Abstract
We apply three different sparse reconstruction techniques to spectral demixing. Endmembers for these signatures are typically highly correlated, with angles near zero between the high-dimensional vectors. As a result, theoretical guarantees on the performance of standard pursuit algorithms like orthogonal matching pursuit (OMP) and basis pursuit (BP) do not apply. We evaluate the performance of OMP, BP, and a third algorithm, sparse demixing (SD), by demixing random sparse mixtures of materials selected from the USGS spectral library (Clark et al., USGS digital spectral library splib06a. U.S. Geological Survey, Digital Data Series 231, 2007). Examining reconstruction sparsity versus accuracy shows clear success of SD and clear failure of BP. We also show that the relative geometry between endmembers creates a bias in BP reconstructions.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Hyperspectral demixing
- Sparse demixing (SD)
- Correlated endmembers
- Basis pursuit (BP)
- Orthogonal matching pursuit (OMP)
1 Introduction
Sparsity arises in many important problems in mathematics and engineering. Recent algorithms for finding sparse representations of signals have achieved success in applications including image processing [15], compression [22], and classification [19]. These algorithms, including basis pursuit (BP) [7] and various matching pursuit (MP) methods [16], are guaranteed to converge to correct solutions for problems that meet criteria established, for example, in [6, 23]. In practice, however, they are often applied to problems that do not meet those criteria. One such example is the source separation problem of spectral demixing of hyperspectral images (HSI).
Spectral demixing is the identification of the materials, called endmembers, comprising a hyperspectral pixel, and their fractional abundances. Images captured by hyperspectral sensors such as airborne visible/infrared imaging spectrometer (AVIRIS) [14], hyperspectral mapper (HyMap) [9], and hyperspectral digital imagery collection experiment (HYDICE) [1] have pixels containing spectral measurements for hundreds of narrowly spaced wavelengths. Ideally these measurements could be used to identify materials by comparing directly with a spectral library, trading a difficult computer vision problem for relatively straightforward spectral analysis. In reality, measured signatures rarely correspond to spectra of pure materials. HSI cameras take images with high spectral resolution at the expense of low spatial resolution; for example, AVIRIS has a 20-m ground resolution when flown at high altitude (20 km) [14]. As a result, measured spectra often correspond to mixtures of many materials.
1.1 The Linear Mixture Model
Even though HSI sensors generally measure nonlinear combinations of the constituent materials’ spectra, HSI analysts often assume linear mixing of pure signals [13, 25]. This assumption holds if the materials occur in spatially separated regions with negligible light scattering.
Let E denote an n-by-k matrix of endmembers \(E ={ \left [{\mathbf{e}}_{i}\right ]}_{1}^{k}.\) The linear mixture model (LMM) assumes that every pixel signature x ∈ ℝ n has an abundance vector α ∈ ℝ k satisfying
where η is a small error term. Ideally, endmembers correspond to pure materials, but they more likely represent common mixtures of materials. The abundance vector α gives the relative quantities of the materials making up the mixture.
We consider three common versions of the LMM, each with its own set of constraints on α and E :
-
(LMM 1)
α i ≥ 0, ∑α i = 1, and rank\(\left (E\right ) \geq k - 1.\)
-
(LMM 2)
α i ≥ 0, ∑α i ≤ 1, and E has full rank.
-
(LMM 3)
α i ≥ 0 and E has full rank.
The rank constraints ensure uniqueness of each signature’s abundance vector. Each of these models has different assumptions on the physical properties of the endmembers in E. For LMM 1, we assume either full illumination of every pixel or that at least one endmember represents shade. LMM (2) assumes that E contains spectral signatures corresponding to materials lit as brightly as the brightest pixels in the image. Darker pixels have abundance vectors with ∑α i < 1. LMM 3 allows pixels brighter than any of the e i . The rank restrictions typically pose no problem, since k < < n in practice.
Each LMM has its own constraint set \(\mathcal{A}\) for the abundance vectors. Define
In LMM2 1 and (2), S describes a simplex. In LMM 1, S is a (k − 1)-dimensional simplex with corners given by the k columns of E. For LMM (2), S is a k-dimensional simplex determined by E and the origin. The two problems are mathematically equivalent: we can write LMM 1 as LMM (2) by translating the origin to an arbitrary endmember in E, then removing that column from E. Similarly, we can write LMM (2) as LMM 1 by adding a column of zeros to E. In LMM 3, S is the wedge determined by the columns of E. Notice that for a given image, we can rescale the columns of E so that ∑α i ≤ 1 for all pixels within the image. We therefore focus on LMMs 1 and (2). In particular, we assume E has full rank.
Demixing often requires learning the endmembers as well as the abundances (blind source separation), but throughout this chapter, we assume known endmembers. See [2, 11, 17, 18, 21, 26] for more information on learning endmembers.
1.2 Sparse Mixtures
Figure 1a shows a standard idealized scatter plot of LMM 1 [3, 4, 13, 26]. In such illustrations, authors typically distribute most pixels throughout the simplex interior. We argue that Fig. 1b illustrates hyperspectral data more accurately. Any interior point of a simplex is a combination of all the endmembers. Physically, this means that the region captured by the pixel contains samples of each endmember within the scene. We suspect that such pixels rarely occur. Instead, most pixels contain a strict subset of the scene’s endmembers and thus lie on the simplex boundary. The simplex interior should be nearly empty.
LMM (2) determines another simplex, but one face of this simplex corresponds to admissible abundance vectors satisfying ∑α i = 1. These vectors are not necessarily sparse. For LMM 3, sparse signals again occur on the boundary of S. Figure 2 illustrates sparse and non-sparse mixtures for LMM 3 with the extra boundary of LMM (2) included for reference.
Traditional pixel demixing algorithms minimize error, using nonnegative least squares (NLS). This might not achieve the most realistic results, however, since we expect some error due to model uncertainty and noise. Assuming that most pixels are made up of only a few endmembers, we may instead seek a balance between error and sparsity. We allow some error in the reconstruction if it comes with a sparser mixture.The ideal mixture is the sparsest mixture with small, but acceptable, error.
1.3 Outline
We evaluate three different algorithms for calculating sparse abundance vectors: a basis pursuit (BP) algorithm that uses the L 1 of the abundance vectors [12], a greedy algorithm called sparse demixing [11], and a natural extension of orthogonal matching pursuit (OMP) to the endmember problem. Theorems guaranteeing the convergence of OMP and BP to accurate sparse mixtures all require low mutual information of the set of vectors being searched over. In practice, they are often used with highly correlated vectors. One such example is hyperspectral demixing.
Section 2 briefly describes the three algorithms and summarizes results from [11] that show that BP preferentially selects endmembers based on the relative geometry between endmembers. Section 4 demonstrates the relative performance of OMP, BP, and SD by demixing spectra with known abundances. We choose a set of endmembers from the USGS Library [8], then randomly select a sparse matrix A. We look at this problem both with and without Gaussian noise. Since these algorithms are intended to find sparse mixtures that accurately approximate the signature, we judge success by examining reconstruction sparsity versus accuracy.
2 Spectral Demixing
Given a matrix of endmembers E and a spectral signature x, HSI analysts typically demix pixels with NLS [13]. The NLS approximation y of x solves y = E α for
Defining S by (2), we rewrite (3) as
This quadratic programming problem has a simple geometric solution. Let \(\mathbf{\hat{x}}\) denote the orthogonal projection of x on the column space of E. For problems LMMs (2) and (3),
Note that since E has full rank, E ⊤ E is invertible. Since \(\mathbf{\hat{x}}\) is the orthogonal projection of x, y solves (4) if and only if
If \(\mathbf{\hat{x}}\) lies in S, then \(\mathbf{y} = \mathbf{\hat{x}}.\) If \(\mathbf{\hat{x}}\) lies outside S, then (5) gives the closest point y to \(\mathbf{\hat{x}}\) on the boundary of S.
3 Applying Sparse Coding to the Demixing Problem
We assume that the abundance vector α satisfies (1) for some η with
Unlike standard demixing, we assume that the correct abundance vector is the sparsest one that gives an approximation within ε of the measured spectrum. Minimizing the L 2 (Euclidean) norm generally does not give sparse mixtures. The NLS constraint y ∈ S, however, automatically enforces sparsity for some mixtures. If a pixel lies exactly on the boundary of S, then NLS correctly recognizes it as a sparse mixture. However, sensor noise, measurement errors, and model inaccuracy likely prevent such cases. Even when a pixel contains only a few endmembers, these errors push the pixel off the boundary of S. If they push the pixel outside S, then NLS gives the correct sparse solution. If they push the pixel inside S, NLS gives a mixture of all the endmembers.
The sparsest abundance vector giving an approximate mixture with error ε is
for ε > 0. The L 0 semi-norm of α, ∥ α ∥ 0, is the number of nonzero components of α. Minimizing the nonconvex L 0 semi-norm is NP-hard, so matching pursuit algorithms only find approximate solutions to the problem. One of these algorithms, OMP has been shown to solve (6) for some matrices E [23]. Minimizing the L 1 norm [defined by (8)] also gives sparse solutions for some matrices E, and it has the mathematical advantage of convexity [5, 10].
In this section, we describe three algorithms for calculating sparse abundance vectors. The first is a basis pursuit (BP) algorithm that uses the L 1 norm. The others, OMP and sparse demixing (SD), find approximate solutions to (6).
3.1 Basis Pursuit
In [12], Guo et al. calculated sparse abundance vectors by minimizing
In each constraint set \(\mathcal{A},\) α i ≥ 0, so
Note that the L 1 term makes no meaningful contribution to (7) for LMM 1, since ∥ α ∥ 1 = 1 for all admissible α. Whenever discussing BP, we assume LMM 2 or LMM 3.
Unfortunately, for general endmember matrices E, (7) does not necessarily give sparse abundance vectors. In fact, Greer[11] shows that (7) gives sparser solutions than NLS only for certain cases. For many other cases, including the L 1 norm reduces sparsity. We briefly describe a rationale for this and refer to [11] for details.
The set \(\mathcal{A}\) of admissible abundance vectors determines a subset S of the column space of E [see (2)]. Since E has full rank, every y ∈ S has a unique abundance vector α satisfying E α = y. In fact, α(y) = Φy for
We call the abundance vector components, α i , coefficient functions. Each coefficient function is linear, with α i (e i ) = 1 and α i (y) = 0 for all y on the (k − 1)-dimension hyperplane determined by e j≠i and the origin.
For y ∈ S define
In particular, ϕ is a linear function of y in S with ∇ ϕ = Φ ⊤ 1, where 1 denotes a column vector of ones. Since ϕ(e i ) = 1 for every column e i of E, and \(\phi \left (0\right ) = 0,\) ∇ϕ is normal to the (k − 1)-dimensional hyperplane determined by the columns of E. Thus the L 1 term’s effect depends entirely on the geometry of the endmembers in E.
Define
Solving (7) is equivalent to solving
The convex function F has a global minimum in ℝ k at
Compare (11) with the minimum of the NLS optimization function,
The minimum of G(s) occurs inside S only when x lies inside S. The minimum of F can occur on the interior of S even for cases where x lies on the exterior—cases where NLS gives a sparse solution. If y lies inside S, then the L 1 reconstruction does not give a sparser representation than NLS. This happens, for example, for any point x on the interior of S that is further than \(\lambda \left \vert \mathbf{\nabla \phi }\right \vert\) from the boundary of S. On the other hand, the L 1 reconstruction of x is less accurate than the NLS solution, which gives x the exact answer. For these points, NLS is clearly the better method.
The L 1 norm’s ability to increase sparsity depends on the sign of ∇α i ⋅∇ϕ for each i. Figure 3 shows the effects of the L 1 norm for a problem with two endmembers giving coefficient functions, α1 and α2, with ∇α 1 ⋅∇ϕ < 0 and ∇α 2 ⋅∇ϕ > 0. Let \({\mathbf{y}}_{\lambda }\left (\mathbf{x}\right )\) solve (11) for given λ and x. Suppose ∇α i ⋅∇ϕ > 0. Then for any given x, \({\alpha }_{i}\left ({\mathbf{y}}_{\lambda }\left (\mathbf{x}\right )\right )\) monotonically decreases to 0 as λ increases. The same does not hold if ∇α i ⋅∇ϕ < 0. In this case, x close to the boundary α i = 0, either inside or outside S, can yield minima \({\mathbf{y}}_{\lambda }\left (\mathbf{x}\right )\) that lie on the interior of S. Section 4.2 will demonstrate how this affects demixing.
3.2 Orthogonal Matching Pursuit
OMP is a greedy algorithm that iteratively increases the number of nonzero components in α while minimizing the approximation’s residual error at each step. The linear mixing model constraints require modifying the OMP algorithm (introduced in [27]). This section describes one natural modification.
Suppose we have a spectral signature x, a set of endmembers \(\left \{{e}_{i}\right \}\) and an error bound ε. For a subset of endmembers, Λ k , let x k be the NLS reconstruction of x over Λ k . Define
and set
Until ∥ r k ∥ 2 < ε, OMP sequentially improves the approximation x k by setting
The standard OMP algorithm uses the absolute value of the inner product, but the LMMs nonnegativity constraint leads to better results without the absolute value.
3.3 Sparse Demixing
Sparse demixing (SD) uses the greedy approach of OMP, but works in the opposite direction: it begins with a representation over all endmembers, then removes endmembers one by one until reaching the sparsest representation within a specified accuracy. SD first performs NLS over the full set of endmembers, then removes the endmember corresponding to the smallest component of α. Next, it performs NLS on the simplex determined by this smaller set of endmembers. This process repeats, in each iteration removing the endmember corresponding to the smallest abundance value, until the approximation leaves the accuracy range specified by ε in (6). See [11] for details on the application of SD to all three LMMs.
SD has some advantage over OMP. Its initial step gives the widely used NLS solution. For examples like the one in Fig. 4, OMP produces mixtures containing all the endmembers, even though sparsity assumptions for (1) place x on the simplex’s left edge. SD gives the desired mixture in two iterations. SD takes advantage of key differences between pixel demixing and standard sparse reconstruction problems. In its intended applications, OMP searches large overcomplete sets of vectors for spanning subsets. SD is impractical for such applications, but HSI demixing usually involves far fewer endmembers than the number of spectral bands. Thus, starting with all the endmembers and sequentially eliminating them is feasible.
Consider the simplex and data point shown in the left figure. This point is nearly a mixture of the two materials forming the left side of the simplex. The middle figure shows the approximations produced in three iterations of OMP. The right shows the approximations produced in three iterations of SD. Notice that the second iteration of SD gives the closest sparse mixture, which OMP never produces
4 Numerical Experiments
To evaluate the three algorithms describe in Sect. 2, we demix random mixtures satisfying LMM 1 and LMM (2). Each mixture consists of a sparse subset of 14 endmembers chosen from the USGS spectral libraries [8]: nine minerals and five vegetation endmembers. Figure 5 shows the spectral signatures of these endmembers. The spectra from these libraries each have 450 bands ranging from 0.395 to 2.56 μm. These materials differ enough to distinguish between them in HSI (see, for example, [20]).
Endmember plots. The spectra used as endmemebrs for the numerical tests. Spectra consist of minerals and vegetation from the USGS spectral library [8]
Although linearly independent, the spectra are highly correlated, with angles between many pairs of the spectra near zero. See Fig. 6 for a histogram of the dot products between the 14 (normalized) spectra. Notice that those dot products are all larger than \(\frac{1} {2}.\) Due to this high correlation, support recovery theory for BP and OMP does not apply [23, 24]. The examples demonstrate the failure of BP.
4.1 LMM 1 ( ∑ \nolimits α i = 1)
The first example uses mixtures satisfying LMM 1. We determined the set of spectra, X, by randomly selecting a 14-by-1, 000 matrix of abundances. Each entry had a 20 % chance of being nonzero, with nonzero entries distributed uniformly between 0 and 1. Columns with all zeros were eliminated. This example has 961 nonzero spectra. Finally, we scaled each column so that its abundances added to 1. Figure 7 shows the distribution of endmembers in X. It also shows the distribution of the number of endmembers comprising each spectral signature in X : each is a mixture of between 1 and 6 endmembers.
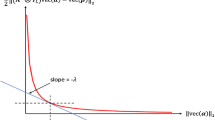
Figure 8 shows results of demixing X with no added noise. In this case, NLS gives the exact abundances of X. As discussed in Sect. 3.1, for all λ, BP gives the same solution as NLS for LMM 1. Each curve in Fig. 11 is parametrized by ε, the amount of error allowed for the sparse approximations [see (6)]. Increasing ε sacrifices some of that accuracy for sparsity. This increase of ε corresponds to more iterations in SD and fewer iterations in OMP. The curves intersect at ε = 0, which is the NLS solution. Curves lying closer to the origin correspond to methods that find sparser solutions with greater accuracy. In this case SD performs better than OMP until both reach the large-error regime—about 9 % relative error.
We next add Gaussian noise with a standard deviation of 0.03 to X. NLS does not give the correct solution for this more realistic scenario. Noise has pushed some of the spectra in X to the interior of the endmember-determined simplex, making NLS choose a mixture that is non-sparse and incorrect. We use the non-noisy signatures in X to calculate errors. Figure 9 shows that SD improves the accuracy of the abundances while simultaneously decreasing the number of endmembers. OMP does not perform as well, but it still shows an initial drop in the number of endmembers with very little increase in error.
4.2 LMM (2) ( ∑ \nolimits α i ≤ 1)
For this example, we used the process in Sect. 4.1 to randomly select a 15-by-1, 500 sparse abundance matrix, with the extra row corresponding to an endmember of all zeros. After scaling and eliminating all-zero columns, we removed the 15th row. The resulting set, X, contains 1,152 spectral signatures satisfying LMM (2). Figure 10 shows the distribution of endmembers across X. Figure 11 shows how BP, OMP, and SD all perform on Xwithout added noise. NLS again gives the exact solution. Both OMP and SD depend on the parameter ε, with ε = 0 giving the NLS solution [see (6)] and increased ε giving sparser solutions. The curve for BP depends on λ, with λ = 0 corresponding to NLS [see (7)]. BP performs poorly. In fact, increasing λ increases both errors and the number of endmembers. The set X contains only exact sparse mixtures that lie on the simplex boundary. As discussed in Sect. 3.1, as λ increases, BP drives many of these spectra to the simplex interior.
BP preferentially selects the ith endmember if ∇ α i ⋅ ∇ ϕ < 0, for abundance α i and ϕ defined by (9). Figure 12 demonstrates this phenomenon. The curve plots the number of BP approximations that contain each given endmember as λ increases. For very large λ, nearly all the BP approximations contain the same endmember.
BP’s selection of endmembers for LMM (2) spectra without noise. Increasing λ should increase sparsity, meaning abundances of endmembers should shift to zero. However, the effect on each endmember e i depends on ∇ α i ⋅ ∇ ϕ for abundance α i and ϕ defined by (9). BP prefers some endmembers over others. In this example, the actual endmembers are distributed nearly uniformly, but BP loses that statistical pattern as λ increases
SD and OMP perform very differently. Figure 13 tracks the selection of endmembers by SD and OMP for each value of ε. SD consistently decreases each endmember’s number of substantiations as ε increases. It shows no obvious bias. On the other hand, OMP treats some endmembers differently, with substantiations of one of the endmembers increasing with ε. Nevertheless, OMP shows far less bias than BP.
We next added Gaussian noise (standard deviation 0.03) to the signatures in X. In this more realistic case, NLS gives incorrect mixtures. All three curves intersect at the NLS solution, \(\lambda = \epsilon = 0.\) Errors are calculated with respect to the exact, non-noisy mixtures. Again SD shows the best performance. Both SD and OMP provide more accurate solutions than NLS. BP does not perform nearly as well. It does, however, improve the sparsity as λ increases. This example shows a greater difference in performance between BP and SD than shown in [11]. This is likely because that paper measures error as the distance between the reconstruction and the pixel, which in this example corresponds to the difference between the reconstruction and the noisy signature (Fig. 14).
5 Conclusions
This chapter evaluates the ability of sparse reconstruction algorithms to find sparse mixtures of endmembers, which are typically highly correlated. Although restricted to hyperspectral demixing, the work may give some insight into the more general problem of sparse reconstruction over coherent sets. In this case, which certainly is not unique to the HSI problem, we have no theory guaranteeing that standard pursuit algorithms will provide sparse and accurate reconstructions. This chapter’s examples show the failure of BP, and some success with OMP, for the endmember problem. It’s natural to wonder about their relative performance for other problems. There may also be other application-specific pursuit algorithms that, like SD, offer superior performance searching for sparse support over sets of correlated vectors.
References
Basedow, R.W., Carmer, D.C., Anderson, M.E.: HYDICE system: implementation and performance. In: Descour, M.R., Mooney, J.M., Perry, D.L., Illing, L.R. (eds.) Proceedings of SPIE, vol. 2480 (1995)
Berman, M., Kiiveri, H., Lagerstrom, R., Ernst, A., Dunne, R., Huntington, J.: Ice: a statistical approach to identifying endmembers. IEEE Trans. Geosci. Remote Sens. 42, 2085–2095 (2004)
Boardman, J.W.: Analysis, understanding and visualization of hyperspectral data as convex sets in n-space. In: Descour, M., Mooney, J., Perry, D., Illing L. (eds.) Proceedings of SPIE, vol. 2480 (1995)
Bowles, J.H., Gillis, D.B.: An optical real-time adaptive spectral identification system (ORASIS). In: Change, C.I. (ed.) Hyperspectral Data Exploitation: Theory and Applications. Wiley, Hoboken (2007)
Candes, E.J., Romberg, J., Tao, T.: Robust uncertainty principles: exact signal reconstruction from highly incomplete frequency information. IEEE Trans. Inform. Theor. 52(2), 489–509 (2006)
Candes, E.J., Romberg, J., Tao, T.: Stable signal recovery from incomplete and inaccurate measurements. Commun. Pure. Appl. Math. 59(8), 1208–1223 (2006)
Chen, S.S., Donoho, D.L., Saunders, M.A.: Atomic decomposition by basis pursuit. SIAM J. Scientific Comput. 20, 33–61 (1998)
Clark, R.N., Swayze, G.A., Wise, R., Livo, E., Hoefen, T., Kokaly, R., Sutley, S.J.: USGS digital spectral library splib06a. U.S. Geological Survey, Digital Data Series 231 (2007)
Cocks, T., Jenssen, R., Stewart, A., Wilson, I., Shields, T.: The HyMap airborne hyperspectral sensor: The system, calibration and performance. In: First EARSEL Workshop on Imaging Spectroscopy. SPIE, Zurich (1998)
Donoho, D.L., Tanner, J.: Sparse non-negative solutions of underdetermined linear equations by linear programming. Proc. Natl. Acad. Sci. 102(27), 9446–9451 (2005)
Greer, J.B.: Sparse demixing of hyperspectral images. IEEE Trans. Image Process. 21(1), 219–228 (2012)
Guo, Z., Wittman, T., Osher, S.: L 1 unmixing and its application to hyperspectral image enhancement. In: Shen, S., Lewis, P. (eds.) Proceedings of SPIE, vol. 7334 (2009)
Keshava, N., Mustard, J.F.: Spectral unmixing. IEEE Signal Process. Mag. 19(1), 44–57 (2002)
Laboratory, J.P.: AVIRIS homepage: http://aviris.jpl.nasa.gov
Mairal, J., Elad, M., Sapiro, G.: Sparse representation for color image restoration. IEEE Trans. Image Process. 17(1), 53–69 (2008)
Mallat, S., Zhang, Z.: Matching pursuits with time-frequency dictionaries. IEEE Trans. Signal Process. 41(12), 3397–3415 (1993)
Moussaoui, S., et al.: On the decomposition of Mars hyperspectral data by ICA and Bayesian positive source separation. Neurocomputing 71, 2194–2208 (2008)
Nascimento, J., Bioucas-Dias, J.: Vertex component analysis: a fast algorithm to unmix hyperspectral data. IEEE Trans. Geosci. Remote Sens. 43(4), 898–910 (2005)
Ramirez, I., Sprechmann, S., Sapiro, G.: Classification and clustering via dictionary learning with structured incoherence and shared features. In: IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (2010)
Resmini, R.G., Kappus, M.E., Aldrich, W.S., Harsanyi, J.C., Anderson, M.: Mineral mapping with hyperspectral digital imagery collection experiment (HYDICE) sensor data at Cuprite, Nevada, U.S.A. Int. J. Remote Sens. 18(7), 1553–1570 (1997)
Robila, S.A., Maciak, L.G.: Considerations on parallelizing nonnegative matrix factorization for hyperspectral data unmixing. IEEE Geosci. Remote Sens. Lett. 6(1), 57–61 (2009)
Taubman, D., Marcellin, M.: JPEG2000: Image Compression Fundamentals, Standards and Practice. Kluwer Academic Publishers, Boston (2001)
Tropp, J.: Greed is good: algorithmic results for sparse approximation. IEEE Trans. Info. Theor. 50(10), 2231–2242 (2004)
Tropp, J.: Just relax: Convex programming methods for identifying sparse signals in noise. IEEE Trans. Info. Theor. 52(3), 1030–1051 (2004)
Verrelst, J., Clevers, J., Schaepman, M.: Merging the Minnaert-k parameter with spectral unmixing to map forest heterogeneity with CHRIS/PROBA data. IEEE Trans. Geosci. Remote Sens. 48(11), 4014–4022 (2010)
Winter, M.E.: N-FINDR: An algorithm for fast autonomous spectral end-member determination in hyperspectral data. In: Descour, M., Shen, S. (eds.) Proceedings of SPIE, vol. 3753 (1999)
Zhang, Z.: Matching pursuit. Ph.D. thesis, Courant Institute, New York University (1993)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Birkhäuser Boston
About this chapter
Cite this chapter
Greer, J.B. (2013). Hyperspectral Demixing: Sparse Recovery of Highly Correlated Endmembers. In: Andrews, T., Balan, R., Benedetto, J., Czaja, W., Okoudjou, K. (eds) Excursions in Harmonic Analysis, Volume 1. Applied and Numerical Harmonic Analysis. Birkhäuser, Boston. https://doi.org/10.1007/978-0-8176-8376-4_10
Download citation
DOI: https://doi.org/10.1007/978-0-8176-8376-4_10
Published:
Publisher Name: Birkhäuser, Boston
Print ISBN: 978-0-8176-8375-7
Online ISBN: 978-0-8176-8376-4
eBook Packages: Mathematics and StatisticsMathematics and Statistics (R0)