Abstract
Protein oxidation is a post-translational modification that can have beneficial or detrimental effects on cells. The interaction of reactive oxygen species (ROS) with proteins leads to their oxidation and ROS may be produced by several different enzymes. The first section of this review examines the major intracellular sources of ROS, with special attention paid to mitochondria and NADPH oxidases. It discusses the different oxidation of amino acid residues with a focus on cysteine oxidation as it is involved in many signaling pathways. Carbonylation and nitrosylation are two other protein modifications that are of particular importance in cellular metabolism. The final section is concerned with the role that protein oxidation plays in disease.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
3.1 Introduction
Protein oxidation occurs when reactive oxygen species (ROS) donate electrons to proteins. ROS are a group of molecules derived from oxygen, with the two main members being superoxide (O •−2 ) and hydrogen peroxide (H2O2). They are generated under many conditions and can be either deleterious or beneficial for cells. If they are produced at high concentrations, cells undergo oxidative stress, which can result in damage to proteins, lipids and DNA and ultimately cause cell death. Consequently, cells have many different systems capable of neutralising ROS, e.g. superoxide dismutase (SOD), catalase and glutathione (Fig. 3.1). Therefore, in order for meaningful signaling to occur through protein oxidation, any ROS produced needs to interact with specific proteins and not result in general, non-specific protein oxidation.
The predominant sources and sinks of ROS in a typical cell. Mitochondria and Nox enzymes generate superoxide (O •−2 ), which can be dismutated to hydrogen peroxide (H2O2) through the action of superoxide dismutase (SOD) and then converted to water by catalase. Xanthine oxidoreductase (XOR) can also generate O •−2 . The glutathione system, consisting of reduced glutathione (GSH), oxidised glutathione (GSSG), and several enzymes such as glutathione peroxidase (GPx), can convert ROS to less reactive molecules. Thioredoxin (Trx) performs a similar function but using a different mechanism. There are also several non-enzymic anti-oxidant compounds in cells that also reduce ROS, such as tocopherol and ascorbate
This review is divided into three main sections. Mitochondria and NADPH oxidase (Nox) proteins are the two primary sources of ROS generation and will be given particular attention (Fig. 3.1). A second focus is the different ways in which proteins can be oxidized, either reversibly which generally allows a signaling pathway to operate, or irreversibly which tends to result in proteasomal degradation. Finally the role of protein oxidation in health and disease is examined.
3.2 Sources of ROS
3.2.1 Mitochondria
Mitochondria are the power-house of cells as they provide most of the energy required by eukaryotic cells. In their production of ATP, a process known as oxidative phosphorylation, they consume large quantities of oxygen and form ROS as a by-product. Oxidative phosphorylation uses the electron transport chain of mitochondria, which consists of several enzyme complexes in the inner mitochondrial membrane, namely: complex I (NADH-CoQ reductase), complex II (succinate-CoQ reductase), complex III (reduced CoQ-cytochrome c reductase), complex IV (cytochrome c oxidase) and complex V (ATP synthase) (Fig. 3.2). Together these generate a proton gradient, which allows the movement of key molecules into and out of the mitochondrial matrix (Kakkar and Singh 2007). Complexes I + III have proven to be primary sources for O •−2 (Turrens and Boveris 1980; Turrens et al. 1985; Barja and Herrero 1998). Therefore, the main sources of ROS in mitochondria are few, but the electron transport chain is in constant use, and so there exists a great potential for ROS production and hence protein oxidation.
The movement of electrons in mitochondria and the locations of superoxide (O •−2 ). Roman numerals indicate complex number. Complexes I, III and IV transport protons out of the mitochondrial matrix, but complex V transports them back into the matrix to enable it to produce ATP. Complexes I and III (shaded in grey) are the primary sources of O •−2 production
ROS produced in mitochondria tend either to be metabolized within these organelles or result in oxidisation of other mitochondrial molecules. The mitochondrial matrix contains manganese SOD (MnSOD) which facilitates the dismutation of O •−2 to H2O2 (Kakkar and Singh 2007), whilst the inter-membrane space contains a different isozyme, CuZnSOD (Okado-Matsumoto and Fridovich 2001). The inter-membrane space also contains high levels of cytochrome c, which can be oxidized by O •−2 regenerating O2 (Butler et al. 1975), and has a low pH, facilitating the spontaneous dismutation of O •−2 (Guidot et al. 1995). Mitochondria contain two other main anti-oxidant systems, those of glutathione and thioredoxin 2 (Trx2) (Go and Jones 2008). Glutathione exists mainly in its reduced form under normal conditions, but can be oxidized when it interacts with ROS. The mitochondrial specific Trx2 acts as all other thioredoxins by reducing oxidized proteins through cysteine thiol-disulfide exchange and has proven to be essential in cellular metabolism (Chen et al. 2002; Tanaka et al. 2002). Through all of these systems, mitochondria generally maintain their redox status, contributing minimally to the overall redox status of cells (St-Pierre et al. 2002).
When any of the systems previously mentioned malfunction, it may lead to the overproduction of ROS by mitochondria which shifts the cellular redox status towards oxidative stress. In fact, mitochondria play a key role in the intrinsic pathway of apoptosis (Fleury et al. 2002). In this pathway, ROS tend to accumulate in mitochondria and once the mitochondrial membrane potential depolarizes, there is a release of ROS and cytochrome c from these organelles and cells undergo apoptosis. This overproduction of ROS by mitochondria leading to increased cell death has been proposed as one of the factors leading to various neurodenegerative diseases, but the link has yet to be conclusively proven (Mancuso et al. 2006). So even though mitochondria generally do not contribute to the overall redox status of cells, their production of ROS can play a vital role to cellular health in certain conditions.
3.2.2 Nox Proteins
As mitochondria generally tend to have a balanced redox status, other sources of ROS need to be considered when discussing protein oxidation. The best known non-mitochondrial source of ROS is the Nox family of proteins. These proteins have 6 transmembrane domains, which form a channel to allow the successive transfer of electrons from NADPH to FAD to heme and then to O2 to make O •−2 (Fig. 3.3) (Sumimoto 2008). In total, there are seven members of the Nox family, Nox1-5 and Dual Oxidases (Duox) 1 and 2. Nox1-4 are similar in sequence (Suh et al. 1999; Shiose et al. 2001), whereas Nox5 is significantly different due to its four EF domains which confer a dependency on calcium (Banfi et al. 2001, 2004a, b). Duox1 and 2 are similar to Nox5, as they also have EF domains (Dupuy et al. 1999), but they stand apart from the others because of having a peroxidase-like domain (Edens et al. 2001), which means they produce H2O2 as their final product (Geiszt et al. 2003). All Nox proteins generate some form of ROS and so can be involved in protein oxidation. Indeed, ROS produced by Nox proteins have been shown to play a role in cell survival through protein oxidation (Vaquero et al. 2004; Mackey et al. 2008).
Nox family members have different tissue expression patterns, which determine where they produce ROS and so which proteins they can oxidize. Nox2, originally called gp91phox, is the prototype member of this family. It is highly expressed in phagocytes where it produces ROS to destroy engulfed pathogens (Iyer et al. 1961; Royerpokora et al. 1986). Nox1 has high expression in the colon (Cheng et al. 2001). Nox3 is expressed predominantly in the inner ear (Banfi et al. 2004a, b) and in some foetal tissues (Cheng et al. 2001). Nox4 has high expression in the kidney (Geiszt et al. 2000), but is found at low levels in many tissues (Cheng et al. 2001). Nox5 is highly expressed in testes, spleen and lymph nodes (Banfi et al. 2001). Duox1 and 2 are mainly found in the thyroid and in airway epithelia (Donko et al. 2005). Due to the widespread distribution of Nox proteins, they may produce ROS in a range of conditions and hence affect protein oxidation in a variety of circumstances.
To understand exactly how Nox proteins produce ROS, which may lead to protein oxidation, it is important to understand their regulation (Lambeth et al. 2007). Nox2 usually resides in the cytoplasmic membrane with p22phox. In the resting cell, the other signaling partners, p47phox, p67phox and p40phox, form a large complex in the cytoplasm (Wientjes et al. 1993). During cellular stimulation, the auto-inhibition of p47phox is removed through phosphorylation (Inanami et al. 1998). This phosphorylation is correlated with recruitment of Rac to this complex and together these four proteins translocate to Nox2 in the membrane resulting in the production of O •−2 . The regulation of the other Nox family members has been less comprehensively investigated, although that of Nox1 appears similar with NoxO1 being homologous to p47phox, and NoxA1 homologous to p67phox. Nox5, Duox1 and 2 do not require any of the known regulatory subunits (Kawahara et al. 2005) and their activity is dependent on calcium (Banfi et al. 2001; Banfi et al. 2004a, b). There is still much debate over the regulation of Nox3 and Nox4 (Bedard and Krause 2007). To date many varied events such as insulin signaling (Mahadev et al. 2004) and adherence to fibronectin (Edderkaoui et al. 2005) have been shown to activate various Noxes upstream of the events detailed and lead to ROS formation. Once the regulation of these proteins is understood fully, better control of overall cellular redox status and hence protein oxidation will be possible.
3.2.3 Other Non-Mitochondrial Sources
There are sources of ROS other than mitochondria and Nox proteins, which can lead to protein oxidation, but much less is known about them. Xanthine oxidoreductase (XOR) is an enzyme that can be found in two forms, xanthine oxidase (XO) and xanthine dehydrogenase (XDH) (Chung et al. 1997). XO is capable of producing O •−2 and uric acid from xanthine and O2 XDH, however, produces NADH and uric acid using xanthine and NAD. The switch between these two forms is complex (Nishino et al. 2008). XOR is found in many mammalian tissues, predominantly in epithelia and in the liver (Chung et al. 1997). Its precise physiological function remains unknown, but it is thought to contribute to liver metabolism through the degradation of xanthine to uric acid and is important in some cardiovascular disorders (Boueiz et al. 2008).
Myeloperoxidase (MPO) is an ROS generating enzyme mainly expressed in macrophages (Malle et al. 2007). It combines H2O2 with chloride (or other halides) to form hypochlorous acid, which is very toxic and acts as an anti-microbial agent. Despite its limited presence amongst cells in the body, MPO can affect many different organs, due to the ability of macrophages to travel and secrete MPO (Miyasaki et al. 1991). Hence, it has been listed as a causative agent in several diseases e.g. renal injury, cancer and multiple sclerosis (Klebanoff 2005).
Other enzymes contribute to ROS production indirectly, through generating different types of reactive species that can eventually interact with O2 to form ROS. The three main families of this type are lipoxygenases (LOX), cyclooxygenases (COX) and nitric oxide synthases (NOS). The LOX family uses O2 and arachidonic acid to form leukotriene, lipoxins and their derivatives (Kuhn and Thiele 1999). During this process unstable intermediaries and by-products can form and these act as ROS (Kim et al. 2008). Some of these products are also known to directly activate Nox proteins and so generate more ROS (Sadok et al. 2008). COX enzymes also use arachidonic acid, but to create prostaglandins again via reactive intermediates (Suleyman et al. 2007). NOS usually generate nitric oxide (NO•) which can interact with O •−2 to form the highly reactive molecule peroxynitrite that is implicated in many cardiovascular diseases (Puddu et al. 2008). Also, if NOS become uncoupled, they no longer transfer electrons to arginine, but to O2 to form O •−2 directly (Vasquez-Vivar et al. 1998). All of the enzymes that contribute indirectly to ROS formation may be the ultimate cause of protein oxidation and they should therefore be given due consideration when examining this post-translational modification.
3.3 Targets of ROS
ROS can react with amino acid and oxidize both the backbone and their side chain leading to a loss of function of proteins and deactivation of enzymes. High concentration of H2O2 may lead to irreversible damage, followed by cell death; however it is not always the case and H2O2 is capable of reversible inhibition of many proteins (i.e. phosphatases) along the main survival pathways.
3.3.1 Amino Acid Oxidation
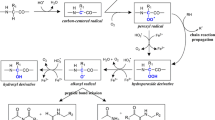
Almost all the amino acids residues in the proteins can be oxidized by ROS. However the sulfur containing amino acids (cysteine and methionine) and the aromatic amino acid (tyrosine and tryptophan) are the most sensitive to oxidation. Oxidation produces hydroxyl and carbonyl groups on proteins and can also induce more significant changes such as intra or inter molecular crosslinking. Secondary oxidations, whose consequences are not necessarily less important, include the lipid oxidation leading to the release of lipid peroxidation products such as the malondialdehyde, particularly reactive with amino acids like cysteine or lysine. In turn, they will form what is called the additions of Michael (for review see Stadtman 2001).
The aromatic amino acid residues of protein are prime target for oxidation by various forms of ROS. As shown in Fig. 3.4 phenylalanine residues are oxidized to ortho- and meta-tyrosine derivatives. Tyrosine residues are converted to the 3, 4-dihydroxy derivative (DOPA) and also to bi-tyrosine cross-linked derivatives. Tryptophan residues are converted to either 2-, 4-, 5-, 6-, or 7-hydroxyl derivatives, and also to N-formylkynurenine. Histidine residues are oxidized to 2-oxohistidine (Stadtman and Levine 2003).
The sulfur containing amino acid residues are particularly susceptible to oxidation by various ROS. Methionine is a sulfur containing amino acid residue, and though its role in proteins is not well defined it is likely that methionine can function as a key component in the regulation of cellular metabolism. Methionine is oxidized to methionine sulfoxide by many different ROS and reactive nitrogen species (RNS). The methionine sulfoxide reductases have the potential to reduce the residue back to methionine. Such reversible modifications have been recognized to provide the mechanistic basis for most cellular regulation (phosphorylation/dephosphorylation being another way of regulation which is well studied). Methionine/methionine sulfoxide interconversion can thus function to regulate biological activity of proteins through modification of the catalytic efficiency or through modification of the surface hydrophobicity of the protein (Levine et al. 1996).
3.3.2 Cysteine Oxidation
Cysteine oxidation is of particular interest because it is present in the active site of several enzymes involved in a cell survival response to stress. For a long time ROS were considered harmful for the cell, but it is increasingly clear ROS (in particular H2O2) plays a significant role as a signaling molecule in different survival pathways.
3.3.2.1 Phosphatases
The regulation of cell function through redox-sensitive cysteine residues has been most convincingly demonstrated in protein tyrosine phosphatases (PTP). The main survival pathways, being the mitogen-activated protein kinase (MAPK) or the Pi3kinase/Akt pathway, transduce their signal for cell survival mainly through phosphorylation of target molecules. The MAPK pathways operate in a cascade fashion with MAPKKK phosphorylating and activating MAPKK which then activates MAPK; the PI3K/Akt pathway operates by regulating the phosphorylation of Akt and GSK3β. As a consequence of the survival pathway transducing its survival signal via phosphorylation of key proteins, phosphatases are potent negative regulators. Multiple steps along the PI3-kinase/Akt survival pathway are negatively regulated by protein phosphatases (PP).
The PP superfamily can be categorized into three smaller subfamilies based on substrate specificity; (a) the classical protein phosphatase which is a tyrosine-specific phosphatase (PTP); (b) the dual-specificity phosphatases (DSP) which can dephosphorylate phospho-tyrosine, phospho-serine and phospho-threonine containing susbstrates; and (c) serine/threonine (Ser/Thr)-specific phosphatases which are further divided into two major classes. Type I phosphatase includes PP1; Type II phosphatase include spontaneously active phosphatase (PP2A), calcium dependent (PP2B) or magnesium dependent (PP2C) classes of phosphatases (for more information regarding classification and actions of protein phosphatases see Chap. 10 of this book).
All PTPs contain an essential cysteine residue in the signature motif C[X]5R that exists as a thiolate anion at neutral pH (Denu and Dixon 1998). This thiolate anion contributes to the formation of a thiol-phosphate intermediate in the catalytic mechanism of PTPs. Oxidation of the active-site cysteine of PTPs to a sulfenic derivative leads to enzymatic inactivation, however this modification can be reversed by incubation with thiol compounds. In some cases, the sulphydryl group is open to further irreversible oxidation if no cysteine derivatives or thiols are close enough to facilitate the formation of a disulphide bridge. The addition of another oxygen molecule or two additional oxygen molecules results in the formation of sulphinic and sulphonic acid, respectively. These oxidative modifications are irreversible and the phosphatase will be unable to become active again even in a reducing environment. It is highly likely that all phosphatases are sensitive to oxidative inhibition to some degree, as they all require a reduced cysteine for catalysis. Reversible oxidation was first demonstrated for PTP-1B during EGF (Lee et al. 1998) and insulin (Mahadev et al. 2001) signaling. The same regulation was then demonstrated for low molecular weigh-PTP (LMW-PTP) during PDGF stimulation (Chiarugi et al. 2001). Both PTP-1B and LMW-PTP rescued their phosphatase activity thanks to a re-reduction 30 min after receptor activation (Barrett et al. 1999; Caselli et al. 1998). Reversible oxidation of the Scr-homology-2 domain (SHP-2) PTP has also been reported (Meng et al. 2002).
The dual specificity phosphatases have also been reported to be oxidized, but in contrast with the classical PTP member like PTP-1B that forms a sulphenyl amide linkage between the active-site cysteine and an adjacent main-chain nitrogen (Salmeen and Barford 2005), oxidation of PTEN in vitro with H2O2 leads to the formation of a disulphide bond between the active site cysteine (Cys-124) and another cysteine residue (Cys-71) which is close by in the three dimensional structure of PTEN; this disulphide bond prevents further irreversible oxidation of the cysteine residues and PTEN can retain its phosphatase ability. PTEN has been demonstrated with many studies to be the main phosphatase negatively regulating the Pi3-kinase/Akt pathway. PTEN can dephosphorylate the lipid PIP3 to PIP2 preventing the recruitment of PH containing proteins to the plasma membrane. This results in a decrease in the survival signal transduced by Akt. PTENs importance in regulating this pathway is highlighted by its classification as a tumour suppressor molecule. Savitsky and Finkel (2002) have provided evidence for the degradation of cdc25 phosphatase (another DSP) which is the result of H2O2-induced disulfide bond formation between the active-site cysteine and another invariant cysteine residue.
The serine/threonine phosphatases [the main members being Protein Phosphatase 1 (PP1), Protein Phosphatase 2A (PP2A), Protein Phosphatase 2B (PP2B) and Protein Phosphatase 2C (PP2C)] dephosphorylate serine and threonine which are the main phosphorylation events in the transduction of the PI3-kinase/Akt survival pathway. Various studies have demonstrated links between these phosphatases and the Pi3-kinase/Akt pathway. For example the calcium activated PP2B has been shown to be a direct phosphatase of Akt, Gsk3β, and Bad (Millward et al. 1999; Klumpp et al. 2003). PP1α has been shown to dephosphorylate Akt and Bad, while PP2A has been demonstrated to be a key Akt phosphatase. PP2A can dephosphorylate Akt on both threonine 308 and serine 473 blocking the Pi3-kinase/Akt pathway. Immunoprecipitation studies have shown that PP2A can co-localize with Akt. Recently a novel phosphatase belonging to the PP2C family of phosphatases has been shown to dephosphorylate Akt on Ser473 only. This novel phosphatase (PHLPP) contains a PH domain that localizes it in the vicinity of activated Akt. Expression of PHLPP in cells derived from a small cell lung cancer that have constitutively active Akt (phosphorylated on Ser473 and Threo409), led to a 50% decrease in phosphoserine-473 Akt levels. This resulted in a correlating decrease in the phosphorylation levels of an Akt substrate, and in increase in the basal levels of apoptosis in this cell line (Gao et al. 2005; Vandermoere et al. 2005). These experiments highlight the importance of the phosphorylated serine residue in Akt signaling and demonstrate a clear link between the increased activity of PHLPP with an induction of apoptosis via the PI3-kinase/Akt pathway.
Inactivation of PP2A has been shown in cells treated with Tumor Necrosis Factor-alpha (TNF-α) or interleukin-1 (Guy et al. 1995) which can both induce H2O2 production. As described for the PTPs and DSP, the Ser/Thr phosphatases are thought to be sensitive to redox modifications. It has been demonstrated in vitro that hydrogen peroxide can reversibly block some of the main Ser/Thr phosphatases like PP2A, and PP1α (Rao and Clayton 2002; O’Loghlen et al. 2003). PP1 and PP2A contain redox-sensitive Cys residues (Fetrow et al. 1999; Guy et al. 1995). Structure based analysis has identified a potential disulfide oxidoreductase active site, Cys-X-X-Cys, in members of the PP1 subfamily (Fetrow et al. 1999). It is still not clear whether the oxidation of this pair of cysteine residues can result in PP1 inactivation in vivo.
3.3.2.2 Caspases
Under certain conditions ROS may directly affect the activity of cell death effector proteins. Several effectors of apoptosis are redox sensitive and their functions can be directly modulated by intracellular ROS, those effectors are caspases, Bcl-2 and cytochrome c.
A hallmark of apoptosis is the activation of caspases which requires sequential proteolysis of the initiator caspases and effector caspases. Stimuli that induce apoptosis can trigger caspase activation either through the extrinsic (death receptor-mediated activation) or the intrinsic (mitochondria mediated activation) pathways. Activation of the caspase cascade ultimately leads to the cleavage of different target proteins such as poly(ADP-ribose) polymerase (PARP) and α-fodrin leading to cell death. ROS can directly affect functions of caspases; the reduced state of the cysteine in the active site is necessary for the catalytic activity of caspases, thus depending on the degree of intracellular oxidative stress caspases can be activated or inhibited. Using different concentration of exogenous H2O2 Hampton et al. demonstrated that a low dose of H2O2 can activate caspases and induce apoptosis, but high dose of H2O2 inhibits caspases and cells undergo necrosis (Hampton and Orrenius 1997).
3.3.2.3 Transcription Factors
Another way for the cell to regulate cell survival through protein oxidation is through activation of transcription factors. Different transcription factors are known to be redox sensitive; these include p53, HIF, AP-1 and NF-κB.
The heterodimeric protein NF-κB is a ubiquitous redox-regulated transcription factor that remains in the cytoplasm as an inactive complex with its inhibitory counterpart IκBα. Exposure to oxidative stimuli leads to phosphorylation and subsequent proteasomal degradation of IκBα, thereby releasing free NF-κB dimers for translocation to the nucleus. Experimental evidence suggests that ROS seem to have paradoxical effects on NF-κB regulation. ROS can either activate or inhibit NF-κB activity depending on the ROS levels, type of stimuli and cell types. Moderate level of ROS generally leads to NF-κB activation. On the contrary high level of ROS could inactivate NF-κB leading to cell death. In the nucleus, direct oxidation of the redox-sensitive Cys62 of the p50 subunit inhibits its availability to bind DNA (Toledano and Leonard 1991). This oxidation is reversible and DNA binding can be restored. Besides direct structural modifications, DNA binding activity of NF-κB can be modulated by chromatin remodelling (Rahman et al. 2004). Thus, the enzyme histone deacetylase (HDAC), which catalyses the removal of an acetyl group from histone can be inactivated by oxidative stress allowing histone acetylation, chromatin uncoiling and increased accessibility for NF-κB (Rahman et al. 2002). In the cytosol, NF-κB is sequestrated as a complex formed with its inhibitor IκB, its activation is then regulated by phosphorylation of NF-κB itself or phosphorylation of its inhibitor. Under certain condition H2O2 is able to directly activate NF-κB activity through phosphorylation of IκB-kinase (IκB) (Kamata et al. 2002) or indirectly through activation of Akt and/or MEKK1 which then phosphorylates and activates IκB. The transactivation of NF-κB induced by Akt or MEKK1 has been shown really important in NF-κB anti-apoptotic effects (Nawata et al. 2003; Vandermoere et al. 2005). Active IκB phosphorylates IκB and liberates active NF-κB from the complex to translocate to the nucleus. Phosphorylated IκB is then degraded by the proteasome. Since the proteasome system is also redox-sensitive, ROS can regulate NF-κB activity by affecting IκB stability.
3.3.3 Carbonylation/Nitrosylation of Proteins
As described above some ROS-induced protein modifications are benign events and can even promote cell survival. However, irreversible modifications can also result in unfolding or alteration of protein structure leading to inactivation of various proteins.
Carbonylation is an irreversible, non enzymatic modification of proteins. The chemistry of the reaction that gives rise to carbonyl groups is well described in reviews by Stadtman and coworkers. Lysine, arginine, proline and threonine residues of proteins are particularly sensitive to metal-catalysed oxidation leading in each case to the formation of carbonyl derivatives (Dalle-Donne et al. 2006). Briefly there are four different oxidative pathways by which carbonyl groups are introduced into proteins: (a) direct oxidation of Lys, Arg, Pro and Thr residues’ side chain especially via metal-catalyzed oxidation; (b) oxidative cleavage of the protein backbone by the α-amidation pathway or by oxidation of glutamyl residues; (c) adduction of reactive aldehyde derived from the metal-catalyzed oxidation of polyunsaturated fatty acids (Lys, His and Cys are the residues reacting preferentially with the lipoxidation products) (Refsgaard et al. 2000); (d) reaction with reactive carbonyl derivatives generated as a consequence of the reaction of reducing sugars or their oxidation products with lysine residue of proteins.
The chemical modification of protein by reactive carbonyl compounds derived from lipid peroxidation reactions results in the formation of advanced lipoxidation end-products (ALEs) while the one derived from sugar oxidation results in advanced glycation-products (AGEs). Whereas moderately carbonylated proteins are degraded by the proteasome, heavily carbonylated proteins tend to form aggregates that are resistant to degradation and accumulate as damaged or unfolded proteins; those aggregates can even lead to an inactivation of the proteasome. The presence of carbonyl groups in proteins is used as a marker of ROS-mediated protein oxidation. Considering the presence of carbonyl groups it has been established that protein oxidation is associated with aging, oxidative stress and a number of diseases; the identification of specific carbonylated proteins should then provide new diagnostic tools for human diseases (Dalle-Donne et al. 2003).
Nitric oxide (NO•) is generated from arginine by the action of nitric oxide synthase. It is an important signaling molecule playing a major role in physiological processes such as smooth muscle relaxation and neurotransmission. However, depending on the redox state of the cell NO• can also induce oxidative stress even though it can not be categorized as a classical ROS it is part of the reactive nitrogen species (RNS) (for a review see Moncada and Erusalimsky 2002). NO• reacts with superoxide to give peroxynitrite (ONOO−) which under physiological conditions can react with CO2 to form nitrosoperoxocarboxylate (NPC or ONOOCO −2 ); both those RNS are particularly harmful for the cell.
Cysteine and methionine residues are particularly sensitive to oxidation by peroxynitrite. Peroxynitrite is responsible for the oxidation of methionine residues to methionine sulfoxide and the nitration of protein sulfhydryl groups to form S-nitrosothiol derivatives (Leseney et al. 1999). The covalent attachment of a nitrogen monoxide group to the thiol side chain of cysteine is called an S-nitrosylation and has emerged as an important post-transcriptional modification of signaling proteins. Both tyrosine and tryptophane residues are selective targets for nitrosoperoxocarboxylate (NPC)-dependent nitration. The nitration of tyrosine residue is particularly important because nitration prevents tyrosine residue to undergo cyclic interconversion between phosphorylated and unphosphorylated forms (Hunter 1995); it is an irreversible process thus locking the targeted enzyme into an inactive configuration. Accordingly transduction signals involving a phospho-tyrosine (such as interferon-α signaling) could be inactivated by nitration of the same tyrosine.
3.4 Protein Oxidation and Disease
Reactive oxygen species such as H2O2, O •−2 and hydroxyl radicals (HO•) are all biologically relevant oxygen radicals which are routinely produced as normal by-products of many different metabolic processes, and small amounts of ROS are needed by cells to act as signaling molecules (Halliwell and Gutteridge 1999; Hensley et al. 2000). However ROS have the potential to induce significant biological damage. If the sensitive redox status of a cell is perturbed in some way this can result in an imbalance in the delicate intracellular oxidant/antioxidant status and can lead to accumulation of ROS and ultimately to oxidative stress-induced injury and cell death. It is the increased concentration and the body’s inability to effectively deal with these ROS that can often lead to the oxidation of various biochemicals, such as lipids, nucleic acids, sugars and proteins.
It is still remains relatively unclear whether oxidative stress is actually the cause of or the consequence of many human pathologies; nevertheless it is now widely accepted that aberrant oxidative stress is intricately linked to the progression of specific diseases such as Alzheimer’s and Parkinson’s. ROS can directly affect proteins by oxidation of both the backbone and amino acid side chains, and can also interact with other biomolecules such as sugars and lipids generating products that can then go on to also interact with proteins. Oxidative modification of proteins can have many detrimental biological affects and as such this post-translational modification often has a role to play in the pathogenesis several diseases.
As protein activity and function are both tightly regulated by protein conformation, oxidative damage of proteins can in some cases lead to abnormal formation of aggregated cross-linked protein which may be resistant to proteinase degradation and thus lead to accumulation of such aggregates (Butterfield and Stadtman 1997). In some cases oxidation can convert proteins into forms that are more susceptible to degradation by proteases (Stadtman 1990). This interference in normal protein turnover and loss of protein and enzyme catalytic function can ultimately impede the normal functioning of a cell and lead to pathological alterations.
Increased levels of oxidised proteins have indeed been associated with a number of diseases including Alzheimer’s disease (AD) (Smith et al. 1991; Markesbery and Lovell 2007), muscular dystrophy (Murphy and Kehrer 1989), cataractogenesis (Davies and Truscott 2001), rheumatoid arthritis (Hitchon and El-Gabalawy 2004), respiratory distress syndrome (Lamb et al. 1999) and progeria (Hutchinson-Gilford progeria syndrome) (Oliver et al. 1987). Although not directly implicated there is also substantial evidence to suggest that oxidatively modified proteins are implicated in the pathogenesis of cystic fibrosis (CF) (Salh et al. 1989; Starosta et al. 2006), Parkinson’s disease (PD) (Danielson and Andersen 2008), diabetes (Cakatay 2005), atherosclerosis (Brennan and Hazen 2005), essential hypertension (Puddu et al. 2008), diabetes (Kubisch et al. 1994) and ulcerative colitis (Babbs 1992).
AD is the most prevalent dementia in the elderly population. It affects approximately 16 million people worldwide and beyond the age of 65 the incidence doubles every 5 years (Goedert and Spillantini 2006). AD brains show evidence of ROS-mediated injury (Praticò and Sung 2004) and there is an increasing body of evidence indicating protein oxidation as one of the main contributors towards Alzheimer’s associated neurofibrillary degeneration and concurrent cognitive deterioration (Butterfield and Stadtman 1997; Aksenov et al. 2001).
The irreversible formation and accumulation of protein carbonyls is very often a strong indicator of severe oxidative protein damage and protein carbonylation is thus the most commonly used marker for protein oxidation (Dalle-Donne et al. 2003). The production of protein carbonyls can arise via the direct oxidation of amino-acid side chains and through oxidative cleavage of proteins. Carbonyl groups can also be added to proteins via reactions of unsaturated aldehydes which are derived from lipid peroxidation, and can also be introduced by addition of reactive carbonyl derivatives produced by the reaction of reducing sugars or their oxidation products (Dalle-Donne et al. 2003).
Carbonylation of proteins often leads to loss of protein function and accumulation of proteolysis-resistant aggregates of carbonylated proteins in tissues has been noted for a large number of neurodegenerative diseases (Dalle-Donne et al. 2006). When ROS attack protein side-chains it can cause specific chemical alterations such as the formation of hydroxyl and carbonyl groups as already discussed. Such changes in turn can lead to loss of protein function, and the identification of specific functionally impaired carbonylated proteins is just one hallmark of the AD brain (Stadtman 1990). This oxidative modification of proteins is pertinent to the pathogenesis of AD and it is these protein carbonyls that have been detected in significantly increased amounts in the AD brain (Markesbery and Carney 1999; Starosta et al. 2006).
Parkinson disease (PD) is the second most common neurodegenerative disorder (Farrer 2006). PD is a severe and debilitating neurodegenerative syndrome caused by various factors including genetic susceptibility, the ageing process and various environmental factors (Danielson and Andersen 2008). There is no cure as yet and the disease affects around 1–2% of the population over 50 years old (Thomas and Beal 2007). The exact molecular pathways involved in the pathological progression of this disease are still obscure. There is substantial evidence however that increased oxidative stress and decreased levels of antioxidants (Pearce et al. 1997) appear to be common underlying factors involved in the loss of dopaminergic neurons from the substantia nigra (SN), the region of the brain most prominently affected by the disease (Farrer 2006). As is the case with Alzheimer’s increased levels of oxidized protein carbonyls have also been detected in the brain of PD sufferers (Alam et al. 1997) and in a more recent study it was demonstrated that antioxidants efficiently prevented protein carbonylate formation (Esteves et al. 2009) suggesting this as a potential treatment.
There are also a number of specific proteins that have been found to be post-translationally modified via oxidation in the brains of PD patients, for example the protein DJ-1 is thought to function as an antioxidant (Taira et al. 2004) and mass spectrometric data have shown increased amounts of methionine oxidized DJ-1 in the brains of patients with sporadic PD thus suggesting a role of methionine oxidation of this particular protein in disease pathogenesis (Choi et al. 2006). Although it is unclear as to whether oxidative stress and subsequent oxidative modification of proteins play a primary role in the initiation of PD or whether it is simply a secondary effect due to the disease progression, the fact that several oxidised proteins have been described in PD brains would suggest that these modifications seem to be highly relevant to and contributors towards PD (Danielson and Andersen 2008).
Not only does protein oxidation seem to play a significant role in the aforementioned neurodegenerative disorders but also in the more common condition of cataractogenesis. Cataract development in the ageing population affects more than 20 million people worldwide and is the leading cause of blindness in developing countries (West 2007). Cataracts form when the lens of the eye becomes opaque and it is believed that oxidative stress is an initiating factor in the development of age-related cataracts (Spector 1995). The protein redox status seems to be crucial to maintain proper function of the lens and lens transparency, and, when this delicate balance is upset it can lead to development of cataracts. In the lens, one particular group of proteins, crystalins, constitute around 90% of the total protein content and are susceptible to age-related oxidative changes (Boscia et al. 2000), including disulfide bond formation, inter- and intramolecular cross-linking, and methionine oxidation, all of which result in production and accumulation of high molecular weight aggregates (Siew et al. 1981). Generally any protein that has been abnormally modified will be degraded via proteasomal degradation (Jung and Grune 2008). However, in the case of cataractogenesis these oxidatively modified proteins are not recognized by the host, are not degraded and thus accumulate in the lens over time causing the formation of cataracts (Spector 1995; Williams 2006).
Cystic fibrosis (CF) is a condition in which protein oxidation is believed to contribute towards disease progression. CF is a chronic, progressive, genetic disease (Schidlow 2000) involving persistent inflammation and continuous periods of infection leading to progressive damage of the lungs and pulmonary fibrosis which is eventually responsible for over 90% of the mortality linked with this disease (Davis 1993). There is now evidence to support the hypothesis that free radical protein damage is involved in the pathogenesis of the disease. In a study by Starosta et al. (2006) the level of oxidative stress was measured in CF patients by assessing protein oxidation from the content of protein carbonyls in their bronchoalveolar fluid. It was found that CF patients had significantly higher levels of protein carbonyls in their bronchoalveolar fluid when compared to healthy test subjects, these data thus supported the long held hypothesis that an over abundance of ROS may be one of the major contributing factors towards the gradual destructive pulmonary damage seen in CF patients (Starosta et al. 2006).
It is widely accepted that protein oxidation has a major role to play in the normal process of aging (Chakravarti and Chakravarti 2007) so it is interesting to note that the premature aging disease progeria is also characterized by increased levels of protein oxidation in the form of protein carbonyls in sufferers of the disease (Oliver et al. 1987). Progeria is a rare autosomal-dominant disorder characterized by the appearance of premature signs of aging (Hennekam 2006; Kieran et al. 2007). In a study by Oliver et al. (1987) the levels of oxidized proteins in cultured human fibroblasts from healthy individuals and from progeria sufferers were determined by measuring carbonyl content. It was discovered that the carbonyl content of the progeria samples was significantly higher than that of the age-matched controls; in fact the levels were comparable to those found in cultured fibroblasts from 80-year-old subjects (Oliver et al. 1987).
Generally the intracellular level of an oxidized protein is determined by the balance between the rate of protein oxidation and the rate at which the oxidized protein is degraded. The two processes, protein oxidation and degradation of oxidized proteins, are multifactorial activities controlled by a number of different events including the production of ROS, the intracellular redox potential and the availability and concentration of antioxidants: It is the fine balance between these myriad of factors that determines the steady state level of oxidized protein in cells (Stadtman and Berlett 1997). As previously stated there is now sufficient evidence to demonstrate that protein oxidation is most likely a key contributor to the dysfunction associated with the aforementioned pathologies although it is important to bear in mind that a definitive causal relationship between protein oxidation and the etiology or progression of the aforementioned diseases has not been thoroughly established. Nevertheless in the cases of neurodegenerative diseases such as AD there is a definite positive correlation between increased amounts of oxidized proteins in the brain of sufferers and increased progression of the disease (Forster et al. 1996; Carney et al. 1991).
3.5 Concluding Remarks
As previously discussed there are many sources of ROS in a biological system, the two primary sources being mitochondria and the Nox proteins. Protein oxidation seems to play a vital role in many different biological functions; the reversible oxidation of proteins which usually takes place under normal physiological conditions allows various signaling pathways to operate, however, during conditions of oxidative stress the irreversible oxidation of proteins is thought to play a significant role in the etiology and or progression of certain diseases. The challenges that lie ahead are to understand the mechanisms that drive disease-associated oxidative stress, to elucidate diagnostic biomarkers for oxidative damage during disease and develop effective therapeutic options such as antioxidant treatments.
Abbreviations
- AD:
-
Alzheimer’s disease
- CF:
-
Cystic fibrosis
- COX:
-
Cyclooxygenase
- DUOX:
-
Dual oxidase
- HO• :
-
Hydroxyl radical
- H2O2 :
-
Hydrogen peroxide
- LOX:
-
Lipoxygenase
- MPO:
-
Myeloperoxidase
- NO• :
-
Nitric oxide
- NOS:
-
Nitric oxide synthase
- Nox:
-
NADPH Oxidase
- O •−2 :
-
Superoxide
- PD:
-
Parkinson’s disease
- RNS:
-
Reactive nitrogen species
- ROS:
-
Reactive oxygen species
- SOD:
-
Superoxide dismutase
- Trx2:
-
Thioredoxin2
- XDH:
-
Xanthine dehydrogenase
- XO:
-
Xanthine oxidase
- XOR:
-
Xanthine oxidoreductase
References
Aksenov, M.Y., Aksenova, M.V., Butterfield, D.A., et al. (2001). Protein oxidation in the brain in Alzheimer’s disease. Neuroscience 103:373–383.
Alam, Z.I., Daniel, S.E., Lees, A.J., et al. (1997). Generalised increase in protein carbonyls in the brain in Parkinson’s but not incidental Lewy body disease. J. Neurochem. 69:1326–1329.
Babbs, C.F. (1992). Oxygen radicals in ulcerative colitis. Free Radical Biol. Med. 13:169-181.
Banfi, B., Molnar, G., Maturana, A., et al. (2001). A Ca2+-activated NADPH oxidase in testis, spleen, and lymph nodes. J. Biol. Chem. 276:37594–37601.
Banfi, B., Malgrange, B., Knisz, J., et al. (2004). NOX3, a superoxide-generating NADPH oxidase of the inner ear. J. Biol. Chem. 279:46065–46072.
Banfi, B., Tirone, F., Durussel, I., et al. (2004). Mechanism of Ca2+ activation of the NADPH oxidase 5 (NOX5). J. Biol. Chem. 279:18583–18591.
Barja, G., and Herrero, A. (1998). Localization at complex I and mechanism of the higher free radical production of brain nonsynaptic mitochondria in the short-lived rat than in the longevous pigeon. J. Bioenerg. Biomembr. 30:235–243.
Barrett, W.C., DeGnore, J.P., Konig, S., et al. (1999). Regulation of PTP1B via glutathionylation of the active site cysteine 215. Biochemistry 38:6699–6705.
Bedard, K., and Krause, K.H. (2007). The NOX family of ROS-generating NADPH oxidases: Physiology and pathophysiology. Physiol. Rev. 87:245–313.
Boscia, F., Grattagliano, I., Vendemiale G., et al. (2000). Protein oxidation and lens opacity in humans. Invest. Ophthalmol. Vis. Sci. 41:2461–2465.
Boueiz, A., Damarla, M., and Hassoun, P.M. (2008). Xanthine oxidoreductase in respiratory and cardiovascular disorders. Am. J. Physiol. Lung Cell. Mol. Physiol. 294:L830–L840.
Brennan, M.L., and Hazen, S.L. (2005). Amino acid and protein oxidation in cardiovascular disease. Amino Acids 25:365–374.
Butler, J., Jayson, G.G., and Swallow, A.J. (1975). Reaction between superoxide anion radical and cytochrome C. Biochim. Biophys. Acta 408:215–222.
Butterfield, D.A., and Stadtman, E.R. (1997). Protein oxidation processes in aging brain. Adv. Cell Aging Gerontol. 2:161–191.
Cakatay, U. (2005). Protein oxidation parameters in type 2 diabetic patients with good and poor glycaemic control. Diabetes Metab. 31:551–557.
Carney, J. M., Starke-Reed, P. E., Oliver, C. N., et al. (1991) Reversal of age-related increase in brain protein oxidation, decrease in enzyme activity loss and loss of temporal and spatial memory by chronic administration of the spin-trapping compound N-tert-butyl-alpha-phenylnitrone. Proc. Natl. Acad. Sci. U.S.A. 88:3633–3636.
Caselli, A., Marzocchini, R., Camici, G., et al. (1998). The inactivation mechanism of low molecular weight phosphotyrosine-protein phosphatase by H2O2. J. Biol. Chem. 273:32554–32560.
Chakravarti, B., and Chakravarti, D.N. (2007). Oxidative modification of proteins: age-related changes. Gerontology 53:128–139.
Chen, Y., Cai, J.Y., Murphy, T.J., et al. (2002). Overexpressed human mitochondrial thioredoxin confers resistance to oxidant-induced apoptosis in human osteosarcoma cells. J. Biol. Chem. 277:33242–33248.
Cheng, G.J., Cao, Z.H., Xu, X.X., et al. (2001). Homologs of gp91phox: cloning and tissue expression of Nox3, Nox4, and Nox5. Gene 269:131–140.
Chiarugi, P., Fiaschi, T., Taddei, M.L., et al. (2001). Two vicinal cysteines confer a peculiar redox regulation to low molecular weight protein tyrosine phosphatase in response to platelet-derived growth factor receptor stimulation. J. Biol. Chem. 276:33478–33487.
Choi, J., Sullards, M.C., Olzmann, J.A., et al. (2006). Oxidative damage of DJ-1 is linked to sporadic Parkinson and Alzheimer diseases. J. Biol. Chem. 281:10816–10824.
Chung, H.Y., Baek, B.S., Song, S.H., et al. (1997). Xanthine dehydrogenase, xanthine oxidase and oxidative stress. Age 20:127–140.
Dalle-Donne, I., Giustarini, D., Colombo, R., et al. (2003). Protein carbonylation in human diseases. Trends Mol. Med. 9:169–176.
Dalle-Donne, I., Aldini, G., Carini, M., et al. (2006). Protein carbonylation, cellular dysfunction, and disease progression. J. Cell. Mol. Med. 10:389–406.
Danielson, S.R., and Andersen, J.K. (2008). Oxidative and nitrative protein modifications in Parkinson’s disease. Free Radical Biol. Med. 44:1787–1794.
Davies, M.J., and Truscott, R.J. (2001). Photo-oxidation of proteins and its role in cataractogenesis. J. Photochem. Photobiol. B 63:114–125.
Davis, P.B.O. Pathophysiology of the lung disease in cystic fibrosis. In: Davis P.B., ed. Cystic fibrosis. New York: Marcel Dekker, 1993, pp. 193–218.
Denu, J.M., and Dixon, J.E. (1998). Protein tyrosine phosphatases: mechanisms of catalysis and regulation. Curr. Opin. Chem. Biol. 2:633–641.
Donko, A., Peterfi, Z., Sum, A., et al. (2005). Dual oxidases. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 360:2301–2308.
Dupuy, C., Ohayon, R., Valent, A., et al. (1999). Purification of a novel flavoprotein involved in the thyroid NADPH oxidase. Cloning of the porcine and human cDNAs. J. Biol. Chem. 274:37265–37269.
Edderkaoui, M., Hong, P., Vaquero, E.C., et al. (2005). Extracellular matrix stimulates reactive oxygen species production and increases pancreatic cancer cell survival through 5-lipoxygenase and NADPH oxidase. Am. J. Physiol. Gastrointest. Liver Physiol. 289:G1137–G1147.
Edens, W.A., Sharling, L., Cheng, G.J., et al. (2001). Tyrosine cross-linking of extracellular matrix is catalyzed by Duox, a multidomain oxidase/peroxidase with homology to the phagocyte oxidase subunit gp91 phox. J. Cell Biol. 154:879–891.
Esteves, A.R., Arduíno, D.M., Swerdlow, R.H., et al. (2009). Oxidative stress involvement in α-synuclein oligomerization in Parkinson disease cybrids. Antioxid. Redox Signal 11:439–448.
Farrer, M.J. (2006). Genetics of Parkinson disease: paradigm shifts and future prospects. Nat. Rev. Genet. 7:306–318.
Fetrow, J.S., Siew, N., Skolnick, J. (1999). Structure-based functional motif identifies a potential disulfide oxidoreductase active site in the serine/threonine protein phosphatase-1 subfamily. FASEB J. 13:1866–1874.
Fleury, C., Mignotte, B., and Vayssiere, J.L. (2002). Mitochondrial reactive oxygen species in cell death signaling. Biochimie 84:131–141.
Forster, M. J., Dubey, A., Dawson, K. M., et al. (1996). Age-related losses of cognitive function and motor skills in mice are associated with oxidative protein damage in the brain. Proc. Natl. Acad. Sci. U.S.A. 93:4765–4769.
Gao, T., Furnari, F., and Newton, A.C. (2005). PHLPP: a phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Mol. Cell 18:13–24.
Geiszt, M., Kopp, J.B., Varnai, P., et al. (2000). Identification of renox, an NAD(P)H oxidase in kidney. Proc. Natl. Acad. Sci. U.S.A. 97:8010–8014.
Geiszt, M., Witta, J., Baffi, J., et al. (2003). Dual oxidases represent novel hydrogen peroxide sources supporting mucosal surface host defense. FASEB J. 17:1502–1504.
Go, Y.M., and Jones, D.P. (2008). Redox compartmentalization in eukaryotic cells. Biochim. Biophys. Acta 1780:1271–1290.
Goedert, M., and Spillantini, M.G. (2006). A century of Alzheimer’s disease. Science 314:777–781.
Guidot, D.M., Repine, J.E., Kitlowski, A.D., et al. (1995). Mitochondrial respiration scavenges extramitochondrial superoxide anion via a nonenzymatic mechanism. J. Clin. Invest. 96:1131–1136.
Guy, G.R., Philp, R., and Tan, Y.H. (1995). Activation of protein kinases and the inactivation of protein phosphatase 2A in tumour necrosis factor and interleukin-1 signal-transduction pathways. Eur. J. Biochem. 229:503–511.
Halliwell, B., and Gutteridge, J. (1999). Free radicals in biology and medicine, 3rd ed. Oxford: Oxford University Press.
Hampton, M.B., and Orrenius, S. (1997). Dual regulation of caspase activity by hydrogen peroxide: implications for apoptosis. FEBS Lett. 414:552–556.
Hennekam, R.C. (2006). Hutchinson-Gilford progeria syndrome: review of the phenotype. Am. J. Med. Genet. A, 140:2603–2624.
Hensley, K., Robinson, K.A., Gabbita, S.P., et al. (2000). Reactive oxygen species, cell signaling, and cell injury. Free Radical Biol. Med. 28:1456–1462.
Hitchon, C.A., and El-Gabalawy, H.S. (2004).Oxidation in rheumatoid arthritis. Arthritis Res. Ther. 6:265–278.
Hunter, T. (1995) Protein kinases and phosphatases: The yin and yang of protein phosphorylation and signalling. Cell 80:225–236.
Inanami, O., Johnson, J.L., McAdara, J.K., et al. (1998). Activation of the leukocyte NADPH oxidase by phorbol ester requires the phosphorylation of p47(PHOX) on serine 303 or 304.J. Biol. Chem. 273:9539–9543.
Iyer, G.Y., Islam, M.F., and Quastel, J.H. (1961). Biochemical aspects of phagocytosis. Nature 192:535–542.
Jung T., and Grune T.T. (2008). The proteasome and its role in the degeneration of oxidised proteins. IUBMB Life 60:743–752.
Kakkar, P., and Singh, B.K. (2007). Mitochondria: a hub of redox activities and cellular distress control. Mol. Cell. Biochem. 305:235–253.
Kamata, H., Manabe, T., Oka, S., et al. (2002). Hydrogen peroxide activates IkappaB kinases through phosphorylation of serine residues in the activation loops. FEBS Lett. 519:231–237.
Kawahara, T., Ritsick, D., Cheng, G.J., et al. (2005). Point mutations in the proline-rich region of p22(phox) are dominant inhibitors of Nox1- and Nox2-dependent reactive oxygen generation. J. Biol. Chem. 280:31859–31869.
Kieran, M.W., Gordon, L., and Kleinman, M. (2007). New approaches to Progeria. Pediatrics 120:834–841.
Kim, C., Kim, J.Y., and Kim, J.H. (2008). Cytosolic phospholipase A(2), lipoxygenase metabolites, and reactive oxygen species. BMB Rep. 41:555–559.
Klebanoff, S.J. (2005). Myeloperoxidase: friend and foe. J. Leukoc. Biol. 77:598–625.
Klumpp, S., Selke, D., and Krieglstein, J. (2003). Protein phosphatase type 2C dephosphorylates BAD. Neurochem. Int. 42:555–560.
Kubisch, H. M., Wang, J., Luche, R., et al. (1994). Transgenetic copper/zinc superoxide dismutase modulates susceptibility to type 1 diabetes. Proc. Natl. Acad. Sci. U.S.A. 91:9956–9959.
Kuhn, H., and Thiele, B.J. (1999). The diversity of the lipoxygenase family: Many sequence data but little information on biological significance. FEBS Lett. 449:7–11.
Lamb, N.J., Gutteridge, J.M., Baker, C., et al. (1999). Oxidative damage to proteins of bronchoalveolar lavage fluid in patients with acute respiratory distress syndrome: Evidence for neutrophil-mediated hydroxylation, nitration, and chlorination. Critical Care Med. 27:1738–1744.
Lambeth, J.D., Kawahara, T., and Diebold, B. (2007). Regulation of Nox and Duox enzymatic activity and expression. Free Radical Biol. Med. 43:319–331.
Lee, S.R., Kwon, K.S., Kim, S.R., et al. (1998). Reversible inactivation of protein-tyrosine phosphatase 1B in A431 cells stimulated with epidermal growth factor. J. Biol. Chem. 273:15366–15372.
Leseney, A.M., Deme, D., Legue, O., et al. (1999). Biochemical characterization of a Ca2+/NAD(P)H-dependent H2O2 generator in human thyroid tissue. Biochimie 81:373–380.
Levine, R.L., Mosoni, L., Berlett, B.S., et al. (1996). Methionine residues as endogenous antioxidants in proteins. Proc. Natl. Acad. Sci. U.S.A. 93:15036–15040.
Mackey, A.M., Sanvicens, N., Groeger, G., et al. (2008). Redox survival signalling in retina-derived 661W cells. Cell Death Differ. 15:1291–1303.
Mahadev, K., Zilbering, A., Zhu, L., et al. (2001). Insulin-stimulated hydrogen peroxide reversibly inhibits protein-tyrosine phosphatase 1B in vivo and enhances the early insulin action cascade. J. Biol. Chem. 276:21938–21942.
Mahadev, K., Motoshima, H., Wu, X.D., et al. (2004). The NAD(P)H oxidase homolog Nox4 modulates insulin-stimulated generation of H2O2 and plays an integral role in insulin signal transduction. Mol. Cell. Biol. 24:1844–1854.
Malle, E., Furtmuller, P.G., Sattler, W., et al. (2007). Myeloperoxidase: a target for new drug development? Br. J. Pharmacol. 152:838–854.
Mancuso, M., Coppede, F., Migliore, L., et al. (2006). Mitochondrial dysfunction, oxidative stress and neurodegeneration. J. Alzheimer’s Dis. 10:59–73.
Markesbery, W.R., and Carney, J.M. (1999). Oxidative alterations in Alzheimer’s disease. Brain Pathol. 9:133–146.
Markesbery, W.R., and Lovell, M.A. (2007). Damage to lipids, proteins, DNA, and RNA in mild cognitive impairment. Arch. Neurol. 64:954–956.
Meng, T.C., Fukada, T., and Tonks, N.K. (2002). Reversible oxidation and inactivation of protein tyrosine phosphatases in vivo. Mol. Cell 9:387–399.
Millward, T.A., Zolnierowicz, S., and Hemmings, B.A. (1999). Regulation of protein kinase cascades by protein phosphatase 2A. Trends Biochem. Sci. 24:186–191.
Miyasaki, K.T., Song, J.P., and Murthy, A.R.K. (1991). Secretion of myeloperoxidase isoforms by human neutrophils. Anal. Biochem. 193:38–44.
Moncada, S., and Erusalimsky, J.D. (2002). Does nitric oxide modulate mitochondrial energy generation and apoptosis? Nat. Rev. Mol. Cell Biol. 3:214–220.
Murphy, M.E., and Kehrer, J.P. (1989). Oxidative stress and muscular dystrophy. Chem. Biol. Interact. 69:101–173.
Nawata, R., Yujiri, T., Nakamura, Y., et al. (2003). MEK kinase 1 mediates the antiapoptotic effect of the Bcr-Abl oncogene through NF-kappaB activation. Oncogene 22:7774–7780.
Nishino, T., Okamoto, K., Eger, B.T., et al. (2008). Mammalian xanthine oxidoreductase. Mechanism of transition from xanthine dehydrogenase to xanthine oxidase. FEBS J. 275:3278–3289.
Okado-Matsumoto, A., and Fridovich, I. (2001). Subcellular distribution of superoxide dismutases (SOD) in rat liver -Cu,Zn-SOD- in mitochondria. J. Biol. Chem. 276:38388–38393.
Oliver, C. N., Ahn, B.W., Moerman, E. J., Goldstein, S., and Stadtman, E. R. (1987). Age-related changes in oxidized proteins. J. Biol. Chem. 262:5488–5491.
O’Loghlen, A., Perez-Morgado, M.I., Salinas, M. et al. (2003). Reversible inhibition of the protein phosphatase 1 by hydrogen peroxide. Potential regulation of eIF2 alpha phosphorylation in differentiated PC12 cells. Arch. Biochem. Biophys. 417:194–202.
Pearce, R.K., Owen, A., Daniel, S., et al. (1997). Alterations in the distribution of glutathione in the substantia nigra in Parkinson’s disease. J. Neural Transm. 104:661–677.
Praticò, D., and Sung, S. (2004). Lipid peroxidation and oxidative imbalance: early functional event in Alzheimer’s disease. J. Alzheimers Dis. 6:171–175.
Puddu, P., Puddu, G.M., Cravero, E., et al. (2008). The molecular sources of reactive oxygen species in hypertension. Blood Press.17:70–77.
Rahman, I., Gilmour, P.S., Jimenez, L.A. et al. (2002). Oxidative stress and TNF-alpha induce histone acetylation and NF-kappaB/AP-1 activation in alveolar epithelial cells: potential mechanism in gene transcription in lung inflammation. Mol. Cell. Biochem. 234–235:239–248.
Rahman, I., Marwick, J., and Kirkham, P. (2004). Redox modulation of chromatin remodeling: impact on histone acetylation and deacetylation, NF-kappaB and pro-inflammatory gene expression. Biochem. Pharmacol. 68:1255–1267.
Rao, R.K., and Clayton, L.W. (2002). Regulation of protein phosphatase 2A by hydrogen peroxide and glutathionylation. Biochem. Biophys. Res. Commun. 293:610–616.
Refsgaard, H.H., Tsai, L., Stadtman, E.R. (2000). Modifications of proteins by polyunsaturated fatty acid peroxidation products. Proc. Natl. Acad. Sci. U.S.A. 97:611–616.
Royerpokora, B., Kunkel, L.M., Monaco, A.P., et al. (1986). Cloning the gene for an inherited human disorder -Chronic Granulomatous-Disease- on the basis of its chromosomal location. Nature 322:32–38.
Sadok, A., Bourgarel-Rey, V., Gattacceca, F., et al. (2008). Nox1-dependent superoxide production controls colon adenocarcinoma cell migration. Biochim. Biophys. Acta 1783:23–33.
Salh, B., Webb, K., Guyan, P.M., et al. (1989). Aberrant free radical activity in cystic fibrosis. Clin. Chim. Acta 181:65–74.
Salmeen, A., and Barford, D. (2005). Functions and mechanisms of redox regulation of cysteine-based phosphatases. Antioxid. Redox Signal. 7:560–577.
Savitsky, P.A., and Finkel, T. (2002). Redox regulation of Cdc25C. J. Biol. Chem. 277:20535–20540.
Schidlow, D.V. (2000). Newer therapies for cystic fibrosis. Paediatr. Respir. Rev. 1:107–113.
Shiose, A., Kuroda, J., Tsuruya, K., et al. (2001). A novel superoxide-producing NAD(P)H oxidase in kidney. J. Biol. Chem. 276:1417–1423.
Siew, E.L., Opalecky, D., and Bettelheim, F.A. (1981). Light scattering of normal human lens. II. Age dependence of the light scattering parameters. Exp. Eye Res. 33:603–614.
Smith, C. D., Carney, J. M., Starke-Reed, P. E., et al. (1991). Excess brain protein oxidation and enzyme dysfunction in normal and Alzheimer’s disease. Proc. Natl. Acad. Sci. U.S.A. 88:10540–10543.
Spector, A. (1995). Oxidative stress-induced cataract: mechanism of action. FASEB J. 9:1173–1182.
Stadtman, E.R. (1990). Metal ion-catalyzed oxidation of proteins: biochemical mechanisms and biological consequences. Free Radical Biol. Med. 9:315–325.
Stadtman, E.R. (2001). Protein oxidation in aging and age-related diseases. Ann. New York Acad. Sci. 928:22–38.
Stadtman, E.R., and Berlett, B.S. (1997). Reactive oxygen-mediated protein oxidation in aging and disease. Chem. Res. Toxicol. 10:485–494.
Stadtman, E.R., and Levine, R.L. (2003). Free radical-mediated oxidation of free amino acids and amino acid residues in proteins. Amino Acids 25:207–218.
Starosta, V., Rietschel, E., Paul, K., et al. (2006). Oxidative changes of bronchoalveolar proteins in cystic fibrosis. Chest 129:431–437.
St-Pierre, J., Buckingham, J.A., Roebuck, S.J., et al. (2002). Topology of superoxide production from different sites in the mitochondrial electron transport chain. J. Biol. Chem. 277:44784–44790.
Suh, Y.A., Arnold, R.S., Lassegue, B., et al. (1999). Cell transformation by the superoxide-generating oxidase Mox1. Nature 401:79–82.
Suleyman, H., Demircan, B., and Karagoz, Y. (2007). Anti-inflammatory and side effects of cyclooxygenase inhibitors. Pharmacol. Rep. 59:247–258.
Sumimoto, H. (2008). Structure, regulation and evolution of Nox-family NADPH oxidases that produce reactive oxygen species. FEBS J. 275:3249–3277.
Taira, T., Saito, Y., Niki, T., et al. (2004). DJ-1 has a role in antioxidative stress to prevent cell death. EMBO Rep. 5:213–218.
Tanaka, T., Hosoi, F., Yamaguchi-Iwai, Y., et al. (2002). Thioredoxin-2 (TRX-2) is an essential gene regulating mitochondria-dependent apoptosis. EMBO J. 21:1695–1703.
Thomas, B., and Beal, M.F. (2007). Parkinson’s disease. Human. Mol. Genet. 16:183–194.
Toledano, M.B., and Leonard, W.J. (1991). Modulation of transcription factor NF-kappa B binding activity by oxidation-reduction in vitro. Proc. Natl. Acad. Sci. U.S.A. 88:4328–4332.
Turrens, J.F., and Boveris, A. (1980). Generation of superoxide anion by the NADH dehydrogenase of bovine heart-mitochondria. Biochem. J. 191:421–427.
Turrens, J.F., Alexandre, A., and Lehninger, A.L. (1985). Ubisemiquinone is the electron-donor for superoxide formation by Complex III of heart-mitochondria. Arch. Biochem. Biophys. 237:408–414.
Vandermoere, F., El Yazidi-Belkoura, I., Adriaenssens, E., et al. (2005). The antiapoptotic effect of fibroblast growth factor-2 is mediated through nuclear factor-kappaB activation induced via interaction between Akt and IkappaB kinase-beta in breast cancer cells. Oncogene 24:5482–5491.
Vaquero, E.C., Edderkaoui, M., Pandol, S.J., et al. (2004). Reactive oxygen species produced by NAD(P)H oxidase inhibit apoptosis in pancreatic cancer cells. J. Biol. Chem. 279:34643–34654.
Vasquez-Vivar, J., Kalyanaraman, B., Martasek, P., et al. (1998). Superoxide generation by endothelial nitric oxide synthase: the influence of cofactors. Proc. Natl. Acad. Sci. U.S.A. 95:9220–9225.
West, S. (2007). Epidemiology of cataract: accomplishments over 25 years and future directions. Ophthalmic Epidemiol. 14:173–178.
Wientjes, F.B., Hsuan, J.J., Totty, N.F., et al. (1993). P40phox, a third cytosolic component of the activation complex of the NADP oxidase to contain Src homology 3 domains. Biochem. J. 296:557–561.
Williams, D.L. (2006). Oxidation, antioxidants and cataract formation: a literature review 1: Vet. Ophthalmol. 9:292–298.
Acknowledgments
This work was supported by Science Foundation Ireland, Children Leukemia Research Project and the Irish Cancer Society.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2011 Springer Science+Business Media, LLC
About this chapter
Cite this chapter
Quiney, C., Finnegan, S., Groeger, G., Cotter, T.G. (2011). Protein Oxidation. In: Vidal, C. (eds) Post-Translational Modifications in Health and Disease. Protein Reviews, vol 13. Springer, New York, NY. https://doi.org/10.1007/978-1-4419-6382-6_3
Download citation
DOI: https://doi.org/10.1007/978-1-4419-6382-6_3
Published:
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-4419-6381-9
Online ISBN: 978-1-4419-6382-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)