinoids [154] that could control intestinal permeability through a GLP-2-dependent mechanism [155]. This is reversed upon treatment with prebiotics [156, 157] or probiotics [107]. The increase of plasma LPS concentration is called metabolic endotoxemia. Lipoproteins bind LPS to attenuate the biological inflammatory response of this bacterial factor [158, 159] through all classes of lipoprotein (chylomicrons, VLDL, LDL, and HDL) [9]. Chylomicrons and VLDL have been shown to reduce LPS-induced toxicity in mice [10]. Similarly, the binding of LPS to LDL reduced endothelial cell activity [9, 11]. In one study performed in 10 individuals, HDL has been shown to be the main LPS carrier holding 60 % of the LPS, with LDL and VLDL carrying, respectively, 25 % and 12 % [14]. Among three ex vivo studies, two have indicated that LPS, in humans, was mainly located in VLDL and LDL [15, 16] and one has more precisely indicated that LDL was the major carrier for LPS [9]. Therefore, it is now evident that nutritional and lipid metabolism are tightly linked to the intestinal absorption of gut bacterial factors including LPS for the triggering of metabolic inflammation. The pharmacological control of intestinal LPS absorption could be of importance for the prevention or treatment of metabolic diseases. A change in nutrition involving an increase in fat content is a risk factor for metabolic endotoxemia as demonstrated in epidemiological studies in humans [151, 160, 161]. In the mouse the ablation of various pattern recognition receptors (PRRs) such as TLR4, CD14, and NLRP3 protects mice from diet-/obesity-induced inflammation and insulin resistance [6, 162–164]. In human myotubes, LPS increased JNK phosphorylation and MCP-1 and IL-6 gene expression [165] and could play a role in the pathogenesis of insulin resistance. Therefore, antagonists of CD14/TLR4 may improve insulin action in type 2 diabetic patients. In the mouse the chronic subcutaneous infusion of LPS at low rates favors liver, adipose tissue, and muscle cytokine production and macrophage accumulation showing that in vivo metabolic endotoxemia is an important risk factor of insulin resistance and hence metabolic diseases. Direct evidence shows that the LPS to ligand crosstalk controls adipose tissue inflammation [6, 162, 164, 166], insulin sensitivity [148, 167], and obesity [6, 168] through molecular mechanisms most likely linked to the activation of NFkB and the transcription of genes coding for cytokines. The latter, such as TNFα, will then interfere with the insulin receptor leading to insulin resistance [169]. LPS are also involved in the reduction of adipogenesis [170], as well as hepatic steatosis [171, 172] again through mechanisms involving TNFα production or SREBP-1 activation [173]. Other bacterial components are involved in the inflammatory process characterizing metabolic inflammation. The peptidoglycan is a complex structure of the bacterial wall found mostly in gram-positive bacteria but also found, to a lower extent, in gram-negative bacteria. Its polysaccharide component is different between the two types of bacteria and binds to pattern recognition receptors such the nucleotide oligomerization domain (NOD)1 and NOD2. NOD1 and NOD2 are currently the only known sensors of bacterial cell wall peptidoglycan (PGN) that elicit inflammation by increasing cytokine production, defensin expression by Paneth cells [174–177], and stress kinase responses [178]. In addition to LPS, PGN could be positioned as a component of metabolic endotoxemia that contributes to inflammation and metabolic defects [27, 179]. This is important because gut-derived LPS alone appears to be insufficient for establishing glucose/insulin tolerance in poorly immune-responding germ-free mice [180]. Furthermore, PGN containing meso-DAP motifs (generally dominant in gram-negative bacteria) caused profound insulin resistance through actions on NOD1 directly in metabolic cells, including adipocytes [179]. The NOD2 activation with the minimal bioactive PGN motif, muramyl dipeptide (MDP), mostly present in gram-positive bacteria, elicited cell autonomous inflammation and impaired insulin action directly in muscle cells [181] and caused acute, peripheral insulin resistance in vivo [179]. Mice lacking NOD2 have improved insulin sensitivity during obesity illustrating the protective role of NOD2 as a sensor of bacterial motifs derived from the gut microbiota on the control of metabolic diseases [27, 179]. However, defects in NOD2 immunity have been associated with promoting other chronic proinflammatory pathologies, and human NOD2 variants have the highest risk associated with Crohn’s disease [182–184]. NOD2 immunity is known to contribute to homeostasis of the gut microbiota demonstrating that the microbiota is in a tight relationship with the immune system that secondarily controls the metabolism [138].
11.3.2.1.2 Metabolic Endotoxemia and Gut Microbiota Induced Hepatic Steatosis
Possible mechanisms leading to hepatosteatosis in obese and type 2 diabetic patients that involve gut microbiota implicate intestinal permeability, low-grade inflammation and immune balance [124], the modulation of dietary choline and bile acid metabolism, and the production of endogenous ethanol [125]. A first hypothesis is proposed regarding the key role of the intestinal epithelium as a barrier between the environment, i.e., the luminal side of the intestine and the body. A leaky gut has been described in patients with NAFLD [185] and linked to a change in intestinal microbiota [122, 186]. A second hypothesis is linked to the role of choline which is a major phospholipid component of the cell membrane involved in signal transduction and the control of lipoprotein metabolism, notably the very-low-lipoprotein assembly [187–189]. Therefore, a choline-deficient diet promotes liver steatosis. Such depletion could be induced by the gut microbiota that would convert dietary choline into toxic methylamines [190, 191]. It was shown that circulating levels of plasma phosphatidylcholine were low, whereas urinary excretion rates of methylamines dimethylamine, trimethylamine, and trimethylamine-N-oxide were high and were coprocessed by symbiotic gut microbiota and mammalian enzyme systems. An inflammatory feature is required to qualify hepatic steatosis (NASH) from a NAFLD phenotype. It could originate from the gut microbiota since it interacts with the overall immunity of the host via PAMPs TLRs and NLRs [175]. Similarly, the role of components from the inflammasome such as NLRP6 and NLRP3 has been demonstrated [124]. These inflammation sensors could modulate the gut microbiota and hence the crosstalk of host immunity and gut microbiota leading to the development of the pathology. Altogether, the triggering of innate immunity could lead to the production of cytokines which contribute to the inflammatory phenotype of NASH through a mechanism similar to that suggested for the development of type 2 diabetes and obesity.
11.3.2.2 The Energy Harvesting, Expenditure, and Short-Chain Fatty Acid Hypotheses
The change in intestinal microbiota (increased Firmicutes and decreased Bacteroidetes), observed during obesity, was associated with an increased efficiency of energy harvest in human and mouse [26, 105]. The fecal content in energy was lower in obese mice than in controls by about 150 kCal/day, suggesting that the energy was better absorbed by the gut. This observation was attributed to the excessive hydrolysis of polysaccharides into monosaccharides indispensable to the body as well as to the production of short-chain fatty acids (SCFA) [26]. However, it seems that the production of SCFA diminished over time during long-term high-fat feeding or aging in ob/ob mice [105]. The role of gut microbiota in the production of SCFA is however strongly supported since it was shown that germ-free mice are devoid of SCFAs [192] and could control AMP-activated protein kinase activity and macrophage infiltration in adipose tissue [193]. Human colonic butyrate producers are phylogenetically diverse, with the two most abundant groups related to E. rectale/Roseburia species and to Faecalibacterium prausnitzii. The precise SCFA responsible for the control of metabolism seems to be multiple with a significant role for acetate. In addition, SCFAs such as propionate can be used for de novo glucose or lipid synthesis and serve as an energy source for the host. It has also been demonstrated that butyrate lowers fatty acid content in liver and plasma, reduces food intake, exerts immunosuppressive actions, and probably improves tissue insulin sensitivity [194, 195]. The mechanism could involve notably the promotion of glucagon-like peptide 1 (GLP-1) secretion via the binding of SCFA to G-protein-coupled receptors such as GPR41/43 at the surface of the enteroendocrine cells [195]. Cascades of events such as the control of glucagon and insulin secretion, gastric emptying, and satiety would secondary to GLP-1 secretion control glucose homeostasis. Butyrate is also involved in the improvement of the overall glucose metabolism as demonstrated by its supplementation to a high-fat diet [196]. Other experiments show that by high-pressure liquid chromatography analysis, authors identified significantly higher concentrations of butyrate and propionate in feces from obese versus normal-weight children [197]. Significantly lower concentrations of intermediate metabolites were detected in obese children suggesting exhaustive substrate utilization by obese gut microbiota [197]. Despite this evidence, the role of SCFA on metabolism still remains unclear. Recent data demonstrate that the activation of GPR43 by SCFA at the surface of adipocytes reduced insulin signaling [198]. This could certainly reduce insulin-induced lipid accumulation, and hence body weight gain, but could also be considered as a factor inducing adipose tissue insulin resistance which could prevent glucose to be taken up by adipocytes and hence accumulate in the blood or the liver to induce hyperglycemia and hepatic steatosis. Thus, data relating to the production of SCFA most likely cover other unidentified mechanisms that need to be identified to fully understand the benefit of polysaccharide fermentation.
The role of gut microbiota could also be on energy expenditure since germ-free mice are leaner despite a dramatically increased food intake [90]. The lean phenotype of these mice is associated with increased skeletal muscle and liver levels of phosphorylated AMP-activated protein kinase (AMPK) and its downstream targets involved in fatty acid oxidation such as acetylCoA carboxylase and carnitine palmitoyltransferase [199]. AMPk is a master switch considered to be a molecule recruited in case of stress and energy deprivation [200, 201]. Bacterial factors could activate this enzyme in muscles and the liver to increase energy expenditure. Conversely, the microbiota inhibits fasting-induced adipose factor (Fiaf) which downregulates circulating lipoprotein lipase. Hence, free fatty acids are stored in the liver of conventional mice which is not the case in germ-free animals [199].
Altogether, numerous eukaryotic targets are currently being identified using germ-free mice. An important matter will be to validate them in physiological conditions such as in conventional mice and in humans.
11.3.3 Gut Microbiota During Obesity Surgery
The last 40 years has seen the emergence of the treatment of obesity by surgery. Several types of surgery involving the stomach and the intestine are used. The Roux-en-Y gastric bypass (RYGB) surgery which is to date the major bariatric intervention to treat morbid obesity involves the direct connection of the jejunum to the stomach so that nutrients are no longer in contact with the duodenum. In addition, 90 % of the stomach is removed. The sleeve gastrectomy involves the removal of a major part of the stomach so that food directly reaches the duodenum. Other types of surgery such as the ileal bypass transposition are being developed. All surgical procedures dramatically affect gut microbiota [202, 203]. A first set of experiments demonstrated that before surgery, Firmicutes were dominant in normal-weight and obese individuals but significantly decreased in post-gastric bypass [204]. Interestingly, the gut microbiota from patients after surgery was not similar to that of lean individuals since it was enriched in Gammaproteobacteria which are H(2)-producing Prevotellaceae [204]. These changes were independent of weight change and caloric restriction, were detectable throughout the length of the gastrointestinal tract, and were most evident in the distal gut, downstream of the surgical manipulation site [205]. A precise study identified 14 discriminant bacterial genera (7 were dominant and 7 were subdominant) and 202 genes changed in the white adipose tissue that correlated with RYGB as well as with both clinical phenotypes [206]. Faecalibacterium prausnitzii species was lower in patients with metabolic diseases but associated negatively with inflammatory markers even throughout the follow-up after surgery and independently with changes in food intake suggesting an imprinting of the microbiota that cannot be easily changed [207].
11.3.3.1 The Bile Acids to Microbiota Hypothesis
Intestinal microbiota also has a role in the metabolism of bile acids, which, with other sterols [208], are important regulators of metabolic diseases, as also described in Chaps. 13 and 14. Bile acid can control insulin secretion [209], GLP-1 secretion [210], energy expenditure [211], as well as atherosclerosis [212]. The role of bile acid is also suspected during bariatric surgery and could be the molecular link between the change in intestinal microbiota [213] and the improvement of metabolism [214] notably through the TGR5 receptor [215]. Bile acids are secreted into the duodenum and work to emulsify liposoluble dietary nutrients to facilitate their digestion and absorption. Studies have demonstrated that bile acid composition and secretion in response to fat intake modifies markedly gut microbiota thereby inducing a dysbiosis [216]. The dietary fat can alter the gut microbiota of mice indirectly by changing the animals’ pool of bile acids and steroids that are produced by the liver and secreted into the intestine [216]. Bile acids are synthesized from cholesterol in the liver and further metabolized by the gut microbiota into secondary bile acids [217] which can change the metabolism by acting on the farnesoid X receptor to exert some negative feedback control. When compared to germ-free mice, the conventionally raised mice are characterized by a dramatic reduction in muricholic acid, but not cholic acid levels [218]. The microbiota controls fibroblast growth factor 15 in the ileum and cholesterol 7α-hydroxylase (CYP7A1) in the liver by FXR-dependent mechanisms thereby influencing the metabolism and secretion of bile acids.
Hence, a new ecology is expected from the host and the microbiota through bile acid metabolism.
11.3.3.2 The Immunomicrobiota Crosstalk and Metabolic Diseases
Metabolic diseases are now considered as low-grade immunomodulatory diseases [58–60]. The origin of the antigen is unknown but has been suspected to be from autoantigens, which notably form the adipose tissue [219–221]. The discovery of intestinal microbiota opens new avenues regarding the origin of the metabolic inflammation. This hypothesis stemmed from the observation that intestinal permeability was increased in high-fat-diet-fed mice, leading to accumulation in the blood of bacterial fragments such as LPS [6, 160]. Whereas, during diabetes and obesity, LPS and peptidoglycan were shown to be transported from the intestinal lumen to the blood through the intestinal epithelium, it was initially thought that whole commensal bacteria would be arrested by the mucosal layer and the immune system within Peyer’s patches or the lamina propria [222, 223], thus preventing translocation across the intestinal epithelial layer [224, 225].
A deficiency in host immune defenses and increased permeability and damage to the intestinal mucosal barrier represent mechanisms through which bacteria of intestinal origin accumulate into the tissues. In the healthy situation macrophages, dendritic cells, and antibodies within the lumen restrain, along with the mucus layer, the bacteria within the luminal side of the gut to ensure a tight intestinal permeability. In the proximal intestinal segments where the microbiota is sparse and the mucus layer is thin or absent, commensal bacteria are in close contact with the epithelial cells, and host immunosuppression synergistically promotes bacterial translocation from the gastrointestinal tract resulting in accumulation of bacteria within the mesenteric lymph node [225]. Therefore, lymphocytes from mesenteric lymph nodes, Peyer’s patches, intraepithelial cells, and the lamina propria are specifically educated to recognize commensal bacteria, which are therefore considered as self-antigens. The change of intestinal microbiota that occurs during a fat-enriched diet [81, 104] leads to the production of new antigens that are no longer recognized as self by the intestinal immune cells [134]. This change in gut microbiota can also be controlled by natural antibiotics such as defensins secreted by Paneth cells [226, 227].
11.3.3.3 Intestinal Bacterial Translocation and Metabolic Diseases
Bacterial translocation is defined as the passage of viable indigenous bacteria from the gastrointestinal tract to extra intestinal sites, such as the mesenteric-lymph-node complex, liver, spleen, and bloodstream [224]. This mechanism is largely observed during intestinal bacteria overgrowth leading to cirrhosis [228] or sepsis [229]. Although it is intuitively considered as deleterious for the organism, it could be suggested that the bacterial translocation helps the immune system to be prepared against infections. Another pathological situation of increased bacterial translocation is AIDS where, due to impaired intestinal immunosuppression, bacteria translocate toward tissues and could lead to inflammation and increased mortality [230]. The immune inhibitory receptor programmed death-1 (PD-1) regulates the function of CD8+ cells and the translocation of bacteria [230]. Importantly AIDS is associated with a dramatic increase in the incidence of diabetes and lipodystrophy [231, 232]. The CD4 T helper and Th17 cells appear to be critical for regulating gut mucosal immune responses to extracellular microbial pathogens and therefore could be involved in bacterial translocation [233]. Hence, an impaired intestinal immune system could lead to bacterial translocation and therefore to metabolic diseases. This hypothesis has been validated during high-fat diet-induced metabolic diseases [27]. First it was observed that a rapid augmentation of bacterial adherence to the intestinal epithelium layer occurred suggesting an impaired mucosal defense. Then CD11c positive phagocytes harvested the transepithelial bacteria and translocated with the live bacteria to adipose tissue. The bacteria were co-localized within the adipose depot with CD11c positive cells and most likely were intracellular. Importantly, the bacterial accumulation was considered as a predictive biomarker of type 2 diabetes [234]. The translocated bacteria were also detected in the blood in humans and conversely to the adipose tissue. The Proteobacteria phylum represented more than 80 % of the blood microbiota as assessed by 16S RNA DNA. Interestingly, other bacterial DNA fragments could predict the onset of cardiovascular events in a large cohort of type 2 diabetic patients [235]. The processes involved in bacterial translocation at the onset of type 2 diabetes could be related to molecular determinants involved in bacterial recognition. The NOD1 and LPS-CD14 bacterial receptors were involved in this translocation mechanism since their deletion dramatically reduced the amount of bacterial DNA present within the adipose tissue under high-fat diet [27]. Interestingly, both deletions were associated with improved insulin sensitivity and reduced glycemia suggesting that the tight control of bacterial translocation could be a master regulator of the onset of insulin resistance and diabetes. Conversely, the deletion of NOD2 induced over-accumulation of bacterial DNA within the tissues suggesting a protective effect. This was also observed in mice treated with NOD2 ligands [179]. The importance of the immune system in the translocation mechanism was also illustrated in mice carrying a deleted MyD88 gene and which were prone to diabetes [236]. These mice also displayed a dramatic accumulation of bacteria in the mesenteric lymph nodes and adipose tissue. Therefore, metabolic regulators of bacterial translocation could be linked to hormones controlling the immune system such as estrogens [237] or leptin [238]. This shift in the paradigm is supported by data which show that cardiovascular disease is associated with the role of microbiota in the control of lipid metabolism leading to the development of atherosclerosis [95, 239, 240]. Surprisingly, microbes associated with periodontitis were at the origin of the bacteria present in the plaques of atherosclerotic patients [240]. Studies in animals have revealed a mechanistic link between intestinal microbial metabolism of the choline moiety in dietary phosphatidylcholine (lecithin) and coronary artery disease through the production of a proatherosclerotic metabolite, trimethylamine-N-oxide (TMAO) [241, 242]. This has been confirmed in humans [243]. Importantly, antibiotic administration reduced the TMAO concentration suggesting that the microbiota was indeed the source of this proatherogenic molecule. Furthermore, in addition to bacterial factors, whole bacteria have been identified within the atherosclerotic plaques. The taxa have been linked to periodontal diseases [240]. This finding bridges the gap between metabolic and cardiovascular diseases by means of opportunistic bacteria from the oral cavity. Its proportion increases in response to a change of diet [244]. In addition, genes closely involved in the regulation of cardiovascular diseases such as angiotensin I-converting enzyme (peptidyl-dipeptidase A) 2 (Ace2), which encodes a key regulatory enzyme of the renin-angiotensin system, also have an impact on gut microbiota leading to inflammation [245]. Furthermore, statins, classical anti-dyslipidemic agents prescribed to reduce the incidence of cardiovascular events, have been shown to be associated with microbial-derived agents that could be responsible for the cholesterol-lowering effect, explaining the efficacy of the treatment [246]. Cholesterol metabolism is indeed regulated by gut microbiota since there is a dramatic reduction of muricholic acid, a farnesoid X receptor (FXR) antagonist in the ileum, in conventional mice when compared with germ-free mice. Therefore, a given microbiota could be important for the efficacy of a drug treatment as well as for the susceptibility of developing cardiometabolic diseases, thus reconciling metabolic and cardiovascular diseases under the paradigm of the gut to tissue microbiota crosstalk.
11.3.4 Tissue Microbiota and Metabolic Diseases: The Paradigm Shift of Bacteria Translocation
Bacterial factors such as LPS, peptidoglycans, and bacterial DNA can be absorbed by the gut and found within the blood. These metafactors could serve as biomarkers and also as regulators since they can then activate cells from the immune system to generate inflammation. Pyrosequencing of the 16S RNA DNA from the stroma vascular fraction of human adipose tissue from lean, overweight, and obese patients showed that the diversity of the tissue microbiota was mainly related to Firmicutes and Proteobacteria (in press). Interestingly, although no changes were observed among the Firmicutes phylum, a major dysbiosis was detected within the Proteobacteria phylum and specifically the Ralstonia genus. Ralstonia was dramatically overrepresented within the family and, furthermore, its presence was proportional to increased BMI. This suggested that this bacterial genus might be causal. The bacterial translocation process also leads to the accumulation of bacterial DNA in the blood fraction [234], suggesting that either bacteria migrate through the blood to reach the tissues or are released by the tissues into the blood.
The role of this tissue microbiota is not known; similarly the proportion of live bacteria compared to dead bacteria or fragments is not perfectly identified and will most likely depend on diet, age, and other genetic and environmental factors. This paradigm shift is promising but will require much work to determine its physiological role.
11.3.4.1 Tissue Microbiota as Biomarkers of Metabolic and Cardiovascular Diseases
The blood tissue microbiota has been recently described in humans and mice [27, 234, 235]. The presence of bacterial DNA has been revealed by qPCR from human cohorts from the general population [234] and from diabetic patients [235]. It could be shown that the increased 16S RNA DNA concentration in blood predicts the onset of diabetes 6–9 years later [234], whereas the quantification of other bacterial factors could predict the onset of cardiovascular events in a population of type 2 diabetics [235]. In both cases it is suggested that the bacterial fragments which accumulate in the blood could be causal of the disease. Since the bacteria are inherited at birth, the accumulation of bacterial DNA initiates at a very early stage, linking the environment with the host genome. In risk situations, such as when feeding on a fat-enriched diet, the bacterial DNA increases reflecting the new nutritional situation and could be involved in the triggering of metabolic adaptation. Adipose tissue is targeted [6, 27, 137] and adipogenesis is induced [170]. Hence, these bacteria can be considered as nutrient sensors informing the tissues.
11.4 Therapeutic Perspectives
The therapeutic perspectives are huge but so far at their infancy, as presented and discussed in Chap. 19. One should separate the nutritional approaches aiming at maintaining health in the general population. This would first involve phenotyping of the human microbiome [21] and second performing studies to demonstrate the prevention of the risk of disease development. A second strategy would involve preventing the disease in patients at risk to develop metabolic diseases. In these patients subpopulations should be defined and studied in order to perfectly adapt the nutritional strategy. This should be a precisely directed strategy to treat patients for the prevention of developing a metabolic disease. This would involve overweight individuals, smokers, sedentary individuals, and hypertensive patients. With this aim pre- and probiotics selected for their capacity to influence all the above physiological mechanisms at the cross road of microbiota and the host should be delineated. Other nutritional approaches could be pursued aiming to target intestinal functions [115] by using antioxidants like polyphenols such as resveratrol. From a therapeutic point of view, pharmacological strategies can be envisaged that involve small molecules to target molecular mechanisms such as the immune system, mucosal defense, bile acid synthesis, incretin secretion, or the production of short-chain fatty acids. Again, they should address well-defined subgroups of patients characterized by their specific microbiota or blood biomarkers. Companion blood bacterial biomarkers could be used to monitor the impact of the intervention over a long-term treatment. This reasoned strategy should help to control metabolic diseases and associated cardiovascular events. The advent of next-generation sequencing strategies along with the development of bioinformatics and biostatistical skills are now available to set up these programs.
11.5 Conclusions
In the quest of a mechanism explaining the pandemic development of metabolic diseases, the consequent cardiovascular events, and further reconciling the key role of the adaptive and innate immune system, the role of the intestinal microbiota has emerged as a very promising candidate. The diversity and huge complexity of the microbiome precludes a rapid and clear identification of the molecular mechanisms at the crosstalk between the host and the microbiota. The molecular hypotheses for the explanation of the metabolic phenotype are numerous and probably related to the large number of molecular origins of the disease. This suggests that patients should be screened for their microbiota to host crosstalk. Thus, therapeutic strategies or preventive programs could be successful. We have entered a new era and one can no longer disregard the microbiome from the gut or other locations since it is now clearly involved at the onset and during the development of metabolic disease.
References
Haffner SM, Ruilope L, Dahlof B, Abadie E, Kupfer S, Zannad F. Metabolic syndrome, new onset diabetes, and new end points in cardiovascular trials. J Cardiovasc Pharmacol. 2006;47(3):469–75.
Juonala M, Magnussen CG, Berenson GS, Venn A, Burns TL, Sabin MA, et al. Childhood adiposity, adult adiposity, and cardiovascular risk factors. N Engl J Med. 2011;365(20):1876–85.
Franks PW, Hanson RL, Knowler WC, Sievers ML, Bennett PH, Looker HC. Childhood obesity, other cardiovascular risk factors, and premature death. N Engl J Med. 2010;362(6):485–93.
Virmani R, Kolodgie FD, Burke AP, Farb A, Schwartz SM. Lessons from sudden coronary death: a comprehensive morphological classification scheme for atherosclerotic lesions. Arterioscler Thromb Vasc Biol. 2000;20(5):1262–75.
Danaei G, Lawes CM, Vander Hoorn S, Murray CJ, Ezzati M. Global and regional mortality from ischaemic heart disease and stroke attributable to higher-than-optimum blood glucose concentration: comparative risk assessment. Lancet. 2006;368(9548):1651–9.
Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56(7):1761–72.
Ford ES, Li C, Sattar N. Metabolic syndrome and incident diabetes: current state of the evidence. Diabetes Care. 2008;31(9):1898–904.
Dethlefsen L, Eckburg PB, Bik EM, Relman DA. Assembly of the human intestinal microbiota. Trends Ecol Evol. 2006;21(9):517–23.
Mackie RI, Sghir A, Gaskins HR. Developmental microbial ecology of the neonatal gastrointestinal tract. Am J Clin Nutr. 1999;69(5):1035S–45.
Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118(2):511–21.
Azad MB, Konya T, Maughan H, Guttman DS, Field CJ, Chari RS, et al. Gut microbiota of healthy Canadian infants: profiles by mode of delivery and infant diet at 4 months. Can Med Assoc J. 2013.
Barrett E, Guinane CM, Ryan CA, Dempsey EM, Murphy BP, O’Toole PW, et al. Microbiota diversity and stability of the preterm neonatal ileum and colon of two infants. Microbiologyopen. 2013;2:215.
Gronlund MM, Lehtonen OP, Eerola E, Kero P. Fecal microflora in healthy infants born by different methods of delivery: permanent changes in intestinal flora after cesarean delivery. J Pediatr Gastroenterol Nutr. 1999;28(1):19–25.
Orrhage K, Nord CE. Factors controlling the bacterial colonization of the intestine in breastfed infants. Acta Paediatr Suppl. 1999;88(430):47–57.
Stratiki Z, Costalos C, Sevastiadou S, Kastanidou O, Skouroliakou M, Giakoumatou A, et al. The effect of a bifidobacter supplemented bovine milk on intestinal permeability of preterm infants. Early Hum Dev. 2007;83(9):575–9.
Claesson MJ, Cusack S, O’Sullivan O, Greene-Diniz R, de Weerd H, Flannery E, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci U S A. 2011;108 Suppl 1:4586–91.
Maslowski KM, Mackay CR. Diet, gut microbiota and immune responses. Nat Immunol. 2011;12(1):5–9.
Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65.
Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490(7418):55–60.
Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449(7164):811–8.
Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449(7164):804–10.
Zoetendal EG, Akkermans AD, De Vos WM. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl Environ Microbiol. 1998;64(10):3854–9.
Biagi E, Candela M, Fairweather-Tait S, Franceschi C, Brigidi P. Aging of the human metaorganism: the microbial counterpart. Age (Dordr). 2012;34(1):247–67.
Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–7.
Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124(4):837–48.
Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–31.
Amar J, Chabo C, Waget A, Klopp P, Vachoux C, Bermudez-Humaran LG, et al. Intestinal mucosal adherence and translocation of commensal bacteria at the early onset of type 2 diabetes: molecular mechanisms and probiotic treatment. EMBO Mol Med. 2011;3(9):559–72.
Chen X, D’Souza R, Hong ST. The role of gut microbiota in the gut-brain axis: current challenges and perspectives. Protein Cell. 2013;4:403.
Lebovitz HE. The relationship of obesity to the metabolic syndrome. Int J Clin Pract Suppl. 2003;134:18–27.
Chen G, Koyama K, Yuan X, Lee Y, Zhou YT, O’Doherty R, et al. Disappearance of body fat in normal rats induced by adenovirus-mediated leptin gene therapy. Proc Natl Acad Sci U S A. 1996;93(25):14795–9.
Shimabukuro M, Koyama K, Chen G, Wang MY, Trieu F, Lee Y, et al. Direct antidiabetic effect of leptin through triglyceride depletion of tissues [see comments]. Proc Natl Acad Sci U S A. 1997;94(9):4637–41.
Zhou YT, Shimabukuro M, Koyama K, Lee Y, Wang MY, Trieu F, et al. Induction by leptin of uncoupling protein-2 and enzymes of fatty acid oxidation. Proc Natl Acad Sci U S A. 1997;94(12):6386–90.
Shepherd PR, Gnudi L, Tozzo E, Yang H, Leach F, Kahn BB. Adipose cell hyperplasia and enhanced glucose disposal in transgenic mice overexpressing GLUT4 selectively in adipose tissue. J Biol Chem. 1993;268(30):22243–6.
Casteilla L, Planat-Benard V, Cousin B, Silvestre JS, Laharrague P, Charriere G, et al. Plasticity of adipose tissue: a promising therapeutic avenue in the treatment of cardiovascular and blood diseases? Arch Mal Coeur Vaiss. 2005;98(9):922–6.
Caspar-Bauguil S, Cousin B, Galinier A, Segafredo C, Nibbelink M, Andre M, et al. Adipose tissues as an ancestral immune organ: site-specific change in obesity. FEBS Lett. 2005;579(17):3487–92.
Galinier A, Carriere A, Fernandez Y, Carpene C, Andre M, Caspar-Bauguil S, et al. Adipose tissue proadipogenic redox changes in obesity. J Biol Chem. 2006;281(18):12682–7.
Sengenes C, Lolmede K, Zakaroff-Girard A, Busse R, Bouloumie A. Preadipocytes in the human subcutaneous adipose tissue display distinct features from the adult mesenchymal and hematopoietic stem cells. J Cell Physiol. 2005;205(1):114–22.
Maumus M, Peyrafitte JA, D’Angelo R, Fournier-Wirth C, Bouloumie A, Casteilla L, et al. Native human adipose stromal cells: localization, morphology and phenotype. Int J Obes (Lond). 2011;35:1141.
Planat-Benard V, Silvestre JS, Cousin B, Andre M, Nibbelink M, Tamarat R, et al. Plasticity of human adipose lineage cells toward endothelial cells: physiological and therapeutic perspectives. Circulation. 2004;109(5):656–63.
Villaret A, Galitzky J, Decaunes P, Esteve D, Marques MA, Sengenes C, et al. Adipose tissue endothelial cells from obese human subjects: differences among depots in angiogenic, metabolic, and inflammatory gene expression and cellular senescence. Diabetes. 2010;59(11):2755–63.
Adachi A, Shimizu N, Oomura Y, Kobashi M. Convergence of hepatoportal glucose-sensitive afferent signals to glucose-sensitive units within the nucleus of the solitary tract. Neurosci Lett. 1984;46(2):215–8.
Burcelin R. The gut-brain axis: a major glucoregulatory player. Diabetes Metab. 2010;36 Suppl 3:S54–8.
Konturek SJ, Konturek JW, Pawlik T, Brzozowski T. Brain-gut axis and its role in the control of food intake. J Physiol Pharmacol. 2004;55(1 Pt 2):137–54.
Holst JJ. The physiology of glucagon-like peptide 1. Physiol Rev. 2007;87(4):1409–39.
Cabou C, Burcelin R. GLP-1, the gut-brain, and brain-periphery axes. Rev Diabet Stud. 2012;8:418–31.
Knauf C, Cani PD, Kim DH, Iglesias MA, Chabo C, Waget A, et al. Role of central nervous system glucagon-like peptide-1 receptors in enteric glucose sensing. Diabetes. 2008;57(10):2603–12.
Collins SM, Surette M, Bercik P. The interplay between the intestinal microbiota and the brain. Nat Rev Microbiol. 2012;10(11):735–42.
Forsythe P, Kunze WA, Bienenstock J. On communication between gut microbes and the brain. Curr Opin Gastroenterol. 2012;28(6):557–62.
Foster JA, McVey Neufeld KA. Gut-brain axis: how the microbiome influences anxiety and depression. Trends Neurosci. 2013;36:305.
DeFronzo RA, Ferrannini E, Simonson DC. Fasting hyperglycemia in non-insulin-dependent diabetes mellitus: contributions of excessive hepatic glucose production and impaired tissue glucose uptake. Metab: Clin Exp. 1989;38(4):387–95.
Baudon MA, Ferre P, Penicaud L, Maulard P, Ktorza A, Castano L, et al. Normal insulin sensitivity during the phase of glucose intolerance but insulin resistance at the onset of diabetes in the spontaneously diabetic BB rat. Diabetologia. 1989;32(12):839–44.
Burcelin R, Eddouks M, Maury J, Kande J, Assan R, Girard J. Excessive glucose production, rather than insulin resistance, accounts for hyperglycaemia in recent-onset streptozotocin-diabetic rats. Diabetologia. 1995;38(3):283–90.
Del Prato S, Bonadonna RC, Bonora E, Gulli G, Solini A, Shank M, et al. Characterization of cellular defects of insulin action in type 2 (non-insulin-dependent) diabetes mellitus. J Clin Invest. 1993;91(2):484–94.
Golay A, Felber JP, Jequier E, DeFronzo RA, Ferrannini E. Metabolic basis of obesity and noninsulin-dependent diabetes mellitus. Diabetes Metab Rev. 1988;4(8):727–47.
Hundal RS, Krssak M, Dufour S, Laurent D, Lebon V, Chandramouli V, et al. Mechanism by which metformin reduces glucose production in type 2 diabetes. Diabetes. 2000;49(12):2063–9.
Martin BC, Warram JH, Krolewski AS, Bergman RN, Soeldner JS, Kahn CR. Role of glucose and insulin resistance in development of type 2 diabetes mellitus: results of a 25-year follow-up study [see comments]. Lancet. 1992;340(8825):925–9.
Bergman RN, Ader M. Free fatty acids and pathogenesis of type 2 diabetes mellitus. Trends Endocrinol Metab. 2000;11(9):351–6.
Pickup JC, Crook MA. Is type II diabetes mellitus a disease of the innate immune system? Diabetologia. 1998;41(10):1241–8.
Shoelson SE, Lee J, Goldfine AB. Inflammation and insulin resistance. J Clin Invest. 2006;116(7):1793–801.
Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444(7121):860–7.
Wellen KE, Hotamisligil GS. Inflammation, stress, and diabetes. J Clin Invest. 2005;115(5):1111–9.
Manco M, Fernandez-Real JM, Equitani F, Vendrell J, Valera Mora ME, Nanni G, et al. Effect of massive weight loss on inflammatory adipocytokines and the innate immune system in morbidly obese women. J Clin Endocrinol Metab. 2007;92(2):483–90.
Dandona P, Aljada A, Bandyopadhyay A. Inflammation: the link between insulin resistance, obesity and diabetes. Trends Immunol. 2004;25(1):4–7.
Nishimura S, Manabe I, Nagasaki M, Eto K, Yamashita H, Ohsugi M, et al. CD8+ effector T cells contribute to macrophage recruitment and adipose tissue inflammation in obesity. Nat Med. 2009;15(8):914–20.
Lumeng CN, Bodzin JL, Saltiel AR. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J Clin Invest. 2007;117(1):175–84.
Nguyen MT, Favelyukis S, Nguyen AK, Reichart D, Scott PA, Jenn A, et al. A subpopulation of macrophages infiltrates hypertrophic adipose tissue and is activated by free fatty acids via Toll-like receptors 2 and 4 and JNK-dependent pathways. J Biol Chem. 2007;282(48):35279–92.
Ueki K, Kondo T, Kahn CR. Suppressor of cytokine signaling 1 (SOCS-1) and SOCS-3 cause insulin resistance through inhibition of tyrosine phosphorylation of insulin receptor substrate proteins by discrete mechanisms. Mol Cell Biol. 2004;24(12):5434–46.
Plomgaard P, Bouzakri K, Krogh-Madsen R, Mittendorfer B, Zierath JR, Pedersen BK. Tumor necrosis factor-alpha induces skeletal muscle insulin resistance in healthy human subjects via inhibition of Akt substrate 160 phosphorylation. Diabetes. 2005;54(10):2939–45.
Federici M, Hribal ML, Menghini R, Kanno H, Marchetti V, Porzio O, et al. Timp3 deficiency in insulin receptor-haploinsufficient mice promotes diabetes and vascular inflammation via increased TNF-alpha. J Clin Invest. 2005;115(12):3494–505.
Tanti J, Gual P, Gremeaux T, Gonzalez T, Barres R, Le Marchand-Brustel Y. Alteration in insulin action: role of IRS-1 serine phosphorylation in the retroregulation of insulin signalling. Ann Endocrinol. 2004;65:43–8.
Bouzakri K, Roques M, Gual P, Espinosa S, Guebre-Egziabher F, Riou JP, et al. Reduced activation of phosphatidylinositol-3 kinase and increased serine 636 phosphorylation of insulin receptor substrate-1 in primary culture of skeletal muscle cells from patients with type 2 diabetes. Diabetes. 2003;52(6):1319–25.
Bouloumie A, Curat CA, Sengenes C, Lolmede K, Miranville A, Busse R. Role of macrophage tissue infiltration in metabolic diseases. Curr Opin Clin Nutr Metab Care. 2005;8(4):347–54.
Kanda H, Tateya S, Tamori Y, Kotani K, Hiasa K, Kitazawa R, et al. MCP-1 contributes to macrophage infiltration into adipose tissue, insulin resistance, and hepatic steatosis in obesity. J Clin Invest. 2006;116(6):1494–505.
Suganami T, Tanimoto-Koyama K, Nishida J, Itoh M, Yuan X, Mizuarai S, et al. Role of the Toll-like receptor 4/NF-kappaB pathway in saturated fatty acid-induced inflammatory changes in the interaction between adipocytes and macrophages. Arterioscler Thromb Vasc Biol. 2007;27(1):84–91.
Weisberg SP, Hunter D, Huber R, Lemieux J, Slaymaker S, Vaddi K, et al. CCR2 modulates inflammatory and metabolic effects of high-fat feeding. J Clin Invest. 2006;116(1):115–24.
Lumeng CN, Deyoung SM, Bodzin JL, Saltiel AR. Increased inflammatory properties of adipose tissue macrophages recruited during diet-induced obesity. Diabetes. 2007;56(1):16–23.
Charriere G, Cousin B, Arnaud E, Andre M, Bacou F, Penicaud L, et al. Preadipocyte conversion to macrophage. Evidence of plasticity. J Biol Chem. 2003;278(11):9850–5.
Charriere GM, Cousin B, Arnaud E, Saillan-Barreau C, Andre M, Massoudi A, et al. Macrophage characteristics of stem cells revealed by transcriptome profiling. Exp Cell Res. 2006;312(17):3205–14.
Winer S, Chan Y, Paltser G, Truong D, Tsui H, Bahrami J, et al. Normalization of obesity-associated insulin resistance through immunotherapy. Nat Med. 2009;15(8):921–9.
Feuerer M, Herrero L, Cipolletta D, Naaz A, Wong J, Nayer A, et al. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat Med. 2009;15(8):930–9.
Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, Delzenne NM, et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57(6):1470–81.
Drong AW, Lindgren CM, McCarthy MI. The genetic and epigenetic basis of type 2 diabetes and obesity. Clin Pharmacol Ther. 2012;92(6):707–15.
Medici F, Hawa M, Ianari A, Pyke DA, Leslie RD. Concordance rate for type II diabetes mellitus in monozygotic twins: actuarial analysis. Diabetologia. 1999;42(2):146–50.
Poulsen P, Grunnet LG, Pilgaard K, Storgaard H, Alibegovic A, Sonne MP, et al. Increased risk of type 2 diabetes in elderly twins. Diabetes. 2009;58(6):1350–5.
Vaarala O, Atkinson MA, Neu J. The “perfect storm” for type 1 diabetes: the complex interplay between intestinal microbiota, gut permeability, and mucosal immunity. Diabetes. 2008;57(10):2555–62.
Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, et al. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature. 2008;455(7216):1109–13.
Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339:1084.
Nicholson JK, Holmes E, Wilson ID. Gut microorganisms, mammalian metabolism and personalized health care. Nat Rev Microbiol. 2005;3(5):431–8.
Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291(5505):881–4.
Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004;101(44):15718–23.
Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292(5519):1115–8.
Serino M, Chabo C, Burcelin R. Intestinal MicrobiOMICS to define health and disease in human and mice. Curr Pharm Biotechnol. 2012;13(5):746–58.
Backhed F. Programming of host metabolism by the gut microbiota. Ann Nutr Metab. 2011;58 Suppl 2:44–52.
Turnbaugh PJ, Gordon JI. The core gut microbiome, energy balance and obesity. J Physiol. 2009;587(Pt 17):4153–8.
Karlsson FH, Fak F, Nookaew I, Tremaroli V, Fagerberg B, Petranovic D, et al. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat Commun. 2012;3:1245.
Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444(7122):1022–3.
Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Human gut microbes associated with obesity. Nature. 2006;444:1022–3.
Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A. 2005;102(31):11070–5.
Karlsson FH, Tremaroli V, Nookaew I, Bergstrom G, Behre CJ, Fagerberg B, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99.
Noble D, Mathur R, Dent T, Meads C, Greenhalgh T. Risk models and scores for type 2 diabetes: systematic review. BMJ. 2011;343:d7163.
van Dieren S, Beulens JW, Kengne AP, Peelen LM, Rutten GE, Woodward M, et al. Prediction models for the risk of cardiovascular disease in patients with type 2 diabetes: a systematic review. Heart. 2012;98(5):360–9.
Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. 2011;9(4):279–90.
Serino M, Luche E, Gres S, Baylac A, Berge M, Cenac C, et al. Metabolic adaptation to a high-fat diet is associated with a change in the gut microbiota. Gut. 2012;61(4):543–53.
Turnbaugh P, Ridaura V, Faith J, Rey F, Knight R, Gordon J. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6–14.
Murphy EF, Cotter PD, Healy S, Marques TM, O’Sullivan O, Fouhy F, et al. Composition and energy harvesting capacity of the gut microbiota: relationship to diet, obesity and time in mouse models. Gut. 2010;59(12):1635–42.
Moreira AP, Texeira TF, Ferreira AB, Peluzio Mdo C, Alfenas RC. Influence of a high-fat diet on gut microbiota, intestinal permeability and metabolic endotoxaemia. Br J Nutr. 2012;108(5):801–9.
Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A. 2013;498:99.
Pedersen R, Andersen AD, Molbak L, Stagsted J, Boye M. Changes in the gut microbiota of cloned and non-cloned control pigs during development of obesity: gut microbiota during development of obesity in cloned pigs. BMC Microbiol. 2013;13:30.
Yan H, Potu R, Lu H, Vezzoni de Almeida V, Stewart T, Ragland D, et al. Dietary fat content and fiber type modulate hind gut microbial community and metabolic markers in the pig. PLoS One. 2013;8(4):e59581.
Park DY, Ahn YT, Park SH, Huh CS, Yoo SR, Yu R, et al. Supplementation of lactobacillus curvatus HY7601 and lactobacillus plantarum KY1032 in diet-induced obese mice is associated with gut microbial changes and reduction in obesity. PLoS One. 2013;8(3):e59470.
Parks BW, Nam E, Org E, Kostem E, Norheim F, Hui ST, et al. Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metab. 2013;17(1):141–52.
Mujico JR, Baccan GC, Gheorghe A, Diaz LE, Marcos A. Changes in gut microbiota due to supplemented fatty acids in diet-induced obese mice. Br J Nutr. 2013;110:1–10.
de Wit N, Derrien M, Bosch-Vermeulen H, Oosterink E, Keshtkar S, Duval C, et al. Saturated fat stimulates obesity and hepatic steatosis and affects gut microbiota composition by an enhanced overflow of dietary fat to the distal intestine. Am J Physiol Gastrointest Liver Physiol. 2012;303(5):G589–99.
Liu T, Hougen H, Vollmer AC, Hiebert SM. Gut bacteria profiles of Mus musculus at the phylum and family levels are influenced by saturation of dietary fatty acids. Anaerobe. 2012;18(3):331–7.
Dao TM, Waget A, Klopp P, Serino M, Vachoux C, Pechere L, et al. Resveratrol increases glucose induced GLP-1 secretion in mice: a mechanism which contributes to the glycemic control. PLoS One. 2011;6(6):e20700.
Neyrinck AM, Van Hee VF, Bindels LB, De Backer F, Cani PD, Delzenne NM. Polyphenol-rich extract of pomegranate peel alleviates tissue inflammation and hypercholesterolaemia in high-fat diet-induced obese mice: potential implication of the gut microbiota. Br J Nutr. 2013;109(5):802–9.
Tachon S, Zhou J, Keenan M, Martin R, Marco ML. The intestinal microbiota in aged mice is modulated by dietary resistant starch and correlated with improvements in host responses. FEMS Microbiol Ecol. 2013;83(2):299–309.
Cani PD, Knauf C, Iglesias MA, Drucker DJ, Delzenne NM, Burcelin R. Improvement of glucose tolerance and hepatic insulin sensitivity by oligofructose requires a functional glucagon-like peptide 1 receptor. Diabetes. 2006;55(5):1484–90.
Serino M, Fernandez-Real JM, Fuentes EG, Queipo-Ortuno M, Moreno-Navarrete JM, Sanchez A, et al. The gut microbiota profile is associated with insulin action in humans. Acta Diabetol. 2012.
Vernon G, Baranova A, Younossi ZM. Systematic review: the epidemiology and natural history of non-alcoholic fatty liver disease and non-alcoholic steatohepatitis in adults. Aliment Pharmacol Ther. 2011;34(3):274–85.
Wong VW, Tse CH, Lam TT, Wong GL, Chim AM, Chu WC, et al. Molecular characterization of the fecal microbiota in patients with nonalcoholic steatohepatitis - a longitudinal study. PLoS One. 2013;8(4):e62885.
Raman M, Ahmed I, Gillevet PM, Probert CS, Ratcliffe NM, Smith S, et al. Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol: Off Clin Pract J Am Gastroenterol Assoc. 2013;11:868.
Le Roy T, Llopis M, Lepage P, Bruneau A, Rabot S, Bevilacqua C, et al. Intestinal microbiota determines development of non-alcoholic fatty liver disease in mice. Gut. 2012.
Henao-Mejia J, Elinav E, Jin C, Hao L, Mehal WZ, Strowig T, et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482(7384):179–85.
Aron-Wisnewsky J, Gaborit B, Dutour A, Clement K. Gut microbiota and non-alcoholic fatty liver disease: new insights. Clin Microbiol Infect. 2013;19(4):338–48.
Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–4.
Newman B, Selby J, King M, Slemenda C, Fabsitz R, Friedman G. Concordance for type 2 (non-insulin-dependent) diabetes mellitus in male twins. Diabetologia. 1987;30:763–8.
Bouchard C, Tremblay A, Despres J, Nadeau A, Lupien P, Theriault G, et al. The response to long-term overfeeding in identical twins. N Engl J Med. 1990;322(21):1477–82. 1477-82.
Bouchard C, Tremblay A, Despres J, Nadeau A, Lupien P, Moorjani S, et al. Overfeeding in identical twins: 5-year postoverfeeding results. Metab: Clin Exp. 1996;45:1042–50.
Turnbaugh PJ, Quince C, Faith JJ, McHardy AC, Yatsunenko T, Niazi F, et al. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc Natl Acad Sci U S A. 2010;107(16):7503–8.
Stahringer SS, Clemente JC, Corley RP, Hewitt J, Knights D, Walters WA, et al. Nurture trumps nature in a longitudinal survey of salivary bacterial communities in twins from early adolescence to early adulthood. Genome Res. 2012;22(11):2146–52.
Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328(5975):228–31.
Round JL, O’Connell RM, Mazmanian SK. Coordination of tolerogenic immune responses by the commensal microbiota. J Autoimmun. 2010;34(3):J220–5.
Burcelin R, Garidou L, Pomie C. Immuno-microbiota cross and talk: the new paradigm of metabolic diseases. Semin Immunol. 2012;24(1):67–74.
Natividad JM, Verdu EF. Modulation of intestinal barrier by intestinal microbiota: pathological and therapeutic implications. Pharmacol Res: Off J Ital Pharmacol Soc. 2013;69(1):42–51.
Caricilli AM, Picardi PK, de Abreu LL, Ueno M, Prada PO, Ropelle ER, et al. Gut microbiota is a key modulator of insulin resistance in TLR 2 knockout mice. PLoS Biol. 2011;9(12):e1001212.
Kim KA, Gu W, Lee IA, Joh EH, Kim DH. High fat diet-induced gut microbiota exacerbates inflammation and obesity in mice via the TLR4 signaling pathway. PLoS One. 2012;7(10):e47713.
Rehman A, Sina C, Gavrilova O, Hasler R, Ott S, Baines JF, et al. Nod2 is essential for temporal development of intestinal microbial communities. Gut. 2011;60(10):1354–62.
Mondot S, Barreau F, Al Nabhani Z, Dussaillant M, Le Roux K, Dore J, et al. Altered gut microbiota composition in immune-impaired Nod2(-/-) mice. Gut. 2012;61(4):634–5.
Rogler G. The importance of gut microbiota in mediating the effect of NOD2 defects in inflammatory bowel disease. Gut. 2010;59(2):153–4.
Robertson SJ, Zhou JY, Geddes K, Rubino SJ, Cho JH, Girardin SE, et al. Nod1 and Nod2 signaling does not alter the composition of intestinal bacterial communities at homeostasis. Gut Microbes. 2013;4(3):222–31.
Palmer C, Bik EM, Digiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5(7):e177.
Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107(26):11971–5.
Ajslev TA, Andersen CS, Gamborg M, Sorensen TI, Jess T. Childhood overweight after establishment of the gut microbiota: the role of delivery mode, pre-pregnancy weight and early administration of antibiotics. Int J Obes (Lond). 2011;35(4):522–9.
Koren O, Goodrich JK, Cullender TC, Spor A, Laitinen K, Backhed HK, et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell. 2012;150(3):470–80.
Collado MC, Isolauri E, Laitinen K, Salminen S. Effect of mother’s weight on infant’s microbiota acquisition, composition, and activity during early infancy: a prospective follow-up study initiated in early pregnancy. Am J Clin Nutr. 2010;92(5):1023–30.
Karlsson CL, Onnerfalt J, Xu J, Molin G, Ahrne S, Thorngren-Jerneck K. The microbiota of the gut in preschool children with normal and excessive body weight. Obesity (Silver Spring). 2012;20:2257.
Song MJ, Kim KH, Yoon JM, Kim JB. Activation of Toll-like receptor 4 is associated with insulin resistance in adipocytes. Biochem Biophys Res Commun. 2006;346(3):739–45.
Zu L, He J, Jiang H, Xu C, Pu S, Xu G. Bacterial endotoxin stimulates adipose lipolysis via toll-like receptor 4 and extracellular signal-regulated kinase pathway. J Biol Chem. 2009;284(9):5915–26.
Pendyala S, Walker J, Holt PR. A high-fat diet is associated with endotoxemia that originates from the gut. Gastroenterology. 2012;142:1100.
Lassenius MI, Pietilainen KH, Kaartinen K, Pussinen PJ, Syrjanen J, Forsblom C, et al. Bacterial endotoxin activity in human serum is associated with dyslipidemia, insulin resistance, obesity, and chronic inflammation. Diabetes Care. 2011;34(8):1809–15.
Ghoshal S, Witta J, Zhong J, de Villiers W, Eckhardt E. Chylomicrons promote intestinal absorption of lipopolysaccharides. J Lipid Res. 2009;50(1):90–7.
Laugerette F, Vors C, Geloen A, Chauvin MA, Soulage C, Lambert-Porcheron S, et al. Emulsified lipids increase endotoxemia: possible role in early postprandial low-grade inflammation. J Nutr Biochem. 2010;22:53.
Geurts L, Lazarevic V, Derrien M, Everard A, Van Roye M, Knauf C, et al. Altered gut microbiota and endocannabinoid system tone in obese and diabetic leptin-resistant mice: impact on apelin regulation in adipose tissue. Front Microbiol. 2011;2:149.
Cani PD, Possemiers S, Van de Wiele T, Guiot Y, Everard A, Rottier O, et al. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut. 2009;58(8):1091–103.
Russo F, Linsalata M, Clemente C, Chiloiro M, Orlando A, Marconi E, et al. Inulin-enriched pasta improves intestinal permeability and modifies the circulating levels of zonulin and glucagon-like peptide 2 in healthy young volunteers. Nutr Res. 2012;32(12):940–6.
Stoidis CN, Misiakos EP, Patapis P, Fotiadis CI, Spyropoulos BG. Potential benefits of pro- and prebiotics on intestinal mucosal immunity and intestinal barrier in short bowel syndrome. Nutr Res Rev. 2010:1–9.
Kitchens RL, Thompson PA, Viriyakosol S, O’Keefe GE, Munford RS. Plasma CD14 decreases monocyte responses to LPS by transferring cell-bound LPS to plasma lipoproteins. J Clin Invest. 2001;108(3):485–93.
Vreugdenhil AC, Rousseau CH, Hartung T, Greve JW, van ’t Veer C, Buurman WA. Lipopolysaccharide (LPS)-binding protein mediates LPS detoxification by chylomicrons. J Immunol. 2003;170(3):1399–405.
Amar J, Burcelin R, Ruidavets JB, Cani PD, Fauvel J, Alessi MC, et al. Energy intake is associated with endotoxemia in apparently healthy men. Am J Clin Nutr. 2008;87(5):1219–23.
Moreno-Navarrete JM, Manco M, Ibanez J, Garcia-Fuentes E, Ortega F, Gorostiaga E, et al. Metabolic endotoxemia and saturated fat contribute to circulating NGAL concentrations in subjects with insulin resistance. Int J Obes (Lond). 2010;34(2):240–9.
Shi H, Kokoeva MV, Inouye K, Tzameli I, Yin H, Flier JS. TLR4 links innate immunity and fatty acid-induced insulin resistance. J Clin Invest. 2006;116(11):3015–25.
Vandanmagsar B, Youm YH, Ravussin A, Galgani JE, Stadler K, Mynatt RL, et al. The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nat Med. 2011;17(2):179–88.
Poggi M, Bastelica D, Gual P, Iglesias MA, Gremeaux T, Knauf C, et al. C3H/HeJ mice carrying a toll-like receptor 4 mutation are protected against the development of insulin resistance in white adipose tissue in response to a high-fat diet. Diabetologia. 2007;50(6):1267–76.
Liang H, Hussey SE, Sanchez-Avila A, Tantiwong P, Musi N. Effect of lipopolysaccharide on inflammation and insulin action in human muscle. PLoS One. 2013;8(5):e63983.
Suganami T, Mieda T, Itoh M, Shimoda Y, Kamei Y, Ogawa Y. Attenuation of obesity-induced adipose tissue inflammation in C3H/HeJ mice carrying a Toll-like receptor 4 mutation. Biochem Biophys Res Commun. 2007;354(1):45–9.
Xu H, Barnes GT, Yang Q, Tan G, Yang D, Chou CJ, et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest. 2003;112(12):1821–30.
Gerhardt CC, Romero IA, Cancello R, Camoin L, Strosberg AD. Chemokines control fat accumulation and leptin secretion by cultured human adipocytes. Mol Cell Endocrinol. 2001;175(1–2):81–92.
Hotamisligil GS, Peraldi P, Budavari A, Ellis R, White MF, Spiegelman BM. IRS-1-mediated inhibition of insulin receptor tyrosine kinase activity in TNF-alpha- and obesity-induced insulin resistance. Science. 1996;271(5249):665–8.
Wang L, Li L, Ran X, Long M, Zhang M, Tao Y, et al. Lipopolysaccharides reduce adipogenesis in 3T3-L1 adipocytes through activation of NF-kappaB pathway and downregulation of AMPK expression. Cardiovasc Toxicol. 2013;13:338.
Gabele E, Dostert K, Hofmann C, Wiest R, Scholmerich J, Hellerbrand C, et al. DSS induced colitis increases portal LPS levels and enhances hepatic inflammation and fibrogenesis in experimental NASH. J Hepatol. 2011;55(6):1391–9.
Tomofuji T, Ekuni D, Yamanaka R, Kusano H, Azuma T, Sanbe T, et al. Chronic administration of lipopolysaccharide and proteases induces periodontal inflammation and hepatic steatosis in rats. J Periodontol. 2007;78(10):1999–2006.
Endo M, Masaki T, Seike M, Yoshimatsu H. TNF-alpha induces hepatic steatosis in mice by enhancing gene expression of sterol regulatory element binding protein-1c (SREBP-1c). Exp Biol Med (Maywood). 2007;232(5):614–21.
Sansonetti P, Di Santo J. Debugging how bacteria manipulate the immune response. Immunity. 2007;26:149–61.
Kufer TA, Sansonetti PJ. Sensing of bacteria: NOD a lonely job. Curr Opin Microbiol. 2007;10(1):62–9.
Sansonetti P. Host-pathogen interactions: the seduction of molecular cross talk. Gut. 2002;50 Suppl 3:III2–8.
Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci U S A. 2002;99(24):15451–5.
Carneiro LA, Magalhaes JG, Tattoli I, Philpott DJ, Travassos LH. Nod-like proteins in inflammation and disease. J Pathol. 2008;214(2):136–48.
Schertzer JD, Tamrakar AK, Magalhaes JG, Pereira S, Bilan PJ, Fullerton MD, et al. NOD1 activators link innate immunity to insulin resistance. Diabetes. 2011;60(9):2206–15.
Caesar R, Reigstad CS, Backhed HK, Reinhardt C, Ketonen M, Ostergren Lunden G, et al. Gut-derived lipopolysaccharide augments adipose macrophage accumulation but is not essential for impaired glucose or insulin tolerance in mice. Gut. 2012;61:1701.
Tamrakar AK, Schertzer JD, Chiu TT, Foley KP, Bilan PJ, Philpott DJ, et al. NOD2 activation induces muscle cell-autonomous innate immune responses and insulin resistance. Endocrinology. 2010;151(12):5624–37.
Ogura Y, Lala S, Xin W, Smith E, Dowds TA, Chen FF, et al. Expression of NOD2 in Paneth cells: a possible link to Crohn’s ileitis. Gut. 2003;52(11):1591–7.
Ogura Y, Saab L, Chen FF, Benito A, Inohara N, Nunez G. Genetic variation and activity of mouse Nod2, a susceptibility gene for Crohn’s disease. Genomics. 2003;81(4):369–77.
Inohara N, Ogura Y, Nunez G. Nods: a family of cytosolic proteins that regulate the host response to pathogens. Curr Opin Microbiol. 2002;5(1):76–80.
Miele L, Valenza V, La Torre G, Montalto M, Cammarota G, Ricci R, et al. Increased intestinal permeability and tight junction alterations in nonalcoholic fatty liver disease. Hepatology. 2009;49(6):1877–87.
Lichtman SN, Keku J, Schwab JH, Sartor RB. Hepatic injury associated with small bowel bacterial overgrowth in rats is prevented by metronidazole and tetracycline. Gastroenterology. 1991;100(2):513–9.
Vance JE, Vance DE. The role of phosphatidylcholine biosynthesis in the secretion of lipoproteins from hepatocytes. Can J Biochem Cell Biol. 1985;63(8):870–81.
Ortu G, Rivabene R, Cantafora A, Bravo E. Age-related variations in hepatic biosynthesis of phosphatidylcholine: a study of choline metabolism with perfused rat liver. Proc Soc Exp Biol Med. 1997;216(1):44–51.
Vance DE. Role of phosphatidylcholine biosynthesis in the regulation of lipoprotein homeostasis. Curr Opin Lipidol. 2008;19(3):229–34.
Dumas ME, Barton RH, Toye A, Cloarec O, Blancher C, Rothwell A, et al. Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc Natl Acad Sci U S A. 2006;103(33):12511–6.
Claus SP, Tsang TM, Wang Y, Cloarec O, Skordi E, Martin FP, et al. Systemic multicompartmental effects of the gut microbiome on mouse metabolic phenotypes. Mol Syst Biol. 2008;4:219.
Martin FP, Wang Y, Sprenger N, Yap IK, Lundstedt T, Lek P, et al. Probiotic modulation of symbiotic gut microbial-host metabolic interactions in a humanized microbiome mouse model. Mol Syst Biol. 2008;4:157.
Carvalho BM, Guadagnini D, Tsukumo DM, Schenka AA, Latuf-Filho P, Vassallo J, et al. Modulation of gut microbiota by antibiotics improves insulin signalling in high-fat fed mice. Diabetologia. 2012;55(10):2823–34.
Lin HV, Frassetto A, Kowalik Jr EJ, Nawrocki AR, Lu MM, Kosinski JR, et al. Butyrate and propionate protect against diet-induced obesity and regulate gut hormones via free fatty acid receptor 3-independent mechanisms. PLoS One. 2012;7(4):e35240.
Al-Lahham SH, Peppelenbosch MP, Roelofsen H, Vonk RJ, Venema K. Biological effects of propionic acid in humans; metabolism, potential applications and underlying mechanisms. Biochim Biophys Acta. 2010;1801(11):1175–83.
Gao Z, Yin J, Zhang J, Ward RE, Martin RJ, Lefevre M, et al. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes. 2009;58(7):1509–17.
Payne AN, Chassard C, Zimmermann M, Muller P, Stinca S, Lacroix C. The metabolic activity of gut microbiota in obese children is increased compared with normal-weight children and exhibits more exhaustive substrate utilization. Nutr Diabetes. 2011;1:e12.
Kimura I, Ozawa K, Inoue D, Imamura T, Kimura K, Maeda T, et al. The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nat Commun. 2013;4:1829.
Backhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci U S A. 2007;104(3):979–84.
Hardie DG, Carling D, Carlson M. The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu Rev Biochem. 1998;67:821–55.
Bergeron R, Russell 3rd RR, Young LH, Ren JM, Marcucci M, Lee A, et al. Effect of AMPK activation on muscle glucose metabolism in conscious rats. Am J Physiol. 1999;276(5 Pt 1):E938–44.
Aron-Wisnewsky J, Dore J, Clement K. The importance of the gut microbiota after bariatric surgery. Nat Rev Gastroenterol Hepatol. 2012;9(10):590–8.
Sweeney TE, Morton JM. The human gut microbiome: a review of the effect of obesity and surgically induced weight loss. JAMA Surg. 2013;148:1–7.
Zhang H, DiBaise JK, Zuccolo A, Kudrna D, Braidotti M, Yu Y, et al. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci U S A. 2009;106(7):2365–70.
Liou AP, Paziuk M, Luevano Jr JM, Machineni S, Turnbaugh PJ, Kaplan LM. Conserved shifts in the gut microbiota due to gastric bypass reduce host weight and adiposity. Sci Transl Med. 2013;5(178):178ra41.
Kong LC, Tap J, Aron-Wisnewsky J, Pelloux V, Basdevant A, Bouillot JL, et al. Gut microbiota after gastric bypass in human obesity: increased richness and associations of bacterial genera with adipose tissue genes. Am J Clin Nutr. 2013.
Furet JP, Kong LC, Tap J, Poitou C, Basdevant A, Bouillot JL, et al. Differential adaptation of human gut microbiota to bariatric surgery-induced weight loss: links with metabolic and low-grade inflammation markers. Diabetes. 2010;59(12):3049–57.
Cummings JH, Macfarlane GT. Role of intestinal bacteria in nutrient metabolism. JPEN J Parenter Enter Nutr. 1997;21(6):357–65.
Seyer P, Vallois D, Poitry-Yamate C, Schutz F, Metref S, Tarussio D, et al. Hepatic glucose sensing is required to preserve beta cell glucose competence. J Clin Invest. 2013;123(4):1662–76.
Zarrinpar A, Loomba R. Review article: the emerging interplay among the gastrointestinal tract, bile acids and incretins in the pathogenesis of diabetes and non-alcoholic fatty liver disease. Aliment Pharmacol Ther. 2012;36(10):909–21.
Watanabe M, Morimoto K, Houten SM, Kaneko-Iwasaki N, Sugizaki T, Horai Y, et al. Bile acid binding resin improves metabolic control through the induction of energy expenditure. PLoS One. 2012;7(8):e38286.
Meissner M, Wolters H, de Boer RA, Havinga R, Boverhof R, Bloks VW, et al. Bile acid sequestration normalizes plasma cholesterol and reduces atherosclerosis in hypercholesterolemic mice. No additional effect of physical activity. Atherosclerosis. 2013;228(1):117–23.
Yokota A, Fukiya S, Islam KB, Ooka T, Ogura Y, Hayashi T, et al. Is bile acid a determinant of the gut microbiota on a high-fat diet? Gut Microbes. 2012;3(5):455–9.
Pournaras DJ, Glicksman C, Vincent RP, Kuganolipava S, Alaghband-Zadeh J, Mahon D, et al. The role of bile after Roux-en-Y gastric bypass in promoting weight loss and improving glycaemic control. Endocrinology. 2012;153(8):3613–9.
Keitel V, Haussinger D. Perspective: TGR5 (Gpbar-1) in liver physiology and disease. Clin Res Hepatol Gastroenterol. 2012;36(5):412–9.
Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature. 2012;487(7405):104–8.
Zhao Y, Wu J, Li JV, Zhou NY, Tang H, Wang Y. Gut microbiota composition modifies fecal metabolic profiles in mice. J Proteome Res. 2013.
Sayin SI, Wahlstrom A, Felin J, Jantti S, Marschall HU, Bamberg K, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17(2):225–35.
Hersoug LG, Linneberg A. The link between the epidemics of obesity and allergic diseases: does obesity induce decreased immune tolerance? Allergy. 2007;62(10):1205–13.
Lolmede K, Duffaut C, Zakaroff-Girard A, Bouloumie A. Immune cells in adipose tissue: key players in metabolic disorders. Diabetes Metab. 2011;37(4):283–90.
Decaunes P, Esteve D, Zakaroff-Girard A, Sengenes C, Galitzky J, Bouloumie A. Adipose-derived stromal cells: cytokine expression and immune cell contaminants. Methods Mol Biol. 2011;702:151–61.
Macpherson AJ, Geuking MB, McCoy KD. Immune responses that adapt the intestinal mucosa to commensal intestinal bacteria. Immunology. 2005;115(2):153–62.
Owen RL, Jones AL. Epithelial cell specialization within human Peyer’s patches: an ultrastructural study of intestinal lymphoid follicles. Gastroenterology. 1974;66(2):189–203.
Berg RD. Bacterial translocation from the gastrointestinal tract. Trends Microbiol. 1995;3(4):149–54.
Berg RD, Wommack E, Deitch EA. Immunosuppression and intestinal bacterial overgrowth synergistically promote bacterial translocation. Arch Surg. 1988;123(11):1359–64.
Ouellette AJ, Bevins CL. Paneth cell defensins and innate immunity of the small bowel. Inflamm Bowel Dis. 2001;7(1):43–50.
Schenk M, Mueller C. The mucosal immune system at the gastrointestinal barrier. Best Pract Res Clin Gastroenterol. 2008;22(3):391–409.
Steed H, Macfarlane GT, Blackett KL, Macfarlane S, Miller MH, Bahrami B, et al. Bacterial translocation in cirrhosis is not caused by an abnormal small bowel gut microbiota. FEMS Immunol Med Microbiol. 2011;63(3):346–54.
Koh IH, Liberatore AM, Menchaca-Diaz JL, Ruiz-Silva M, Vilela-Oliveira L, Watanabe AY, et al. Bacterial translocation, microcirculation injury and sepsis. Endocr Metab Immune Disord Drug Targets. 2006;6(2):143–50.
Klatt NR, Funderburg NT, Brenchley JM. Microbial translocation, immune activation, and HIV disease. Trends Microbiol. 2013;21(1):6–13.
Jevtovic DJ, Dragovic G, Salemovic D, Ranin J, Djurkovic-Djakovic O. The metabolic syndrome, an epidemic among HIV-infected patients on HAART. BioMed Pharmacother. 2009;63(5):337–42.
Potthoff A, Brockmeyer NH, Gelbrich G, Neuhaus K, Esser S, Reinsch N, et al. Lipodystrophy – a sign for metabolic syndrome in patients of the HIV-HEART study. J Dtsch Dermatol Ges. 2010;8(2):92–8.
Dandekar S, George MD, Baumler AJ. Th17 cells, HIV and the gut mucosal barrier. Curr Opin HIV AIDS. 2010;5(2):173–8.
Amar J, Serino M, Lange C, Chabo C, Iacovoni J, Mondot S, et al. Involvement of tissue bacteria in the onset of diabetes in humans: evidence for a concept. Diabetologia. 2011;54(12):3055–61.
Amar J, Lange C, Payros G, Garret C, Chabo C, Lantieri O, et al. Blood microbiota dysbiosis is associated with the onset of cardiovascular events in a large general population: the D.E.S.I.R. study. PLoS One. 2013;8(1):e54461.
Hosoi T, Yokoyama S, Matsuo S, Akira S, Ozawa K. Myeloid differentiation factor 88 (MyD88)-deficiency increases risk of diabetes in mice. PLoS One. 2010;5:9.
Riant E, Waget A, Cogo H, Arnal JF, Burcelin R, Gourdy P. Estrogens protect against high-fat diet-induced insulin resistance and glucose intolerance in mice. Endocrinology. 2009;150(5):2109–17.
Otero M, Lago R, Gomez R, Lago F, Gomez-Reino JJ, Gualillo O. Leptin: a metabolic hormone that functions like a proinflammatory adipokine. Drug News Perspect. 2006;19(1):21–6.
Caesar R, Fak F, Backhed F. Effects of gut microbiota on obesity and atherosclerosis via modulation of inflammation and lipid metabolism. J Intern Med. 2010;268(4):320–8.
Koren O, Spor A, Felin J, Fak F, Stombaugh J, Tremaroli V, et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc Natl Acad Sci U S A. 2011;108 Suppl 1:4592–8.
Griffin JL, Scott J, Nicholson JK. The influence of pharmacogenetics on fatty liver disease in the wistar and kyoto rats: a combined transcriptomic and metabonomic study. J Proteome Res. 2007;6(1):54–61.
Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472(7341):57–63.
Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. 2013;368(17):1575–84.
Blasco-Baque V, Serino M, Vergnes JN, Riant E, Loubieres P, Arnal JF, et al. High-fat diet induces periodontitis in mice through lipopolysaccharides (LPS) receptor signaling: protective action of estrogens. PLoS One. 2012;7(11):e48220.
Hashimoto T, Perlot T, Rehman A, Trichereau J, Ishiguro H, Paolino M, et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature. 2012;487(7408):477–81.
Kaddurah-Daouk R, Baillie RA, Zhu H, Zeng ZB, Wiest MM, Nguyen UT, et al. Enteric microbiome metabolites correlate with response to simvastatin treatment. PLoS One. 2011;6(10):e25482.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer-Verlag London
About this chapter
Cite this chapter
Burcelin, R., Courtney, M., Amar, J. (2015). Gut Microbiota and Metabolic Diseases: From Pathogenesis to Therapeutic Perspective. In: Kochhar, S., Martin, FP. (eds) Metabonomics and Gut Microbiota in Nutrition and Disease. Molecular and Integrative Toxicology. Springer, London. https://doi.org/10.1007/978-1-4471-6539-2_11
Download citation
DOI: https://doi.org/10.1007/978-1-4471-6539-2_11
Published:
Publisher Name: Springer, London
Print ISBN: 978-1-4471-6538-5
Online ISBN: 978-1-4471-6539-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)