Abstract
Systems biology has introduced new paradigms in science by switching from a reductionist point of view to a more integrative approach toward the study of systems. As researchers over the past years have produced an extraordinary wealth of knowledge on human physiology, we now aim at integrating this knowledge to decipher the intimate relationships between the different components and scales that form the delicate balance in physiological systems. Our aim is to study the heart.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
12.1 Introduction
Systems biology has introduced new paradigms in science by switching from a reductionist point of view to a more integrative approach toward the study of systems. As researchers over the past years have produced an extraordinary wealth of knowledge on human physiology, we now aim at integrating this knowledge to decipher the intimate relationships between the different components and scales that form the delicate balance in physiological systems. Our aim is to study the heart.
The heart has always produced great interest to scientists since ancient times. Aristotle was one of the first philosophers to acknowledge that “the heart is the beginning and origin of life, and without it, no part can live” [29]. Ever since, anatomists, physiologist, mathematicians, physicists, biologists, biochemists, and geneticists have studied the cardiovascular system, reducing it to its smallest components. Integrative physiology aims to put all these parts together, and the tool employed is computational modeling. The aim of Cardiac Computational Modeling is to create a multi-scale framework to understand the heart physiology, from genes to the whole cardiovascular function. The task is not an easy one. The heart physiology is widely complex, and different tools and algorithms have to be created to intertwine the different systems acting at different levels.
Mathematics is the quantitative tool to represent reality and analyze the physical and biological world around us. One of the best examples of a mathematical model is the one created by Hodgkin and Huxley, which earned them the Nobel Prize in Medicine in 1963, in which they describe the action potential generation quantitatively using voltage–current–capacitance relationships and voltage-dependent conductances of distinct ions. This pivotal work that implements mathematical models to describe ionic flow across excitable membranes is still integrated conceptually in various cellular models of electrophysiology (Chap. 3). By the estimation of the conductance and the capacitance, this model is able to capture the kinetics of sodium and potassium ion channels of neurons [10]. Mathematical descriptions of the myocytes have followed so that generic models of propagation have been modified and used to simulate the electrical propagation in the heart [26]. Highly detailed models have now been used successfully, like the one published by Greenstein [13] and modified by Flaim [11] to include myocyte heterogeneities. High-performance computing is a major determinant for the implementation of computational costly models, Graphic Processing Units (GPUs) can be used to accelerate processing times of parallel problems [17], while existing software and technologies can serve as plug-in applications to a flexible architecture that can integrate these tools to solve multi-scale problems.
Data richness drives biomedicine. Data sharing and collaborations are of utmost importance in our globalized world. Improvements in technology on biology, engineering, physics, biochemistry, mathematics, and computing impact the production and accessibility of data. Particularly, developments in imaging and their extended use in the clinical setting provide large amounts of data at fine resolution. Computed Tomography (CT) and Magnetic Resonance Imaging (MRI) are now common tools used to improve the diagnoses in patients; however, their full potential has not yet been exploited.
We have approached a new era in which medicine can be personalized, and the existent knowledge on mathematical models of physiology can be put into use for the benefit of patients using flexible, easy-to-use, fast, modular computational tools for bedside predictions in the cardiology units.
12.2 Multi-Scale Framework of Cardiac Modeling
Various tools for authoring computational models have been created; namely, Continuity (http://www.continuity.ucsd.edu), OpenCell (http://www.opencellproject.org), Open CMISS and CMGUI (http://www.cmiss.org), GIMIAS (http://www.gimias.org/), OpenSIM (http://www.simtk.org/home/opensim), JSim (http://www.nsr.bioeng.washington.edu/jsim), SOFA (http://www.sofa-framework.org/home), SBML (http://www.sbml.org/Main_Page), etc. These tools use different structures, languages, and architectures to create multi-scale models. We will base herein on the general experience gathered by developing the tool named Continuity. The basic structure for a software program to create patient-specific multi-scale models requires ease of use; it needs to be computationally efficient, reliable, and mathematically correct, so that modelers and nonmodelers can use it to its full extent in the clinical setting. Environments for problem-solving multi-scale physics need to be modular, user-friendly and with an architecture that enables data and model sharing, so that databases are publicly accessible and models are reused and collaboration for further scientific advancement is ensured. Accessibility, compatibility to various operating systems (OSs), and ease of model generation are a must. Various of the multi-scale modeling tools use Extensible Markup Language (XML) for standardizing the ways of encoding the mathematical models; however, this makes it complicated for nonprogrammers to easily generate their own authored models and integrate them into their multi-scale simulations because the markup specification language can become lengthy, and it is not easily readable to nonexperts. Therefore, the use of simple, straightforward, symbolic, mathematical, authorable model editors is preferred, so that the generation of the procedural code is automatic, optimal, and efficient.
Multi-scale modeling tools require an ease of data handling throughout the whole work pipeline to create a model. Imaging technologies in the clinic and multi-scale physics environments require flexible and compatible data handling tools for pre-and post-processing information.
12.3 Input Data Pipeline for Patient-Specific Multi-Scale Cardiac Modeling
In order to generate a patient-specific model of the heart, a series of measurements should be performed by the physicians [16, 21] (Fig. 12.1), see also Chap. 2, 8–10. One of the first main steps toward a patient-specific model is to acquire a detailed, patient-specific geometrical description of the anatomy using CT or MRI imaging. Boundary conditions to a cardiac model relate to all of the adjacent structures or systems interacting with the organ of study. Therefore, measures of hemodynamics and electrophysiology are also important to set up a personalized model, but they require to be selected appropriately according to the nature of the simulation.
12.3.1 Ventricular Anatomy and Fiber Architecture
The anatomy can be obtained using CT and contrast MRI. The images require processing for segmentation and registration to generate a mesh (see Sect. 12.9). Data such as fiber architecture cannot generally be obtained in a patient-specific manner; however, studies have shown that fiber architecture is highly conserved among individuals and species after accounting for the geometric variations between the anatomies [14].
It is also possible to locate scarred or ischemic tissue using measures from gadolinium-enhanced MRI, to determine as detailed as possible the patient’s physio-logy, as part of the mesh generation and parameter estimation for baseline simulations.
12.3.2 Hemodynamics
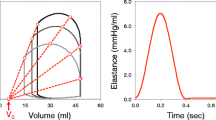
The use of hemodynamic data allows the estimation of patient-specific material properties of the myocardium and parameters for the circulatory system model. Cardiac ultrasound techniques provide approximations to volumes and pressures [19]. Measures of aortic pressure, left and right ventricular pressure and volumes during both systole and diastole, stroke volume, and ejection fraction help the user to define the appropriate circulatory boundary conditions. Pericardial pressure, for example, can be selected and included in the model from existing data and would not require to be measured since it is not easily obtained in a patient-specific manner. Ventricular pressure and volume can be used to obtain an elastance model for the myocardium using the end-diastolic pressure–volume relationship (EDPVR) and end-systolic pressure–volume relationship (ESPVR) if measures of a couple of beats are obtained at different preload conditions. This is generally achieved clinically by inducing a premature ventricular contraction in patients with a pacemaker. With both EDPVR and ESPVR, the passive and active material properties of the myocardium can be estimated (Chap. 8) [32, 33]. The 3D geometrical reconstruction of the ventricles is then inflated to its passive (EDPVR) and active (ESPVR) levels. This will yield the model properties, by estimating the passive stress-scaling factor, passive exponential shape coefficient, and the active stress-scaling factor through minimizing the difference between the model and the measured patient-specific pressure–volume relations.
12.3.3 Electrophysiology
Electrophysiological data are also necessary to replicate as closely as possible the patient’s heart electrical activity for the baseline simulation. The acquisition of the ECG is simple, and it is used to obtain the total ventricular activation time using the QRS complex, and it can be used to roughly estimate the myocardial conductivity. However, the best measure of electrical endocardial activity is obtained with electroanatomic mapping tools, to set up the stimulation pattern endocardially and estimate the electrical properties of the tissue [34] (Chap. 10). The conductivity and the stimuli can be adjusted in the model to approximate the measurements taken from the patient. The earliest activated regions will provide the stimuli information, while matching the activation patterns will provide the conductivity. Unfortunately, this measurement is quite invasive, and sometimes, it is not convenient to perform.
It is also possible to solve the inverse problem by measuring the epicardial activation from body surface potentials [24], but a model of the torso is necessary to solve the inverse problem.
12.4 Software Architecture
The structure suggested for a multi-scale physics environment is shown in Fig. 12.2. An adequate architecture can comprise three major components: a Database Server, a Solver Client, and a Solver Server and any number of Plug-in applications. The Client serves as a graphical user interface (GUI), and the Server carries out all the numerical calculations for solving the problem. This architecture allows the user to launch the program as a client or a server or both. In this way, the user can locally run a client session, while a remote server or host does all the calculations; otherwise, it can also be used as a whole application together on the local machine. There should be no need for client and server to run in the same OS.
12.5 Database Server
The aim for technology development and collaborative research in cardiac modeling is to have access to services, web services, training, and dissemination to enable, catalyze, and conduct medical research by combining technologies and focusing them on targeted translational and multi-scale challenges in the biomedical arena. The National Biomedical Computation Resource (NBCR) (http://www.nbcr.net) provides the support and infrastructure to the developers of Continuity to achieve that goal. The objective for the development of computational cardiac modeling is to improve treatment planning for heart disease through individual patient modeling, by providing the framework to incorporate personalized data into new or existing models to aid in an accurate, personalized diagnosis and predict outcomes of existent therapies (i.e., cardiac resynchronization therapy). In order to achieve this aim, a multi-scale framework requires access to a database or library.
The Database is designed to facilitate model sharing, reusing, and classification, by making the models fully annotatable (Fig. 12.3). The owner of the model can choose to share a model with the public or the public can own the model or the owner can lock its contents for personal use only. The database models should be anonymized appropriately and backed up frequently for security. The database is configured to maximize its functionality, rather than just becoming a storage space. The library can be used to store only the small changes on an already existing model. For example, if the user changes a small part of a model, like the initial conditions, then the library will only store and organize the objects changed instead of duplicating the rest of the model content. It also contains all the information related to the model like date of production, description, basis functions, node labels, electrophysiology model, circulatory model, etc. Therefore, all the objects in the library are easily accessible since the user can search on the database by title, a description of the model, the metadata, or any biological attributes defined (i.e., species, organ-related labels). The user can also retrieve whole models or only specific objects from different models. Attributes can be added to any model, class, object, or subobject, as well as any other file or script necessary to pre- or postprocess data.
The database is a place where users can share complete models, for example, a full model that has been published, so that the scientific audience can reproduce the author’s results. The users should be registered to use the Database for a better control over its contents.
The design of the database described mimics the structure of a Continuity file (*.cont6). The hierarchic structure of the database is shown in Fig. 12.3. At the top end, there are owners, who can have any number of models that are publicly available, or private. Each model has a title, description, date and time of creation, version, variation, and any number of classes. The classes are any loadable modules from the Solver Server. Below the classes are Objects, which contain the data from the Editors’ forms including coordinate system, basis function, elements, material coordinate equations, ionic model, fitting constraints, images, scripts, etc. (Fig. 12.4). Objects like the nodes, initial conditions in biomechanics, and fitting constraints have subobjects. These subobjects provide a different attribute to their parent object (i.e., nodes of a human model with fitted fibers from a different species). Every model, class, object, and subobject can have either the same or different attributes (i.e., species, organ system, organ, tissues, reference author, reference title, reference date, reference publication). Every hierarchy of the classification is useful for retrieving data in the whole database structure.
A user can deposit a model and different versions of it. As a rule, a new version of the model saves only the changes to the model and references back to the original model for the unchanged information. To create a new model, a new unique title has to be used. Only the owner of a confidential model can delete its files and all its contents, while the public cannot erase public models for safety.
12.6 Solver Client
The client serves as a graphic user interface and includes a viewer framework with dynamically loadable menus, commands and data file editing for Input and Output files, viewers, including 3D visualization and animation, rendering controls, graphical plotting and image analysis, a scripting shell, and message area. It also provides the interface to web services, compiling, and registration of users to access the software and the database. Toolbars are commonly used and create an easy, straightforward interaction to the solver. Menus should also contain commands for setting and adjusting visualization parameters and for saving images, animations, or simulations. Important components of the Solver Client are the model editors that form part of each module. The graphic user interface also provides the framework to support external plug-in applications or libraries. For example, the imaging module in Continuity includes a heart-wall marker, some tracking methods, image registration, and segmentation methods; it can also access a library from an Imaging ToolKit (ITK Snap library) for image registration, segmentation, and automatic mesh-building (http://www.itksnap.org/pmwiki/pmwiki.php). A framework that provides access to plug-in applications makes it compatible with other solution methods or pre-/postprocessing applications.
12.7 Model Editors
Any software platform that uses symbolic, mathematical, authorable model editors makes the generation of procedural code automatic and efficient. They are built to facilitate modelers and nonprogrammers the implementation of new mathematical models, or reuse existing ones. One language used for the implementation of the mathematical models is a symbolic mathematics library for python called “SymPy” (http://www.docs.sympy.org/index.html). This python library works as a computer algebra system to make coding as simple as possible and comprehensible. The use of symbolic mathematics makes it easier for the users to directly read and understand the model, and they are not required to learn a programming language to create new models. Online tutorials make it easy to understand and use SymPy. Therefore, each model editor provides the framework to compile the mathematical model using a web service, or if compilers are available in the host computer, then the user can compile his own models. Therefore, the output generated can actually be obtained in any format necessary, that is, C, Python, Fortran, CUDA, or even XML. An XML input can also be translated into readable python, C, or SymPy code.
12.8 Solver Server
The server carries out the numerical calculations for simulation and problem solving. It has dynamically compilable and loadable numerical analysis functions for finite element modeling, nonlinear elasticity and biomechanics, reaction–diffusion systems and electrophysiology, and transport processes and biophysics for cell systems modeling.
The common methods used for cardiac multi-scale modeling are finite element methods (FEMs) for nonlinear mechanics. FEMs are numerical analysis tools to solve partial differential equations (PDEs) in a complex domain (see “Preface” to the book). It is convenient to employ a modular software that includes the anatomy for fitting and creating a mesh; the biomechanics where material properties and circulatory boundaries can be set; the electrophysiology where membrane kinetics are modeled; biotransport to model reaction–diffusion problems; and finally, but most importantly, a method for specifying the interaction and feedback between modules.
More specifically, we can set a system of ordinary differential equations (ODEs) at a particular point in a physical domain called node, defined in a nodes form (Fig. 12.5a). The mathematical model introduced at this level is the smallest scale to be used in our simulation, and it represents a network of biochemical or biophysical interactions at the subcellular and cellular scale. To spatially couple this pointwise network, we include constitutive laws that represent the physical properties of the system. The solution in the mesh is approximated by linear or higher order functions defined element-wise (Fig. 12.5b). By doing so, we construct the substrate with all the properties of the next scale; this is a detailed arrangement at the tissue level that includes the tissue anisotropy/orthotropy, result of the fibrous and laminar construct of the myocardium. The tissue structure is a major determinant of both the contraction and propagation of the action potential. PDEs are used to establish the space and time overall behavior and the conservation laws governing the physics over the whole geometry or anatomy of the system. It is also important to establish the adequate boundary conditions that represent the interactions of the heart with neighboring structures. Hemodynamic models are necessary to establish an adequate arterioventricular coupling. Lumped parameter models of the circulation are generally used for this purpose. The modeler can also author the tight relationships between the mathematical models for excitation–contraction coupling (ECC) and their interactions. Information between modules can be passed in a coordinated and efficient manner by scripting the different time steps at which each system requires feedback.
In this way, we can create highly detailed, fully-coupled, patient-specific anatomic models of electromechanics of the heart, setting up boundary conditions in a patient-specific manner.
12.9 Imaging and Fitting Modules
The imaging and fitting modules allow the user to create patient-specific finite element meshes of the heart. The imaging module includes segmentation (ITK Snap library) and automatic mesh-building tools. The user can import a stack of MR, CT, or other types of volumetric imaging data, apply scaling, rotation, or translation to position the stack in the model space, and manually or automatically segment different contours of interest. The pixels that define the segmented contours are sampled as the data to which the initial mesh will be fit. A rough initial mesh is first built in the imaging module using segmented images as a guide. The data points for each contour can be identified with a label to easily distinguish each contour. Once contours have been differentiated, an initial mesh can be conveniently built.
Each contour of the heart (typically left ventricular cavity, right ventricular cavity, epicardium, septum, etc.) is represented by hollow volumes with nodes as vertices. The volumes together form the entire mesh. The user can place these nodes directly on to the segmented image data at the desired slices to create 2D surfaces of each contour. This allows the user to build an initial mesh that already closely approximates the real geometry.
Another rough geometrically approximated mesh created in parallel is built using the fitting module. The rough mesh is fitted next to the segmented data for a more accurate representation of the geometry. The fitting process minimizes the difference between the coordinates of the segmented data and their corresponding interpolated coordinates on the mesh. The corresponding interpolated coordinate data on the mesh are defined by projecting each data point onto the closest surface of the segmented mesh. The maximum projection and angle with the surface can be controlled to filter out noisy data. The user can define fitting constraints on the coordinates and derivatives of the nodes to have control of the fit. Smoothing weights can also be applied to penalize for large changes in stretching and bending of the mesh surface in the process of fitting; they allow the user to fine-tune the smoothness of the fit if data is sparse or noisy. Finally, data points from all contours can be fitted to their corresponding mesh surfaces simultaneously; this can be done using appropriate labels that distinguish between data points of different contours and their matching surface elements. The fitting module can also fit fields such as fiber angles to the geometry. Fiber direction is generally defined nodewise in the nodes form.
The myofiber architecture is an important component of multi-scale models of the heart. The heart has a characteristic arrangement of fibers that is roughly conserved among individuals [14]. It is possible to obtain accurate measurements of the myofiber orientation using diffusion tensor MRI (DTMRI), or histologically in postmortem hearts; however, there exist some databases (like the Johns Hopkins University public database, http://www.ccbm.jhu.edu/research/dSets.php) where cardiac fiber angles can be obtained and morphed into a new ventricular geometry for patient-specific applications [3] (Chap. 9). In the human and the dog left ventricle, the muscle fiber angle typically varies continuously from about −60° at the epicardium to about +70° at the endocardium, with a higher rate of change at the epicardium [30].
12.10 Mesh Module
The mesh is the geometrical representation of the anatomy to be modeled. It is the frame that holds together the models of different scales. The mesh is obtained from image segmentation from CT or MRI scans and fitted adequately to create a structure that will be discretized into a collection of a finite number of points (nodes), which conform the vertices to the various subdomains (elements). The definition of nodes and elements is a requirement for creating the mesh. The nodes are discrete points in the surfaces of the desired anatomy that are specified on a coordinate system (Fig. 12.5a). The elements establish the connection pattern between the nodes, to dictate the ordered location and connectivity of the elements (Fig. 12.5b). The mesh locates the anatomy into a coordinate system. It is advantageous to have the ability to use various coordinate systems: rectangular cartesian, cylindrical polar, spherical polar, prolate spheroidal, or oblate spheroidal. This variety of coordinate systems allows the user to easily construct a mesh by making use of its geometrical features and using the appropriate coordinate references (for use in Fitting). The left ventricular geometry can be initially approximated by nested ellipses of revolution, that is, two fitted truncated ellipses revolving on the major axis, that is, a prolate spheroidal coordinate system [9]. Cylindrical coordinates facilitate the description of tubular objects, while the spherical polar is useful to describe sphere-shaped geometries [8].
One characteristic of FEMs is the approximation of the continuum using a finite number of functions (finite element interpolation), so the construction of the elements is approximated by basis functions, and their parameters are defined at the nodes. Continuity supports a variety of basis functions for isoparametric finite element interpolation, generally 1D, 2D, and 3D hexahedral Lagrangian and Hermite piecewise polynomial functions as well as Gauss quadrature integration. In a Mesh module, the user can define numerical and graphical functions associated with nodes; use finite element operations and mathematical functions associated with the mesh; use dependent variables including interpolation, global-element mapping, coordinate transformations, and computations of metric tensors and related quantities such as arc lengths and areas. The Mesh module interfaces problem definitions and numerical solutions. It includes algorithms for solution of nonlinear equations, element and global equation assembly, residual calculations, least squares, eigenvalue and time-stepping algorithms, general utility operations, basic numerical algorithms. The simplest type of elements are piecewise linear Lagrange elements. Basis functions are defined in terms of independent variables, that is, spatial coordinates, so that various combinations of interpolation can be used to create the desired geometry, that is, by taking tensor products to create 1D, 2D, or 3D geometries. Both kinds of interpolation can be estimated with two or three collocation points. Collocation points are distinct points in the element trajectory for which the solution satisfies the initial condition. These points are then used to estimate the solution using their derivatives.
The common structure for a complete mesh for a FEM would include four coordinate systems: a rectangular Cartesian global reference coordinate system, an orthogonal curvilinear coordinate system to describe the geometry and deformation, a general curvilinear finite element coordinates (at each element), and a local orthonormal body coordinates, defining the material structure or fiber and sheet angles.
Every component in the software requires a user-friendly interface editor. The nodes and element forms are the mostly used ones to set up a computational model (Fig. 12.5). In the nodes form, we can define the coordinates for the nodes, the interpolation required for each coordinate, and the fiber angle information; it also contains Fields that can be used to define any other parameters required for the simulation. These Fields can be assigned and used inside any model editor.
12.11 Biomechanics
The structural organization of the myocardium is an important determinant of the cardiac mechanics and its material properties. The passive mechanical properties of tissue have mostly been studied on isolated, arrested whole hearts, or on tissue preparations; however, the total stress in the heart is generally considered as the sum of passive stress, when the tissue is at rest, and active stress generated by the contraction of the myocytes.
Governing equations that relate material properties to continuum tissue behavior must be set in the biomechanics module. The myocardial stress and strain distributions are needed to characterize the regional ventricular function, particularly in pathological conditions like ischemia or scarring. The stress is related to the forces exerted at a specific location in the myocardium and is normally calculated with respect to an orthogonal system to the fiber orientations. The strain refers to the deformation, or local shape change, as a result of the applied forces. The passive ventricle can be defined by an exponential strain–energy function, which treats the myocardium as nonlinear, anisotropic, and nearly incompressible material. Various parameters in the models can be used to describe the scaling factors for the stress magnitude, incompressibility, and anisotropy of the tissue. The active stress can be described using a time-varying elastic model and can be dependent on time, sarcomere length, and intracellular calcium concentration [20]; with Hill-type models, where the active fiber stress development is modified by shortening or lengthening according to the force–velocity relation, so that fiber tension is reduced by increased shortening velocity or fully history-dependent models that are more complex, based on cross-bridge theory [6].
In the biomechanics module, the Circulatory model is set as a boundary condition to the mechanics. Various types of circulatory models are available in the literature to set the conditions necessary for the simulation [1].
12.12 Electrophysiology Module
The cell electrophysiology is a complex biological system, determined by ion movement across membranes, changing ionic concentrations in different cellular compartments in a nonlinear fashion. The modification of the function of ion channels by genetic mutation, infarcts, scars, or remodeling can create blocks, cardiac arrhythmias, or even sudden cardiac death. The main aim of computational models of electrophysiology is to integrate the vast knowledge on ion channel properties, generally studied in a single-channel fashion, into full cellular models with feedback interactions to relate the molecular-level dynamics to the whole cell function and their clinical phenotype. ECC occurs at the molecular level, particularly involving Ca2+ ion movement in various cellular compartments to create a transient rise of intracellular Ca2+, which activates contractile proteins of the cell. Consequently, the heart mechanics also have feedback effects on ion dynamics [25, 26].
Various models of electrophysiology for various species (human, mouse, rabbit, dog, guinea pig, etc.) with different complexity have been published. Various reviews have been published on the subject [7, 28]. The biggest source of mathematical models of electrophysiology available online is the one provided in the CellML repository (http://www.models.cellml.org/electrophysiology). Most of the models contained in this repository use the XML and have various levels of curation. Models of electrophysiology range from a simple two-equation model that describes the propagation wave in the tissue [26] to models with much higher number of ODEs to be solved and that account for cellular heterogeneities in the myocardium, like the model by Flaim et al. [11].
The electrophysiology module in Continuity was created to solve Monodomain problems, that is, a unitary reaction–diffusion equation, a single compartment, instead of two that represent the intracellular and extracellular spaces, called Bidomain. Comparisons between both methods have been fully studied, and results are highly correlated (0.9971 for an orthotropic conduction), with the advantage that the monodomain solution is less computationally demanding [12]. In the electrophysiology model editor, all of the parameters can be set in a nodewise fashion in a field in the nodes form, so that a stimuli current can be applied at defined regions in the mesh. A set of initial values can be assigned to distinct elements using labels, so that regions of heterogeneous cell kinetics can be simulated. The purpose is to approximate normal or pathological tissue as close as possible to its natural physiology. For example, by labeling elements containing endocardial, midmyocardial, and epicardial cells, we can simulate their heterogeneous behavior [5]. Furthermore, every model can access a set of Parameters defined in the nodes form as fields. These parameters can be used in the model to set up gradients and nodewise variations in the tissue (i.e., apex-to-base cell heterogeneity, ischemic regions).
Direction of propagation in the myocardium depends on fiber rotation and the electrical properties of the tissue determined by its conductivity and capacitance to simulate the myocardium as an orthotropic substrate [4]. These myocardium propagation properties are determined by a diffusion tensor. All the matrix transformations and rotations should be conveniently set in the software, so that the user can easily create the required diffusion tensor in the electrophysiology module by just adding a diffusion coefficient for each orthogonal direction to the fiber orientation.
In general, the electrophysiology model used for each simulation is dependent on the objectives and requirements of the study. Simple models have proven to provide good approximations for excitation propagation in the myocardium, like the model of Beeler and Reuter with only eight ODEs [2], with the advantage of requiring little computational power. Larger models with a greater number of ODEs describe various ion channels and Ca2+ kinetics using Markov chain models or including gene mutations [7]. These models are well suited for more detailed analysis using meshes with larger number of elements, which increases computer-processing time, but it is up to the users to choose the electrophysiology model that would provide the best information for their simulations.
12.13 Fully Coupled Electromechanics Models
ECC is a complex mechanism that occurs at the subcellular level, where local changes of membrane potential lead to the release of Ca2+ and hence production of force by the myofilaments. Cells are tightly coupled and have a well-structured arrangement in the myocardium. The ionic currents propagate through gap junctions allowing the activation of the whole myocardium. The mechanical coupling of the myocytes is directly linked to cytoskeletal structures, the extracellular matrix, and the contiguous myocytes. The ventricular wall deforms during the cardiac cycle; this deformation is influenced by the myocardium architecture, which in turn, influences the excitation propagation of the wave.
A fully-coupled, electromechanic model is able to set the interaction between the action potential propagation, the contractile force generation, and the myocardial deformation under realistic hemodynamic boundary conditions in an anatomically accurate finite element mesh of the left ventricle [5]. The degree of coupling can be set up through scripting. This means that the algorithms for coupling various models are fully authorable by the user [23]. This form of coupling is useful for investigating questions related to mechanoelectric feedback [22] at physiological and pathological conditions.
Electrophysiology models with various degrees of complexity can be used to simulate the action potential, ionic channels, and Ca2+ ion dynamics. The active tension model is generally the intermediate model between the biomechanics and electrophysiology. The active tension model is able to reproduce the dynamic response of cardiac muscle to time-varying inputs of sarcomere length and cytosolic Ca2+ [25].
Different schemes for fully coupled electromechanics can be implemented; however, the challenge is to create stable numerical algorithms that are able to interchange information between the different modules at various time steps. In general, electrophysiology contains the smaller scale in the simulations (subcellular and cellular), so the time steps to solve the electrophysiology are small. After a few small time steps, the larger scales, like mechanics can be updated using the Ca2+ transient generated by the cell model. This will update the change of shape of the geometry due to the contraction, using the sarcomere length as a parameter, for example.
12.14 Plug-in Applications
The software architecture allows for the implementation and data communication with useful plug-in applications for pre- or postprocessing implementations. One of the most notable plug-in applications is the implementation of Non-Uniform Rational B-Splines (NURBS) methods. NURBS is a convenient tool that is frequently used for commercial purposes by industry and entertainment. NURBS is a method for parameterizing curved surfaces and volumes through tensor products of polynomials (Fig. 12.6a). Finite element modeling, utilizing NURBS, is convenient because these surfaces can be bended and deformed in a user-friendly “point-and-click” manner. Existent methods are able to convert imaging data to NURBS surfaces [35] in a quick, efficient manner and to reparameterize them to finite element meshes with a different set of polynomial basis functions. The resulting NURBS surface or volumetric object can be used in finite element analysis directly (Fig. 12.6b). This implementation makes the process of geometrical reconstruction less time consuming.
12.15 Computational Requirements
As the complexity of the subcellular and cellular models is increasing, the computational frameworks used become challenged. Particularly, systems like the cell pose a great number of ODEs and states to the numerical calculations, which need to be manipulated by processors. Computational power and tools keep increasing exponentially, so a patient-specific computational framework should be able to follow and apply the latest improvements on data processing. In 2002, a coupled model of 3D cardiac electromechanics in an anatomically detailed canine heart with 19,200 degrees of freedom [31] had a total computation time of 78 h and 20 min on a single processor, of which the mechanics problem with 1,960 degrees of freedom took about 6½ h. Soon after, most of the computationally challenging problems were parallelized, so that a similar mechanics problem in 2008 [15] took 40 min per cardiac cycle on 12 nodes of a cluster. These processing times were still long to be used for patient-specific modeling, given that a cluster was available for dedicated modeling in the clinic, and the patient-specific mesh was already fitted and set to solve the problem. The recent extended usage of GPUs for numerical processing purposes is setting new trends for computationally expensive applications. Nowadays, the use of GPUs has been extended to computational models of electromechanics [18], so that using an nVidia GTX-295 GPU (of about US$ 500) for a desktop computer can improve the performance and can speed up calculations over a cluster by 91 times, and 134 times over standard communication interfaces. This work opens up the possibility for real-time simulations for diagnostic purposes in the clinical setting. Furthermore, this kind of application was designed to be used by nonexpert programmers and is easily implemented as part of the computational framework.
The GPU usage in Continuity is invisible to the user. The workflow for the simulations begins with a python program that specifies the mathematical model using the model editor with the SymPy Python library code input. This input is translated into a naïve CUDA C, which is later optimized using source-to-source transformations [17]. The resultant code is compiled with CUDA C, which manages the registers and handles the memory in the GPU to introduce all the thread parallelism to solve the ODEs. PDE calculation is still handled by the CPU, but the computational time required is not as big as the ODEs (before the GPU optimization). However, PDE computations in the GPUs are still part of future work to further speed up computation times. In general, the use of a GPU significantly reduces the computational bottleneck in an 87 ODEs electrophysiology model [11], to simulate a single heartbeat from 4.5 h on a 48 core Opteron cluster to 12.7 min on a desktop workstation without the knowledge of the GPU or CUDA programming.
12.16 Limitations
A mathematical model is an approximation to reality; therefore, all models are inherently wrong. However, mathematical models are still extremely useful to understand the knowledge acquired through experimentation and the components missing to explain the differences between the theoretical knowledge and the actual physiological function. Multi-scale computational models have been thoroughly validated in animal models [15] and may prove useful for prediction and understanding of CRT responder and nonresponder cases (Chaps. 1 and 10). There is a great number of mathematical models for cardiac models of animal physiology, mainly mouse, rat, dog, rabbit, etc. However, human physiology is somewhat different from animal physiology. Accessibility to human heart tissue is limited due to ethical constraints, so new methods and approaches to obtain human cardiac tissue are being implemented to enlarge our knowledge on human cardiac subcellular, cellular, and organ physiology in health and various stages of disease [27]. However, all the gaps on our knowledge on human physiology can be complemented with animal models. Studies on cardiac electrophysiology and arrhythmogenesis have been extensive in animal tissues and have provided a wealth of knowledge, particularly regarding genetic manipulation, mutations, and physiopathologies. For patient-specific applications in the clinical setting, it is important to take into account the objectives of the simulation, so that reduced or minimal models with validated accuracy can be used to minimize the computation times, so that real-time simulations are feasible.
An important limitation to real-time simulations is the reconstruction of the geometrical data from measurements of patient-specific geometries in the clinic. Better tools for automatic segmentation of CT and MRI imaging, and generation of geometrical representations of the anatomy (the finite element mesh) are required. In general, this process is still lengthy and time consuming and may still pose a challenge for the use of computational modeling in the clinical setting.
References
Arts, T, T Delhaas, P Bovendeerd, X Verbeek, and F W Prinzen. “Adaptation to mechanical load determines shape and properties of heart and circulation: the CircAdapt model.” American Journal of Physiology, Heart Circulation Physiology 288 (2005): 1943–1954.
Beeler, G W, and H Reuter. “Reconstruction of the action potential of ventricular myocardial fibres.” The Journal of Physiology 268 (June 1977): 177–210.
Beg, Mirza Faisal, Patrick A Helm, Elliot McVeigh, Michael I Miller, and Raimond L Winslow. “Computational cardiac anatomy using MRI.” Magnetic Resonance in Medicine 52, no. 5 (November 2004): 1167–1174.
Caldwell, Bryan J, Mark L Trew, Gregory B Sands, Darren A Hooks, Ian J LeGrice, and Bruce H Smaill. “Three distinct directions of intramural activation reveal nonuniform side-to-side electrical coupling of ventricular myocytes.” Circulation: Arrhythmia and Electrophysiology 2 (2009): 433–440.
Campbell, Stuart G, et al. “Effect of transmurally heterogenous myocyte excitation–contraction coupling on canine left ventricular electromechanics.” Experimental Physiology 94, no. 5 (May 2009): 541–552.
Campbell, Stuart G, Sarah N Flaim, Chae H Leem, and Andrew D McCulloch. “Mechanisms of transmurally-varying myocyte electromechanics in an integrated computational model.” Philosophical Transactions of the Royal Society A 366, no. 1879 (September 2008): 3361–3380.
Clancy, C E, Z I Zhu, and Y Rudy. “Pharmacogenetics and anti-arrhythmic drug therapy: a theoretical investigation.” American Journal of Physiology, Heart and Circulatory Physiology 292, no. 1 (January 2007): 66–75.
Costa, K D, P J Hunter, J M Rogers, J M Guccione, L K Waldman, and A C McCulloch. “A three-dimensional finite element method for large elastic deformations of ventricular myocaridum: I – cylindrical and spherical polar coordinates.” Transactions of the ASME, Journal of Biomechanical Engineering (ASME) 118 (November 1996): 452–463.
Costa, K D, P J Hunter, J S Wayne, L K Waldman, J M Guccione, and A D McCulloch. “A three-dimensional finite element method for large elastic deformations of ventricular myocardium: II – prolate spheroidal coordinates.” Transactions of the ASME, Journal of Biomechanical Engineering (ASME) 118 (November 1996): 464–472.
FitzHugh, R. “Impulses and physiological states in theoretical models of nerve membrane.” Biophysical Journal 1, no. 6 (July 1961): 445–466.
Flaim, Sarah N, Giles R Wayne, and Andrew D McCulloch. “Contributions of sustained INA and IKv43 to transmural heterogeneity of early repolarization and arrhythmogenesis in canine left ventricular myocytes.” American Journal of Physiology Heart Circulatory Physiology 291 (2006): 2617–2629.
Franzone, Colli P, L F Pavarino, and B Taccardi. “Simulating patterns of excitationm, repolarization and action potential duration with cardiac bidomain and monodomain models.” Mathematical Biosciences 197, no. 1 (September 2005): 35–66.
Greenstein, Joseph L, Robert Hinch, and Raimond L Winslow. “Mechanisms of excitation–contraction coupling in an integrative model of the cardiac ventricular myocyte.” Biophysical Journal 90 (January 2006): 77–91.
Helm, Patrick A, et al. “Evidence of structural remodeling in the dyssynchronous failing heart.” Circulation Research 98 (2006): 125–132.
Kerckhoffs, Roy CP, Andrew D McCulloch, Jeffrey H Omens, and Lawrence J Mulligan. “Effects of biventricular pacing and scar size in a computational model of the failing heart with left bundle branch block.” Medical Image Analysis 13, no. 2 (April 2009): 362–369.
Kerckhoffs, Roy CP, Sanjiv M Narayan, Jeffrey H Omens, Lawrence J Mulligan, and Andrew D McCulloch. “Computational modeling for bedside application.” Heart Failure Clinics 4 (2008): 371–378.
Lionetti, F, Andrew D McCulloch, and Scott B Baden. “GPU accelerated solvers for ODEs describing cardiac membrane equations.” nVidia GPU Technology Conference. San Jose, CA: nVidia, 2009. 34.
Lionetti, Fred. GPU accelerated cardiac electrophysiology. La Jolla, CA: UCSD, Master’s thesis, 2010.
Malm, S, S Frigstad, E Sagberg, H Larsson, and T Skjaerpe. “Accurate and reproducible measurement of left ventricular volume and ejection fraction by contrast echocardiography: a comparison with magnetic resonance imaging.” Journal of the American College of Cardiology 44 (2004): 1030–1035.
McCulloch, Andrew D. Cardiac biomechanics, vol. I, in The Biomedical Engineering Handbook, by Joseph D Bronzino, 28:1–26. Boca Raton, FL: CRC Press, 2000.
Neal, Maxwell Lewis, and Roy CP Kerckhoffs. “Current progress in patient-specific modeling.” Briefings in Bioinformatics 11, no. 1 (January 2010): 111–126.
Niederer, S A, and N P Smith. “A mathematical model of the slow force response to stretch in rat ventricular myocytes.” Biophysical Journal 92, no. 11 (June 2007): 4030–4044.
Niederer, Steven A, and Nicolas P Smith. “An improved numerical method for strong coupling of excitation and contraction models in the heart.” Progress in Biophysics and Molecular Biology 96, no. 1–3 (January–April 2008): 90–111.
Ramanathan, Jia P, R N Ghanem, P Jia, K Ryu, and Y Rudy. “Noninvasive electrocardiographic imaging for cardiac electrophysiology and arrhythmia.” Nature Medicine 10 (2004): 422–428.
Rice, J J, F Wang, D M Bers, and P P de Tombe. “Approximate model of cooperative activation and crossbridge cycling in cardiac muscle using ordinary differential equations.” Biophysical Journal 95, no. 5 (September 2008): 2368–2390.
Rogers, J M, and A D McCulloch. “A collocation-Galerkin finite element model of cardiac action potential propagation.” IEEE Transactions on Biomedical Engineering (IEEE EMBS) 41, no. 8 (August 1994): 743–757.
Rudy, Yoram, et al. “Systems approach to understanding electromechanical activity in the human heart.” Circulation (A National Heart, Lung, and Blood Institute Workshop Summary) 118 (2008): 1202–1211.
Rudy, Yoram, and Jonathan R Silva. “Computational biology in the study of cardiac ion channels and cell electrophysiology.” Quarterly Reviews of Biophysics 39, 1, (2006): 57–116.
Shaw, James Rochester. “Models for cardiac structure and function in Aristotle.” Journal of the History of Biology 5, no. 2 (September 1972): 355–388.
Streeter, D D Jr. “Gross morphology and fiber geometry of the heart.” In Handbook of physiology, section 2: the cardiovascular system, chapter 4, by M. B. R., 61–112. Bethesda, MD: American Physiological Society, 1979.
Usyk, Taras P, Ian J LeGrice, and Andrew D McCulloch. “Computational model of three-dimensional cardiac electromechanics.” Computing and Visualization in Science 4 (2002): 249–257.
Walker, J C, et al. “MRI-based finite-element analysis of left ventricular aneurysm.” American Journal of Physiology, Heart and Circulation Physiology 289 (2005): 692–700.
Watanabe, H, S Sugiura, H Kafuky, and T Hisada. “Multiphysics simulation of left ventricular filling dynamics using fluid–structure interaction finite element method.” Biophysical Journal 87 (2004): 2074–2085.
Yue, A M, M R Franz, P R Roberts, and J M Morgan. “Global endocardial electrical restitution in human right and left ventricles determined by noncontact mapping.” Journal of the American College of Cardiology 46 (2005): 1067–1075.
Zhang, Yongjie, Yuri Balzilevs, Samrat Goswami, Chandrajit L Bajaj, and Thomas JR Hughes. “Patient-specific vascular NURBS modeling for isogeometric analysis of blood flow.” Computer Methods in Applied Mechanics and Engineering 196, no. 29–30 (May 2007): 2943–2959.
Acknowledgements
The authors acknowledge the support of NIH grants P41 RR08605, 1R01 HL96544, 1R01 HL086400, 1R01 HL091036, 1R01 HL083359, NSF grant BES-0506252, and UC Discovery grant it106-10159. ADM is a cofounder of Insilicomed, a licensee of UCSD software developed in this research. Insilicomed was not involved and did not support this research.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2010 Springer Science+Business Media, LLC
About this chapter
Cite this chapter
Aguado-Sierra, J. et al. (2010). A Computational Framework for Patient-Specific Multi-Scale Cardiac Modeling. In: Kerckhoffs, R. (eds) Patient-Specific Modeling of the Cardiovascular System. Springer, New York, NY. https://doi.org/10.1007/978-1-4419-6691-9_12
Download citation
DOI: https://doi.org/10.1007/978-1-4419-6691-9_12
Published:
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-4419-6690-2
Online ISBN: 978-1-4419-6691-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)