Abstract
Artificial intelligence has rapidly become an area of intense research within medical informatics. In particular, machine learning has shown promise in assisting radiologists to prioritize studies and avoid observation errors. Other applications, such as radiomics, may allow evaluation of imaging features that are too subtle for the human eye to detect. But this new technology also has drawbacks and caveats that must be understood as it becomes more extensively deployed.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Artificial intelligence encompasses a wide variety of fields. Recent developments and specifically advances in computer vision have led to a rise in interest in medical images. New machine learning strategies and neural networks (NN) are being developed to help clinical imagers detect and characterize lesions on routine medical images. Other applications include improving image quality using NN-based reconstruction, facilitating clinical workflow, and extracting and analyzing large volumes of quantitative data from images.
DEFINITION: Artificial Intelligence
A field of computer science focused on creating programs that perform tasks normally assigned to human intelligence, thus simulating human intelligence.
OUR EXPERIENCE: The Neural Network Revolution
Huge Internet databases of pictures of animals (cats, dogs, etc.) and objects (cars, toasters, etc.) have allowed “data challenges” open to the whole world, so that teams (industrial, academic, independent “geeks”) can compete to test their image recognition algorithms. The ImageNet Large Scale Visual Recognition Challenge (ILSVRC; now hosted on Kaggle), the best known challenge containing some ten million images, was shaken in 2012 by a Canadian academic team, which, thanks to a neural network, improved by 10% the performance of the best algorithms, which had plateaued at around a 25% error rate until then. These performances, which continue to improve (<5% errors in 2015), have made it possible to consider applications that until then had been science fiction, such as the self-driving car. It took only a small leap to imagine that the algorithms able to detect or recognize an object in an image could be applied to medical imaging to detect (normal/abnormal), localize, or characterize (benign/malignant) lesions.
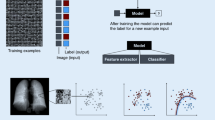
2 Machine Learning and Deep Learning
Within the field of artificial intelligence, machine learning includes methods where machines automatically learn from experience and acquire information without being explicitly programmed. Briefly, an example dataset is submitted to the machine, which determines which function can best predict an output (supervised) or find structure to the data (unsupervised). It will perform the task iteratively until the best solution is found. The function learned will then be applied to new data to perform the given task. A subset of machine learning is deep learning, where neural networks are used to solve a problem instead of using human-engineered equations.
2.1 “Traditional” Machine Learning
DEFINITION: Machine Learning
A field of study in which a machine learns by itself and elaborates a model from a training database.
-
This term refers to machine learning methods other than deep learning. Among numerous traditional machine learning methods used for supervised learning in medical imaging, one can cite random forest, support vector machine (SVM), and LASSO (least absolute shrinkage and selection operator ). Some traditional machine learning methods allow feature selection, which can be used to reduce the number of features to be included in the model. This is especially interesting for radiomics studies where a lot of features are extracted for evaluation. Unsupervised learning for cluster analysis includes methods such as k-means clustering, but unsupervised techniques are currently less used.
2.2 Deep Learning and Neural Networks
KEY CONCEPT: Deep Learning
Deep learning is an end-to-end computer-driven approach, meaning that human-guided steps such as pre-processing and feature extraction are bypassed. The computer decides what aspects of the data are important.
-
Deep learning refers to deep neural networks , which are a subtype of NN that have recently become popular, thanks to the development of high-performance graphics processing unit (GPUs).
-
The basic principle of neural networks is to mimic the functioning of the human brain by combining artificial neurons conceived on the model of biological neurons. Deep learning is defined by the use of architectures combining several hidden layers of artificial neurons . Each neuron layer is composed of several neurons that combine the information transmitted by the neurons of the previous layer by assigning them different weights. During training, the weights are progressively adjusted, thanks to the backpropagation of error.
-
A common approach for medical image analysis is the use of convolutional neural networks (CNN) in which artificial neurons are replaced by a series of successive convolutions applied to the image. During the learning phase, the algorithm progressively tweaks all the successive convolution kernels to improve the model.
-
Many different deep learning architectures are available such as encoder-decoder and atrous convolution architectures for segmentation with CNN.
-
Deep learning often outperforms classical machine learning methods. However, the amount of data and computing power required to train a deep learning algorithm are much greater than those required by traditional machine learning methods.
-
Deep learning models behave like black boxes, which prevents their understanding by the human mind. It is important to keep in mind that deep learning does not use the same rules as the human brain for reading images, and this can result in what will be nonsensical errors to humans.
FURTHER READING: Convolutional Neural Networks
Soffer S, Ben-Cohen A, Shimon O, Amitai MM, Greenspan H, Klang E. Convolutional Neural Networks for Radiologic Images: A Radiologist’s Guide. Radiology. 2019 Mar;290(3):590–606. https://doi.org/10.1148/radiol.2018180547.
DEFINITION: Segmentation
The ability to outline and identify specific anatomic structures or pathology on a radiologic image.
2.3 Supervised vs. Unsupervised Tasks
KEY CONCEPT: Classification vs. Regression
If a deep learning model predicts real numerical values (e.g., a severity score), it is solving a regression equation.
If a model predicts that an image belongs in a certain category (e.g., hematoma vs. calcification), it is solving a classification problem.
-
Supervised algorithms aim to predict a given output, such as class (normal vs. abnormal, benign vs. malignant, disease 1 vs. disease 2, etc.), localization (identifying where an abnormality is located), or a continuous variable (e.g., survival). These algorithms are trained on labeled data , i.e., data that are already confidently attributed to a given class/localization/value. Examples of supervised machine learning are classification such as determining if a lesion is benign or malignant and regression such as determining a severity score.
-
Unsupervised algorithms are trained on non-labeled data. Its aim is to identify subgroups of data sharing common characteristics such as different phenotypes of a single disease. Clustering is the most representative example of unsupervised machine learning. It can also be used in data reduction strategies. Annotated and non-annotated data can be combined in semi-supervised methods .
-
There are different levels of labeling or annotation depending on the intended use of the data. The simplest annotation process is the labeling process in which images are classified according to their content (pneumothorax: yes/no), but the location of the anomaly is not provided within the image. These annotations can be used to build classifiers. The next level of annotation process is the use of boundary boxes to indicate the region of interest, and the most time-consuming annotation process is the delineation of the anomalies on the image. The latter is used for segmentation tasks.
2.4 Datasets
-
The dataset is usually split into three subsets used for training, validation, and testing. The training set is used to build the model. The validation set is used to evaluate the models during the training phase and to select the best one. An alternative to this dataset splitting is the method of k-fold cross validation which allows training and validating on the same dataset. The test set is used at the end of the process to assess the performances of the selected model. It may be an internal test set, which is set aside before training and validation of the algorithm and is used to test the performance of an established algorithm.
-
The terms “validation set” and “test set” are often used loosely, with an independent test set used to clinically validate a model.
-
The final test set must not contain any of the data used to train the algorithm.
-
In order to maximize model performance, it is important to have a training dataset of sufficient size and quality. As the number of available data is often limited in medical imaging, data augmentation methods can be used. Similarly, it is important to take into account the balance between classes in the training dataset. In case of an unbalanced dataset, class balancing methods should be used to properly represent the minority class, and make sure the model has “seen” enough cases of each.
-
Conversely, the prevalence of each class in the test dataset must be representative of the target population in order to avoid overestimating performance of the model, especially for models used in screening.
-
The quality of the dataset is essential to train the model and guarantee the capacity for the model to perform equally well on cases not seen during training (generalizability). It is important to have a dataset that is representative of the diversity of disease, the variations of normal, and the different acquisition techniques/scanners so that the algorithm has been trained to recognize the different presentations.
-
In supervised models, the quality of the initial segmentation or classification must be trustworthy to produce a reliable outcome (“garbage in; garbage out”).
2.5 Validation Metrics
-
The choice of validation metrics is important to estimate the performance of the model compared to a standard of reference.
-
For segmentation tasks, the Dice similarity coefficient and the Hausdorff distance are commonly used. They allow quantification of the similarity between the obtained segmentation and the ground truth.
-
For classification tasks, accuracy, sensitivity, specificity, and positive and negative predictive values are typical validation metrics for comparing binary predictions (e.g., malignant or benign) with the gold standard. However, before being binarized, the output of the algorithm is usually expressed as a probability. To evaluate the performance of the model in this setting, a ROC curve analysis with calculation of the AUC is the method of choice.
KEY CONCEPT: Validation of AI Algorithms
Methods to validate an AI algorithm remain debated. Though accuracy and area under the ROC curve give an indication on gross performance, it may not reflect “real-life” performance when used on other datasets or when integrated in the workflow.
FURTHER READING: ROC Curves
Eng J. Receiver operating characteristic analysis: a primer. Acad Radiol, 12 (2005), pp. 909–916. https://pubmed.ncbi.nlm.nih.gov/16039544/
3 Applications
3.1 Detecting, Classifying, Localizing, and Segmenting
-
Image recognition algorithms can be used on medical images for three different tasks: detection (is there a lesion in an image?), classification (is this lesion normal/abnormal, or benign/malignant?), and localization (where is the lesion in the image?). These tasks may be combined, for example, in a mammography CAD where lesions are detected, classified with a % chance of being cancer, and localized with a marker on the image, to be validated or invalidated by the radiologist.
-
Segmentation (delineation of an organ/tissue/lesion of interest) is another task where deep learning-based algorithms are superior to traditional image processing methods. Applications include segmentation of ventricles and myocardium from a dynamic cardiac MRI at the systolic and diastolic phases to calculate the ejection fraction or thickness of the myocardium. Spinal vertebrae can be automatically identified and numbered, and the loss of height of one or more vertebrae by compression fracture can be detected. The amount of pulmonary emphysema, the volume of renal parenchyma, the loss of muscle mass (sarcopenia), coronary calcifications, etc can be quantified. These segmentations enable the extraction of quantitative biomarkers that can be used to automatically assess disease severity or health status, in addition to visual interpretation of the patient’s target disease.
-
Elastic registration can also be performed using deep learning methods.
FURTHER READING: Image Segmentation
Cardenas CE, Yang J, Anderson BM, Court LE, Brock KB. Advances in Auto-Segmentation. Semin Radiat Oncol. 2019;29(3):185–197. https://doi.org/10.1016/j.semradonc.2019.02.001.
DEFINITION: Registration
A single anatomic structure may appear in a different place on the image when a study is repeated, or from patient to patient. Registration is a means of identifying specific anatomic points so that two images can be “lined up” with each other and more easily compared.
3.2 Enhancing Image Quality
KEY CONCEPT: Advantages of Deep Learning Image Reconstruction
-
Reduce X-ray dose
-
Reduce volume of contrast agent injected
-
Reduce image acquisition time
-
Image denoising and deep learning-based reconstruction algorithms are already developed and implemented on new scanners. The algorithm reconstructs a high spatial- and/or contrast-resolution image from a noisy undersampled image. Patients could have more comfortable, less irradiating exams, and with reduced doses of contrast agent, thereby decreasing potential side effects. Successful examinations could be performed even in disoriented, pediatric, and claustrophobic patients or patients in pain during the examination.
-
Deep learning can be used to enhance spatial resolution of the original medical images (super resolution). This technique can be used to increase spatial resolution or to decrease acquisition time in MRI.
-
Real-time image quality can also be monitored by AI. Automatic slice placement allows for reproducible image orientation. Detection of anatomical coverage allows real-time alerts in case of insufficient coverage of regions such as the lung apices. Image nonconformities such as incorrect patient position, inadequate exposure, and artifacts could be brought to the attention of the radiographer during conventional radiography acquisition.
3.3 Improving the Workflow
-
“Intelligent” imaging appointment management software could allow identification of patients at risk for no-show and plan reminders or automatically reschedule an appointment when the treatment regimen is delayed, or if a patient is hospitalized. The earliest availability for scheduling could be suggested automatically taking into account the presence of the expert imager or radiographer according to the type of examination.
-
The presentation of the scan for interpretation (hanging protocol) can be improved by automated algorithms displaying the most appropriate series according to the indication of the exam. Natural language processing software could retrieve and display the relevant information from the patient’s medical record while the exam is being read to, for example, highlight a history of surgery.
-
In the emergency context, image analysis software is currently able to detect the most urgent pathologies (intracerebral bleeding, pneumoperitoneum, fracture, pulmonary embolism, etc.). They prioritize abnormal examinations for immediate interpretation. This will avoid diagnostic delays for patients with urgent pathologies.
-
Communication with patients and colleagues may also be improved by AI. Alerts could be automatically sent to the referring clinical in case of life-threatening anomalies mentioned in the report. Automatic translation of report text into structured and standardized terms and format would allow both quality control of report content and further use for research or educational purposes. Communication with patients could be facilitated by translating medical terminology into lay terms and supplementing the report with schematics.
3.4 Guiding Interventional Radiology
-
Analysis of previous procedures may allow guiding the choice of equipment and selecting the patients most likely to benefit from a procedure.
-
Image segmentation and registration between two modalities will allow precise real-time identification of the organ and the lesion to be treated. Indeed, high-resolution images or images allowing visualization of the lesion, such as MRI or PET scans, can be merged with images from planar detectors or conventional radiology, to guide the radiologist during the procedure (e.g., fusion of MRI onto live ultrasound during prostate biopsy).
-
Improved guidance to the target and real-time tracking should result in a reduced dose of X-rays and injected contrast agent.
-
Developments in the field of robotics are expected with prototypes of ultrasensitive and miniaturized sensors or intravascular micro-robots allowing procedures similar to robotic surgery, which have become possible, thanks to computer-aided vision techniques and technological advances in highly reliable and latency-free communication networks to produce a signal, allowing an immersive visual and tactile experience.
3.5 Extracting More Information with Radiomics
-
Radiomics is a high-throughput process that uses traditional machine learning or deep learning methods to extract a high number of features from images which will then be correlated to a desired outcome (when supervised). It is a process centered on discovery of new biomarkers.
DEFINITION: Radiomics
A deep learning algorithm extracts numerous features from a radiologic examination, many of which are not evident to a human observer. The resulting imaging biomarkers may provide novel diagnostic or prognostic information. The term “radiomics” is derived from the term “genomics,” which is a battery of genetic biomarkers.
STEP-BY-STEP: Radiomics Process (Machine Learning)
-
1.
Constitution of dataset representing the disease and acquisition variety of the future population on which the discovered signature is meant to be applied. The data acquisition can be prospective or (more often) retrospective.
-
2.
Segmentation to delineate the region of interest in which the features will be extracted.
-
3.
Pre-processing of images to prepare for quantification normalizing signal intensity.
-
4.
Feature extraction yielding parameters quantifying or representing the signal intensity content of the image.
-
5.
Feature reduction, an optional step which can reduce data dimensionality by eliminating nonreproducible or redundant parameters or clustering similar parameters to combine them into a single one.
-
6.
Feature selection performed by correlating to the desired target (outcome or biological marker).
-
7.
Validation in an independent dataset.
-
Radiomics can be used to predict a biological correlate such as histological type or grade, receptor expression, gene expression or genetic mutation, etc., or an outcome such as survival or treatment response.
-
The output of radiomics can be a single feature which is found to be the most correlated to the desired outcome. But more often, it will be a radiomics signature , i.e., a combination of features similarly to genetic signatures.
-
The radiomics process can be performed using traditional machine learning methods, but deep learning can also be used, either as an end-to-end process or only for certain steps such as feature extraction and feature selection.
-
In traditional machine learning radiomics, features are human-engineered. They are most often separated in three categories: shape descriptors , histogram-derived parameters describing signal intensity content of voxels, and texture features describing spatial distribution of signal intensities.
-
Some limitations are specific to the radiomics process . The radiomics signature discovered and its performance will be impacted by choices regarding pre-processing of images, or strategies for feature extraction and feature reduction, reducing the reproducibility of findings. There is also variability in reporting of radiomics results. The same feature name may refer to different concepts in different publications, and the same concept may have different feature names according to research teams and software. Standardization of imaging across scanner manufacturers adds variability. Finally, correlation of the radiomics signature to the target does not prove a causal relationship.
FURTHER READING: Standardising Radiomics Metrology
Zwanenburg A et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology. 2020 May;295(2):328–338. https://doi.org/10.1148/radiol.2020191145. Epub 2020 Mar 10. PMID: 32154773; PMCID: PMC7193906.
4 Limitations and Challenges
4.1 Data
-
Large quantities of labeled data are needed to train ML/DL algorithms. According to the task, volumes of data required may range from several tens to tens of thousands. This data is both time-consuming and costly to obtain.
4.2 Generalizability
KEY CONCEPT: Overfitting and Underfitting
Overfitting occurs when an algorithm fits too closely or exactly to the data in the training set, and therefore does not perform well on new data. Conversely, underfitting is when an algorithm fails to fit the data in the training dataset and therefore fails to learn.
-
Generalizability is the capacity for a model to maintain its performance when applied to new cases, unseen during training. The generalizability of a model must be evaluated on an external dataset. When a model is highly specific to the training dataset (data overfitting), there is a drop in performance when applied to previously unseen data.
4.3 Explainability or Interpretability
-
A major drawback of deep learning is the black box effect , which precludes human understanding of the rules used by the algorithm. While the quality of the segmentations can be easily verified, reporting predictions that cannot be verified is more problematic. This is a current limit for the adoption of these tools and only partially applies to machine learning where features are human-engineered and therefore mathematically explainable. Some of these machine learning features are difficult for humans to conceptualize, such as texture features.
4.4 Detection of Errors
-
Deep learning models have been shown to make what seems to the human mind to be aberrant errors . There are currently no efficient methods for DL models to integrate a detection of errors, but some of the paths pursued are the capacity for an algorithm to detect that a new image is too different from the ones included in its training and validation datasets.
-
The growing popularity of using generative adversarial networks (GANs) has created adversarial images , a new challenge for image analysis tasks. Adversarial images are images for which the classification seems obvious to a human but cause massive failures in a deep neural network. These can be used to challenge models and improve their performance. GANs are thus contributing to the development of semi-supervised learning and possibly paving the path to future unsupervised learning.
4.5 Automation Bias
-
When humans are placed into an environment where automated systems provide guidance, they behave as if they are in a low-risk environment and become less cautious. Thus, radiologists may become overly trustful of machine learning algorithms and interpret the examinations less carefully.
FURTHER READING
“Fundamentals of AI and Machine Learning for Healthcare”. https://www.coursera.org/learn/fundamental-machine-learning-healthcare
PEARLS
-
Artificial intelligence encompasses a large variety of fields, including machine learning and deep learning used for medical images.
-
Convolutional neural networks are the most frequent deep learning neural network used to analyze medical images.
-
Three datasets are necessary to develop AI models: training sets and validation sets, which allow the selection of the best model, and an independent test set on which the performance of the selected model can be evaluated.
-
Applications include detecting and characterizing lesions in medical images, but also performing segmentation and registration, improving image quality, facilitating clinical workflow, and extracting and analyzing large volumes of quantitative data from images, or guiding interventional radiology procedures.
-
Radiomics is a discovery-centered, data-driven approach for extracting large sets of complex descriptors from clinical images. It uses machine learning and/or deep learning methods to extract features and correlate them to a desired target.
-
Limits of AI methods include lack of generalizability of models, the black box effect which limits the human understanding of models and results, and the lack of detection and management of errors of the models.
Self-Assessment Questions
-
1.
Which of the following applies to deep learning?
-
A.
Neural networks are used to develop deep learning models.
-
B.
Deep learning can only perform supervised learning.
-
C.
Deep learning models are complex to interpret.
-
D.
Deep learning extracts shape, histogram, and texture features.
-
A.
-
2.
Which of the following are required for datasets used for developing machine or deep learning models?
-
A.
The test set is a subset of the initial data used to train the model.
-
B.
A sufficient number of examples of each class should be present for the model in the training dataset.
-
C.
The number of cases must reflect clinical prevalence in the test dataset.
-
D.
Acquisition parameters need to be standardized in a dataset to train a model.
-
A.
-
3.
Which of the following steps are a part of the radiomics process?
-
A.
Segmentation
-
B.
Feature extraction
-
C.
Pre-processing
-
D.
Feature reduction
-
E.
All of the above
-
A.
-
4.
Which of the following tasks can be improved using deep learning?
-
A.
Lesion detection
-
B.
Image segmentation
-
C.
Image reconstruction
-
D.
Increasing spatial resolution
-
E.
All of the above
-
A.
-
5.
Which combination of machine learning approach and annotation level is the most suitable to develop a segmentation algorithm?
-
A.
Supervised learning/labeled images
-
B.
Supervised learning/boundary boxes drawn on images
-
C.
Supervised learning/delineated areas on images
-
D.
Unsupervised learning/labeled images
-
E.
Unsupervised learning/non-annotated data
-
A.
-
6.
What are the advantages of deep learning over traditional machine learning methods?
-
A.
It is not based on human-engineered features.
-
B.
Models are easier to explain.
-
C.
It often performs better.
-
D.
Less data is required to train.
-
E.
It does not require feature extraction.
-
A.
-
7.
Cite different fields of application of AI methods in medical imaging.
-
8.
List some challenges to the development and adoption of AI tools in medical imaging.
-
9.
You are installing AI decision support for your radiologists to help them identify some specific emergent diagnoses. What warnings or caveats would you give them before deployment?
Further Reading
LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–44. https://doi.org/10.1038/nature14539.
Do S, Song KD, Chung JW. Basics of deep learning: a radiologist's guide to understanding published radiology articles on deep learning. Korean J Radiol. 2020;21(1):33–41. https://doi.org/10.3348/kjr.2019.0312.
Bluemke DA, Moy L, Bredella MA, Ertl-Wagner BB, Fowler KJ, Goh VJ, Halpern EF, Hess CP, Schiebler ML, Weiss CR. Assessing radiology research on artificial intelligence: a brief guide for authors, reviewers, and readers-from the radiology editorial board. Radiology. 2020;294(3):487–9. https://doi.org/10.1148/radiol.2019192515.
Geis JR, Brady AP, Wu CC, Spencer J, Ranschaert E, Jaremko JL, Langer SG, Borondy Kitts A, Birch J, Shields WF, van den Hoven van Genderen R, Kotter E, Wawira Gichoya J, Cook TS, Morgan MB, Tang A, Safdar NM, Kohli M. Ethics of artificial intelligence in radiology: summary of the joint European and North American Multisociety Statement. Radiology. 2019;293(2):436–40. https://doi.org/10.1148/radiol.2019191586.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this chapter
Cite this chapter
Fournier, L., Chassagnon, G. (2021). Artificial Intelligence and Machine Learning. In: Branstetter IV, B.F. (eds) Practical Imaging Informatics. Springer, New York, NY. https://doi.org/10.1007/978-1-0716-1756-4_14
Download citation
DOI: https://doi.org/10.1007/978-1-0716-1756-4_14
Published:
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-0716-1755-7
Online ISBN: 978-1-0716-1756-4
eBook Packages: MedicineMedicine (R0)