Abstract
Depression is a severe and life-threatening psychiatric illness whose pathogenesis is still essentially unknown. Proteomic analysis of synaptic terminals (synaptoproteomics) in animal models of depression is a powerful approach to gain insight into the molecular mechanisms underlying vulnerability to mood disorders and the long-term action of drug treatments. Here, we employed two different animal models of depression, the Learned Helplessness rats (a classical behavioral model of depression) and a new model of depression with gene—environment interaction (Flinders Sensitive Line rats subjected to early life stress). Both animal models were treated with the antidepressant escitalopram. Analysis of their synaptoproteomic profile revealed a number of protein spots differently regulated by basic vulnerability and/or early life stress. Using this approach, we obtained information regarding biomarkers that may represent predictors of pathology or response/resistance to drug treatment, as well as potential targets for novel pharmacological and therapeutic strategies.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
8.1 8.1 Introduction
The available prescription drugs for the treatment of depression alleviate symptoms in only about 60–70% of patients and many patients experience adverse effects that cause discontinuation leading to cessation of the treatment (Keller and Boland, 1998). Moreover, there is a time lag of 3–4 weeks before the onset of the therapeutic effects can be appreciated. Indeed, individuals respond differently to antidepressant drugs, and often the effects are unpredictable (efficacy, onset of action, adverse effects, placebo effects). Therefore, to optimize response to antidepressant treatments and to minimize adverse effects, it is necessary to identify biomarkers of individual susceptibility and response to drugs. Proteins are the actual effectors of biological functions, and since mRNA and protein expression levels are often not related (Anderson and Seilhamer, 1997; Paulson et al., 2003), information provided by transcriptional studies cannot entirely account for functional changes. Therefore, global analysis of protein expression is a powerful approach to gain insight into the molecular mechanisms underlying vulnerability to psychiatric disorders and the long-term action of drug treatments (Rohlff and Hollis, 2003; Fountoulakis, 2004; Kramer and Cohen, 2004, Vercauteren et al., 2004).
Animal models are important tools for the study of mood disorders because they allow the employment of behavioral and biochemical research methods that cannot be used, for evident ethical reasons, in humans. Therefore, scientists have developed several animal models to evaluate the validity and consistency of diverse experimental approaches to the study of psychiatric diseases. An appropriate model for human mood disorders should accomplish the main criteria of construct validity (how closely the model is consistent with the theoretical rationale), face validity (how closely the model reproduces symptoms of the pathology) and predictive validity (how well the model responds to pharmacological treatment) (Hitzemann, 2000; Urani et al., 2005). These criteria have been used in particular to validate the most important animal models for major depression. In this work we have applied global analysis of protein expression to the study of two animal models. The models employed are a classical behavioral animal model of depression (Learned Helplessness; LH) and a new animal model of depression with gene—environment interaction (Flinders Sensitive Line rats subjected to early life stress).
8.2 8.2 The Learned Helplessness Rat Model of Depression
Clinical studies indicate that a broad range of genetic and environmental influences, through combined action in vulnerable individuals, are involved in the pathophysiology of depression (Matthews et al., 2005). Therefore it is reasonable that an external stress may result in changes in behavior in a laboratory animal. For these reasons, one of the most significant and validated animal models of depression applies an uncontrollable stress, which in a number of animals leads to a depressive-like behavioral phenotype (Vollmayr and Henn, 2003). The LH protocol was initially elaborated by Seligman and Overmier (Overmier and Seligman, 1967) and induces the behavioral condition through the application of acute uncontrollable stress. Usually the aversive stimuli are administered by foot shock, but tail shock and loud acoustic sound have also been used. Originally the procedure was carried out with dogs but the paradigm was subsequently investigated and completely developed in rats. The procedure we used consists of a 40-min session of inescapable shock (0.8 mA; single shocks randomized by computer with inter-shock time lasting from 5 to 15 s) and the total shock duration is 20 min (Vollmayr and Henn, 2001). Twenty-four hours after the application of inescapable stress, the animals are tested in a conditioning chamber. The same current is applied with 15 shocks lasting 60 s each with 24 s time interval. The current is accompanied by a light pointed to a lever in order to facilitate its detection and the animals can stop the shock by pressing the lever. Thus, a computer records the time necessary for each animal to terminate each trial, in order to classify the animal behavior. More than 10 failures identify the animals as “learned helpless”, whereas with less than five failures they are considered “non-learned helpless” (nLH) (Table 8.1 ). Generally with Sprague-Dawley rats we found about 20% of LH in such test runs (Vollmayr and Henn, 2001). The LH rats show significant face, construct and predictive validity: they have weight loss, sleep changes, decreased libido, anhedonia (monitored using sucrose preference test) (Willner, 1990; Willner, 1995), and hypothalamic—pituitary—adrenal (HPA) axis alterations. Moreover, various antidepressants are able to reverse helpless behavior: tricyclics, norepinephrine reuptake inhibitors, and serotonin reuptake inhibitors (Vollmayr and Henn, 2001). Therefore, LH animals display the most important behavioral and symptomatological changes associated with depression; the rats maintain helpless behavior for about 2 weeks after acquisition (Henn et al., 2002; Vollmayr et al., 2003).
8.3 8.3 The Flinders Sensitive Line (FSL) and the Flinders Resistant Line (FRL) Rat Model of Depression
The Flinders Lines of rats were established by selective breeding of Sprague-Dawley rats for their sensitivity to cholinergic agents in behavioral paradigms. The breeding program developed two separate lines, FSL and FRL, distinct for their sensitivity to the effects of the anticholinesterase agent diisopropyl fluorophosphate (Overstreet and Russell, 1982).
The FSL rats are a well-validated animal model of depression carrying genetic vulnerability associated with distinct features of the pathology and responsiveness to antidepressant drugs. The FSL rats fulfill the major validation criteria of construct, face, and predictive validity for a good animal model (Yadid et al., 2000; Overstreet et al., 2005). Indeed, it has been shown that a subset of depressed individuals display supersensitivity to cholinergic agonists (Janowsky et al., 1980, 1994); therefore, the FSL model is consistent with the cholinergic model of depression (good construct validity). Moreover, this model has many behavioral similarities with depressed individuals, e.g., reduced general activity, decreased appetite, decreased sexual activity, reduced latency of REM sleep and increased length of REM sleep episodes, anhedonia (after chronic mild stress) (Pucilowski et al., 1993), and serotonergic abnormalities (good face validity) (see Table 8.2 ). In addition, FSL rats show an increased immobility in the Porsolt swim test (a widely used test for behavioral despair), which is reversible by chronic but not acute treatment with antidepressant drugs (good predictive validity).
8.4 8.4 Gene—Environment Interaction: Developing a new Animal Model of Depression
The pathogenesis of depression is still essentially unknown. Genetic epidemiological studies found no evidence of the classic Mendelian inheritance in humans but showed that interaction between multiple genes of modest effect and environmental factors (G × E) confers vulnerability to the disease (Lesch, 2004). Environmental risk factors include maternal stress during pregnancy, deprivation of parental care during childhood, childhood neglect or childhood abuse, parental loss, substance abuse, toxic exposure, and head injury (Caspi and Moffitt, 2006). Indeed, the experience of stressful events in childhood (early life stress) was found to increase the risk for the development of mood disorders in adult life (Heim and Nemeroff, 2001; Caspi et al., 2003).
We employed an innovative experimental design to reproduce in an animal model the interaction between environmental adverse events and genetic susceptibility. To reproduce early life stress events, the FSL rats and their control FRL rats were subjected to a standard maternal separation protocol from postnatal day 2 (PND-2) to PND-14 (Plotsky and Meaney, 1993). Indeed, maternal separation has been shown to induce HPA axis alteration such as increase in both adrenocorticotropic hormone and corticosterone plasma levels (Vazquez et al., 2000). Moreover, rats subjected to maternal separation exhibit behavioral abnormalities such as decrease in saccharin intake (anhedonia), increased startle response, and reduced exploration in a novel open field (Caldji et al., 2000; Ladd et al., 2000).
We began recently to characterize this new animal model of depression behaviorally and biochemically. It has been shown that FSL rats subjected to maternal separation have lower body weight and higher immobility time in the Porsolt swim test than FRL maternally separated rats. Indeed, lower body weight and lower motor activity are two features that can be found in depressed individuals. Moreover, treatment with escitalopram (ESC) was found to increase swim duration in FSL and FSL subjected to maternal separation but not in FRL (El Khoury et al., 2006). A strong inhibition of long-term potentiation induction (a cellular model of synaptic plasticity) and marked reduction of N-methyl-D-aspartate (NMDA) receptor synaptic expression were found in the hippocampus of the FSL rats. Early life stress exacerbated depressive behavioral phenotype and enhanced synaptic plasticity only in FSL rats. This was accompanied by marked increase of synaptic NMDA receptors, abnormal regulation of presynaptic mechanisms, altered response of synaptic extracellular signal-regulated mitogen-activated protein kinases, and reduced behavioral response to antidepressant treatment (Musazzi et al., submitted). These data suggest that early gene—environment interactions cause lifelong synaptic changes in FSL by inducing maladaptive plasticity, affecting both the course of depressive-like behavior and response to drugs.
8.5 8.5 Optimization of Synaptoproteomic Analysis
Two-dimensional (2D) gel electrophoresis coupled with mass spectrometry is a powerful technique that allows the separation and identification of thousands of proteins in a single experiment. 2D electrophoresis consists of two sequential separation steps: the first dimension resolves proteins on the basis of their isoelectric point (isoelectric focusing, IEF) and the second dimension separates the focused proteins on the basis of their molecular mass by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). In the last decade, both the development of specific software able to simultaneously analyze thousands of proteins in many gels and the improvement of mass spectrometer sensitivity, boosted the proteomic technology potentials. Nevertheless, the number of proteins expressed in a particular cell or tissue has been estimated to be more than 100,000 (including post-translational variants), making the separation and identification of all the proteins in the sample problematic. Moreover, the presence of highly expressed proteins such as cytoskeletal or housekeeping proteins could mask the detection of proteins expressed at a lower level. A strategy to reduce the complexity of the proteome and to enrich for less abundant proteins is to prefractionate the sample examined (Fountoulakis, 2004; Stasyk and Huber, 2004; Vercauteren et al., 2004).
Synaptosomes are isolated nerve endings obtained by differential centrifugation of brain tissue. They contain the presynaptic terminal, including mitochondria and synaptic vesicles, and part of the postsynaptic site, including the postsynaptic density. Therefore, this subcellular fraction is greatly enriched in proteins involved in synaptic functions. Furthermore, the main biological targets of antidepressant drugs are located at synaptic sites. A few proteomics studies have been carried out on subcellular synaptic fractions (Satoh et al., 2002; Li et al., 2004; Liberatori et al., 2004; Boyd-Kimball et al., 2005; Witzmann et al., 2005; Mello et al., 2007), but noone has thus far attempted the characterization of the synaptic proteome in animal models of depression.
Here we give an overview of the 2D electrophoresis protocol that we are currently using in our laboratory to perform synaptoproteomic studies.
8.5.1 8.5.1 Sample Preparation
Synaptosomes were purified by discontinuous Percoll gradient procedure according to Dunkley and colleagues (Dunkley et al., 1986) with minor modifications (Bonanno et al., 2005). This technique has several advantages over the Ficoll or sucrose gradient methods, i.e., isotonicity can be maintained throughout the isolation; synaptosomes are free of myelin and extrasynaptosomial mitochondria and are functional, viable, and characterized as protein markers (Dunkley et al., 1986; Raiteri and Raiteri, 2000, Barbiero et al., 2007). Briefly, pooled hippocampi (HC) and prefrontal/frontal cortex (P/FC) were homogenized in 10 vol of 0.32 M sucrose, and buffered at pH 7.4 with Tris, using a glass/teflon tissue grinder (clearance, 0.25 mm). The homogenate was centrifuged at 1000g to remove nuclei and debris, and the supernatant was gently stratified on a discontinuous Percoll gradient (2, 6, 10, and 20% v/v in Tris-buffered sucrose) and centrifuged at 33,500g for 5 min. The layer between 10 and 20% Percoll (synaptosomal fraction) was collected and washed by centrifugation and the resulting pellet was stored at −80°C.
Transmembrane proteins or membrane-associated proteins present in the synaptosomal fraction are difficult to solubilize. Therefore, to ensure optimal protein solubilization and denaturation, and to minimize horizontal streaking due to protein—protein interaction, synaptosome pellets were dissolved in an IEF buffer containing 7 M urea, 2 M thiourea, 40 mM Tris, 3 mM tributylphosphine, 2% CHAPS, and 1% carrier ampholytes plus protease inhibitors (Carboni et al., 2002). Indeed, in order to obtain solubilization of cytosolic and membrane proteins in a single step, we decided to use CHAPS, a surfactant that performed better than others employed in different buffers. Protein content was measured by BCA assay (Pierce Chemical, Rockford, IL, USA) after dialysis in 1% SDS in water. Salts, small ionic molecules, or other contaminants present in the synaptosomes can potentially interfere with the first-dimension focusing. If necessary, the sample can be subjected to an additional desalting step prior to IEF by means of protein precipitation (2D Clean-Up Kit, GE Healthcare), following the manufacturer's instructions.
8.5.2 8.5.2 Isoelectric Focusing
Immobilized pH gradient (IPG) strips are made by acrylamide derivatives containing either a free carboxylic acid or a tertiary amino group that is copolymerized with acrylamide and bis-acrylamide and immobilized on a plastic sheet. Therefore, the pH gradient is stable and allows better reproducibility between experiments. Since 70% of brain proteins have an isoelectric point (pI) between 3 and 10, with most having a pI range of 4–7, non-linear-gradient pH 3–10 IPG strips were chosen, in contrast to some other groups, to allow better resolution of proteins with pI values between pH 4 and 7. Moreover, we chose to employ 7 cm IPG strips, because this shorter length allows the loading of smaller amounts of synaptosomes without compromising protein resolution. Therefore, a 7-cm nonlinear IPG strip (Bio-Rad) of pH 3–10 was rehydrated for 16 h with 125 μL of IEF buffer containing 115 μg of synaptosomes, 10 mM iodoacetamide as alkylating agent, and a trace of bromophenol blue. IEF was performed at 15°C at a maximum of 4000 V for a total of 28,000 Vh using a Protean IEF Cell (Bio-Rad). The Protean IEF Cell allows the isoelectric focusing of up to 12 strips at the same time. The voltage was increased stepwise from 100 to 4000 V (see Table 8.3 for running conditions).
8.5.3 8.5.3 SDS-PAGE and Gel Staining
Prior to the second dimension, the IPG strips were equilibrated for 25 min in a solution containing 6 M urea; 2% SDS; 375 mM Tris pH 8.8; and 4 mM tributylphosphine. A multiple mini vertical slab gel system (Bio-Rad's Mini-PROTEAN 3 Dodeca Cell) was chosen to perform the SDS-PAGE because it allows the analysis of small amounts of samples while ensuring protein spot resolution. An 8–18%T gradient polyacrylamide gel was used in order to obtain better resolution of proteins ranging 10–250 kD. After the equilibration step, the IPG strips were placed on top of the gradient gels and sealed with 0.5% agarose in running buffer (192 mM glycine, 15 mM Tris, 0.1% SDS plus bromophenol blue). Gels were run at a constant temperature of 15°C at 5 mA per gel for 1 h and 10 mA per gel until the bromophenol blue front reached the bottom of the gel.
Gels with synaptosomal proteins resolved by 2D electrophoresis were then fixed in 10% ethanol and 7% acetic acid for 4 h. SYPRO Ruby fluorescent dye was used to stain proteins in the gels. Indeed, in contrast to the other staining methods, SYPRO Ruby gives very little background is very sensitive (1 ng of protein), and the staining is linear over 3 orders of magnitude. The result is a significant improvement in terms of resolution and number of spots of the maps. The gels were stained for 20 h with SYPRO Ruby and destained for 1 h in a 10% ethanol and 7% acetic acid solution. Afterwards, gels were stored in distilled water until protein spots were excised from the gel.
8.5.4 8.5.4 Two-dimensional Image Analysis
Gel images were acquired by using the Quantity One software and the VersaDoc imaging system (Bio-Rad). Spot detection and map comparison (matchset analysis) were carried out with the PDQuest 7.3 software (Bio-Rad). In brief, the PDQuest software is able to automatically detect protein spots in the acquired 2D maps.
The software creates a set of three images: the original gel image (Figs. 8.1 and 8.2 ), the filtered image, and the Gaussian image (a synthetic image in which each spot has been remodeled to fit a Gaussian curve, thus representing an ideal spot). These images were then landmarked and matched to allow the comparison of protein expression patterns across the experimental samples (Figs. 8.3 and 8.4 ). Tables 8.4 and 8.5 summarize the analysis of spot-matching between 2D gels from different experimental groups. Statistical analysis (t-test) was carried out by PDQuest software to compare replicate groups and identify sets of proteins that show statistically significant difference with a confidence level of 0.05.
8.6 8.6 Experimental Design and Results in the LH-nLH Animal Model
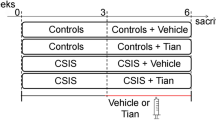
In the study on LH-nLH animal model of depression, we investigated, by using synaptoproteomics, the basal synaptic difference between LH and nLH rats and the changes in the proteome induced by antidepressant treatment in the two animal groups. Both LH and nLH rats were also subchronically treated for 6 days with the selective serotonin reuptake inhibitor ESC (25 mg kg−1 day−1). Thus four different experimental groups were prepared:
-
1.
LH + vehicle
-
2.
nLH + vehicle
-
3.
LH + ESC
-
4.
nLH + ESC
On the last day of drug administration the animals were sacrificed and HC and P/FC were dissected. After preparation of synaptosomes from these brain regions, the “synaptic map” from each group was obtained using the experimental procedures described above (Fig. 8.2). Three comparisons were then performed:
-
1.
(LH + vehicle) vs. (nLH + vehicle)
-
2.
(LH + ESC) vs. (LH + vehicle)
-
3.
(nLH + ESC) vs. (nLH + vehicle)
Maps of both brain areasP/FC and HC showed a considerable number of protein spots whose expression levels were differently regulated in the two ratLH and nLH groups and after antidepressant treatment (Table 8.5).
As an example, with regard to HC synaptosomes (Figs. 8.3 and 8.4) we found 11 proteins differently expressed between basal LH and nLH groups. In detail, nine spots were upregulated and two were downregulated in LH compared to nLH. Following antidepressant treatment, the expression levels of 17 synaptic proteins were modified in LH rats (eight upregulated and nine downregulated). Moreover, 10 proteins were modified by drug administration in nLH rats (six upregulated and only two downregulated by ESC) (Table 8.6 ).
Interestingly, we observed that three proteins that were differently regulated between basal LH and nLH were also modulated by the pharmacological treatment. Similar results were found in the P/FC area.
8.7 8.7 Experimental Design and Results in FSL/FRL Animal Models
To mimic early life adverse events, FSL/FRL rats were subjected to a standard maternal separation protocol according to Plotsky and Meaney (1993). Briefly, FSL/FRL pups and dam were removed from the home cage, and the pups were placed in two distinct temperature-controlled cage (30°C) incubators in separate rooms so that no (ultrasonic) communication was possible between the dam and pups. The separation procedure lasted from PND-2 to PND-14 for 3 h daily. At the end of the separation period, both pups and dam were put back in the home cage. FSL/FRL pups were then left undisturbed with their mother until PND-23 when they were weaned.
Treatment with the selective serotonin reuptake inhibitor ESC (25 mg kg−1 day−1) was carried out from PND-43 to PND-73 by using drug-containing dietary chow. A week before the end of the treatment, rats were subjected to the Porsolt test to check for drug efficacy (El Khoury et al., 2006). At PND-73, P/FC and HC regions were removed and synaptosomes prepared. The experimental groups were as follows:
-
1.
FSL rats treated with vehicle (FSL veh)
-
2.
FRL rats treated with vehicle (FRL veh)
-
3.
FSL rats subjected to ESC treatment (FSL ESC)
-
4.
FSL rats subjected to maternal separation (FSL MS)
-
5.
FRL rats subjected to maternal separation (FRL MS)
-
6.
FSL rats subjected to maternal separation and ESC treatment (FSL MS ESC)
Proteomic analysis was performed for selected comparisons (Tables 8.6–8.8) and high-quality protein maps were generated (Fig. 8.4).
8.7.1 8.7.1 Results of Synaptoproteomics of Prefrontal/Frontal Cortex
Statistical analysis of 2D maps from P/FC synaptosomes revealed 37 proteins differently regulated in basal FSL vs. FRL rats. Stress of maternal separation significantly dysregulated 48 proteins in FSL and 24 proteins in FRL P/FC synaptosomes. In basal FSL the chronic ESC treatment differently regulated 33 protein spots. In FSL subjected to the stress of maternal separation, chronic ESC treatment modulated seven spots (Table 8.7 ).
Interestingly, in FSL twice as many proteins as in FRL were dysregulated by maternal separation, thereby suggesting an increased basal vulnerability of FSL rats to early life stress. In addition, three proteins downregulated by maternal separation in FSL were upregulated by the subsequent treatment with ESC. Indeed, these three proteins, regulated in opposite directions by maternal separation and antidepressant treatment, may represent biomarkers of both vulnerability to stress and response to drug. Therefore, these three proteins (or respective pathways involved) could represent putative targets for the development of novel drugs with antidepressant action. Protein spots differently regulated in FSL vs. FRL by either treatment or maternal separation were excised from gels and submitted to identification by mass spectrometry.
8.7.2 8.7.2 Results of Synaptoproteomics of Hippocampus
HC synaptosomes were prepared from the same animal groups. 2D maps were generated and analyzed as before. Statistical analysis revealed 15 protein spots differently regulated in FSL vs. FRL rats. ESC regulated 13 spots in basal FSL. Early life stress regulated 17 spots in FRL and 14 in FSL; subsequent pharmacological treatment with ESC regulated in the FSL rats maternally separated, 12 protein spots (Table 8.8).
Interestingly, fewer protein spots were significantly up- or downregulated in the different experimental groups: fewer in HC compared to P/FC synaptosomes. Contrary to P/FC, stress of maternal separation differently regulated in HC almost the same number of proteins in FRL and FSL. However, similar to the results obtained in P/FC, maternal separation regulated different sets of proteins in the two rat lines.
8.8 8.8 In silico Analysis of Protein Pathways
The results obtained by synaptoproteomic studies in both animal models will be further analyzed using available bioinformatics platforms. These “in silico” tools allow investigation of gene ontology (GO) terms and help to derive more information about potential cellular pathways that may be modified by stress susceptibility or altered by antidepressant treatments. Several software are available online, among them FatiGO (www.babelomics.org), Ingenuity Pathways Analysis (IPA) (www.ingenuity.com), and DAVID (Database for Annotation, Visualization and Integrated Discovery) (http://david.abcc.ncifcrf.gov/home.jsp).
The main aim of these pathway analysis studies will be a functional classification of the proteins according to GO terms, disease terms, and signaling pathways, in order to understand the biological processes operating behind them using different procedures to scan an apparently disjointed list of proteins.
FatiGO is an application used to find GO terms that are over- and under-represented in a set of proteins and therefore characterizes the biological processes responsible for their differential expressions (Al-Shahrour et al., 2004; Al-Shahrour et al., 2005).
IPA software system is one of the most complete tools that allow obtaining potential pathways theoretically modified by experimental procedures by analyzing protein changes in the experiments. The program is able to generate networks of proteins that are ranked with a score to facilitate data interpretation. The score allows estimating the level of the result obtained, to plan a following trial. Recently, this application was used by Wishart et al. to elaborate the results obtained by 2D-electrophoresis analysis of synaptic proteins isolated from mouse striatum (Wishart et al., 2007).
DAVID is an in silico discovery system developed to facilitate the analysis of large lists of proteins to highlight the potential correlations concerning functional classification, biochemical pathway maps, and conserved protein domains. This web-accessible application operates similar to FatiGO, but additionally allows the simultaneous view of different annotations regarding biological processes, molecular function, and cellular components (Dennis et al., 2003).
8.9 8.9 Summary and Perspectives
It is becoming apparent that proteomic analysis of synaptic terminal fractions, with its powerful investigative potential of the neurotransmitter release machinery and synaptic function, performed on animal models with different combinations of genetic background and environmental manipulation, may represent an unprecedented opportunity for research on biomarkers in psychiatric disorders. Recently, we observed an increased interest in the field of biomarker research, owing to their potential use in early diagnosis and prognosis of disease, and in the development of new drug targets. Moreover, biomarkers could provide clinicians with the means to tailor a pharmacological therapy to an individual patient on the basis of the particular endophenotype or vulnerability to adverse effects. Therefore, validated biomarkers could greatly reduce both the individual suffering and the enormous costs of the affective disorders to society. In particular, biomarkers associated with mood disorders are needed, as diagnoses are mainly based on phenotypical categorizing signs and symptoms. A biomarker may be a genetic trait or a biochemical change, such as the expression of proteins, and their study could be accomplished by using different methods or strategies. In this chapter, we have described one of these strategies, consisting of synaptoproteomics applied to different animal models of depression. LH and FSL are, as described above, two well-validated models that replicate symptoms of the pathology. Both animal models were treated with the same antidepressant and their synaptoproteomic profile was characterized to identify molecular correlates of pathology and response to drug and the resulting altered neurobiological pathways. Using this approach, we plan to obtain at the same time information concerning biomarkers that may be used as predictors of pathology and new targets of drug action, available for detection of treatment efficacy as well as for development of novel pharmacological and therapeutic strategies. Moreover, this approach, in combination with functional proteomics and trascriptomics, could provide a powerful strategy for the identification of the complex pathophysiological mechanisms in mood disorders. Indeed, proteome and transcriptome analyses could suggest gene and protein clusters corresponding to the pathological phenotype (“trait biomarkers”) and clusters corresponding to modification of phenotype by treatment (“state biomarkers”).
Abbreviations
- 2D:
-
Two-dimensional
- 5-HT1A :
-
Serotonin 1A receptor
- BCA:
-
Bicinchoninic Acid
- CHAPS:
-
(3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate)
- DA:
-
Dopamine
- DAVID:
-
Database for Annotation, Visualization and Integrated Discovery
- ESC:
-
Escitalopram
- FRL:
-
Flinders sensitive line
- FSL:
-
Flinders resistant line
- G × E:
-
Gene—environment interaction
- GO:
-
Gene Ontology
- HC:
-
Hippocampus
- HPA:
-
Hypothalamic—pituitary—adrenal
- IBS:
-
Inflammatory bowel syndrome
- IEF:
-
Isoelectric focusing
- IPA:
-
Ingenuity pathways analysis
- IPG:
-
Immobilized pH gradient
- LH:
-
Learned helplessness
- mRNA:
-
Messenger RNA
- MS:
-
Maternal separation
- nLH:
-
Non-learned helplessness
- NMDA:
-
N-methyl-D-aspartate
- P/FC:
-
Prefrontal/frontal cortex
- pI:
-
Isoelectric point
- PND:
-
Post natal day
- REM:
-
Random eyes movement
- SDS:
-
Sodium dodecyl sulfate
- SDS-PAGE:
-
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- Tris:
-
Tris(hydroxymethyl)aminomethane
- Veh:
-
Vehicle
References
Al-Shahrour F, Diaz-Uriarte R, Dopazo J (2004) FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics 20:578–580
Al-Shahrour F, Minguez P, Vaquerizas JM, Conde L, Dopazo J (2005) BABELOMICS: a suit of web tools for functional annotation and analysis of groups of genes in high-throughput experiments. Nucleic Acids Res 33:W460–W464
Anderson L, Seilhamer J (1997) A comparison of selected mRNA and protein abundances in human liver. Electrophoresis 18:533–537
Barbiero VS, Giambelli R, Musazzi L, Tiraboschi E, Tardito D, Perez J, Drago F, Racagni G, Popoli M (2007) Chronic antidepressants induce redistribution and differential activation of alphaCaM Kinase II between presynaptic compartments. Neuropsychopharmacology advance online publication, 14 March 2007; doi:10.1038/sj.npp.1301378
Bonanno G, Giambelli R, Raiteri L, Tiraboschi E, Zappettini S, Musazzi L, Raiteri M, Racagni G, Popoli M (2005) Chronic antidepressants reduce depolarization-evoked glutamate release and protein interactions favoring formation of SNARE complex in hippocampus. J Neurosci 25:3270–3279
Boyd-Kimball D, Castegna A, Sultana R, Poon HF, Petroze R, Lynn BC, Klein JB, Butterfield DA (2005) Proteomic identification of proteins oxidized by Abeta(1–42) in synaptosomes: implications for Alzheimer's disease. Brain Res 1044:206–215
Caldji C, Francis D, Sharma S, Plotsky PM, Meaney MJ (2000) The effects of early rearing environment on the devepopment of GABAA and central benzodiazepine receptor levels and novelty-induced fearfulness in the rat. Neuropsychopharmacology 22:219–229
Caspi A, Moffitt TE (2006) Gene-environment interactions in psychiatry: joining forces with neuroscience. Nat Rev Neurosci 7:583–590
Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, McClay J, Mill J, Martin J, Braithwaite A, Poulton R (2003) Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science 301:386–389
Carboni L, Piubelli C, Righetti PG, Jansson B, Domenici E (2002) Proteomic analysis of rat brain tissue: comparison of protocols for two-dimensional gel electrophoresis analysis based on different solubilizing agents. Electrophoresis 23:4132–4141
Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA (2003) DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4:P3
Dunkley PR, Jarvie PE, Heath JW, Kidd GJ, Rostas JA (1986) A rapid method for isolation of synaptosomes on Percoll gradients. Brain Res 372:115–129
El Khoury A, Gruber SHM, Mϕrk A, Mathé AA (2006) Adult life behavioral consequences of early maternal separation are alleviated by escitalopram treatment in a rat model of depression. Prog Neuropsychopharmacol Biol Psychiatr 30:535–540
Fountoulakis M (2004) Application of proteomiics technologies in the investigation of the brain. Mass Spectrom Rev 23:231–258
Heim C, Nemeroff CB (2001) The role of childhood trauma in the neurobiology of mood and anxiety disorders: preclinical and clinical studies. Biol Psychiatr 49:1023–1039
Henn FA, Edwards E, Anderson D, Vollmayr B (2002) Psychotherapy and antidepressant treatment of depression: evidence for similar neurobiological mechanism. World Psychiatr 1:115–117
Hitzemann R (2000) Animal models of psychiatric disorders and their relevance to alcoholism. Alcohol Res Healt 3:149–158
Janowsky DS, Risch SC, Parker D, Huey L, Judd L (1980) Increased vulnerability to cholinergic stimulation in affect disorder patients. Psychopharmacol Bull 16:29–31
Janowsky DS, Overstreet DH, Nurnberger JI Jr (1994) Is cholinergic sensitivity a genetic marker for the affective disorders? Am J Med Genet 54:335–344
Keller MB, Boland RJ (1998) Implications of failing to achieve successful long-term maintenance treatment of recurrent unipolar major depression. Biol Psychiatr 44:348–360
Kramer R, Cohen D (2004) Functional genomics to new drug targets. Nat Rev Drug Discov 3:965–72
Ladd CO, Huot RL, Thrivikraman KV, Nemeroff CB, Meaney MJ, Plotsky PM (2000) Long-term behavioural and neuroendocrine adaptations to adverse early experience. Prog Brain Res 122:81–103
Lesch KP (2004) Gene-environment interaction and the genetics of depression. J Psychiatr Neurosci 29:174–84
Li KW, Hornshaw MP, Van Der Schors RC, Watson R, Tate S, Casetta B, Jimenez CR, Gouwenberg Y, Gundelfinger ED, Smalla KH, Smit AB (2004) Proteomics analysis of rat brain postsynaptic density. Implications of the diverse protein functional groups for the integration of synaptic physiology. J Biol Chem 279:987–1002
Liberatori S, Canas B, Tani C, Bini L, Buonocore G, Godovac-Zimmermann J, Mishra OP, Delivoria-Papadopoulos M, Bracci R, Pallini V (2004) Proteomic approach to the identification of voltage-dependent anion channel protein isoforms in guinea pig brain synaptosomes. Proteomics 4:1335–1340
Matthews K, Christmas D, Swan J, Sorrel E (2005) Animal models of depression: navigating through the clinical fog. Neurosci Biobehav Rev 29:503–513
Mello CF, Sultana R, Piroddi M, Cai J, Pierce WM, Klein JB, Butterfield DA (2007) Acrolein induces selective protein carbonylation in synaptosomes. Neuroscience 147:674–679
Musazzi L, Ryan B, Barbiero VS, Mallei A, Giambelli R, Tardito D, Gruber SHM, El Khoury A, Andersson W, Anwyl R, Racagni G, Mathé AA, Rowan MJ, Popoli M Early-life stress enhances NMDA receptor-dependent synaptic plasticity in a gene-environment rat model of depression. Submitted Proc Natl Acad Sci USA 2007
Overmier JB, Seligman ME (1967) Effects of inescapable shock upon subsequent escape and avoidance responding. J Comp Physiol Psychol 63:28–33
Overstreet DH, Russell RW (1982) Selective breeding for diisopropyl fuorophosphate-sensitivity: behavioural effects of cholinergic agonists and antagonists. Psychopharmacology (Berl.) 78:150–155
Overstreet DH, Friedman E, Mathé AA, Yadid G (2005) The Flinders Sensitive Line rat: a selectively bred putative animal model of depression. Neurosci Biobehav Rev 29:739–759
Paulson L, Martin P, Persson A, Nilsson CL, Ljung E, Westman-Brinkmalm A, Eriksson PS, Blennow K, Davidsson P (2003) Comparative genome- and proteome analysis of cerebral cortex from MK-801-treated rats. J Neurosci Res 71:526–533
Plotsky PM, Meaney MJ (1993) Early, postnatal experience alters hypothalamic corticotropin-releasing factor (CRF) mRNA, median eminence CRF content and stress-induced release in adult rats. Brain Res Mol Brain Res 18:195–200
Pucilowski O, Overstreet DH, Rezvani AH, Janowsky DS (1993) Chronic mild stress-induced anhedonia: greater effect in a genetic rat model of depression. Physiol Behav 54:1215–1220
Raiteri L, Raiteri M (2000) Synaptosomes still viable after 25 years of superfusion. Neurochem Res 25:1265–1274
Rohlff C, Hollis K (2003) Modern proteomic strategies in the study of complex neuropsychiatric disorders. Biol Psychiatr 53:847–853
Russel RW, Overstreet DH (1987) Mechanisms underlying sensitivity to organophosphororus anticholinesterase compounds. Prog Neurobiol 28:97–129
Stasyk T, Huber LA (2004) Zooming in: fractionation strategies in proteomics. Proteomics 4:3704–3716
Satoh K, Takeuchi M, Oda Y, Deguchi-Tawarada M, Sakamoto Y, Matsubara K, Nagasu T, Takai Y (2002) Identification of activity-regulated proteins in the postsynaptic density fraction. Genes Cells 7:187–197
Urani A, Chourbaji S, Gass P (2005) Mutant mouse models of depression: candidate genes and current mouse lines. Neurosci Biobehav Rev 29:805–828
Vazquez DM, Lopez JF, Van Hoers H, Watson SJ, Levine S (2000) Maternal deprivation regulates serotonin 1A and 2A receptors in the infant rat. Brain Res 855:76–82
Vercauteren FG, Bergeron JJ, Vandesande F, Arckens L, Quirion R (2004) Proteomic approaches in brain research and neuropharmacology. Eur J Pharmacol 500:385–398
Vollmayr B, Henn FA (2001) Learned helplessness in the rat: improvements in validity and reliability. Brain Res Protoc 8:1–7
Vollmayr B, Henn FA (2003) Stress models of depression. Clin Neurosci Res 3:245–251
Vollmayr B, Simonis C, Weber S, Gass P, Henn FA (2003) Reduced cell proliferation in the dentate gyrus is not correlated with the development of learned helplessness. Biol Psychiatry 54:1035–1040
Wasinger VC, Cordwell SJ, Cerpa-Poljak A, Yan JX, Gooley AA, Wilkins MR, Duncan MW, Harris R, Williams KL, Humphery-Smith I (1995) Progress with gene-product mapping of the Mollicutes: Mycoplasma genitalium. Electrophoresis 16:1090–1094
Willner P (1990) Animal models of depression: an overview. Pharmacol Ther 45:425–455
Willner P (1995) Animal models of depression: validity and applications. Adv Biochem Psychopharmacol 49:19–41
Wishart TM, Paterson JM, Short DM, Meredith S, Robertson KA, Sutherland C, Cousin MA, Dutia MB, Gillingwater TH (2007) Differential proteomic analysis of synaptic proteins identifies potential cellular targets and protein mediators of synaptic neuroprotection conferred by the slow Wallerian degeneration (Wld) gene. Mol Cell Proteomics 8:1318–1330
Witzmann FA, Arnold RJ, Bai F, Hrncirova P, Kimpel MW, Mechref YS, McBride WJ, Novotny MV, Pedrick NM, Ringham HN, Simon JR (2005) A proteomic survey of rat cerebral cortical synaptosomes. Proteomics 5:2177–2201
Yadid G, Nakash R, Deri I, Tamar G, Kinor N, Gispan I, Zangen A (2000) Elucidation of the neurobiology of depression: insights from a novel genetic animal model. Prog Neurobiol 62:353–378
Acknowledgments
This work was funded by a European Union (6th Framework Program) grant for project GENDEP (contract no. LSHB-CT-2003-503428) and by the Swedish Medical Research Council grant 10414 (to AAM).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2008 Springer Science+Business Media, LLC
About this chapter
Cite this chapter
Mallei, A. et al. (2008). Synaptoproteomics of Existing and new Animal Models of Depression. In: Turck, C. (eds) Biomarkers for Psychiatric Disorders. Springer, Boston, MA. https://doi.org/10.1007/978-0-387-79251-4_8
Download citation
DOI: https://doi.org/10.1007/978-0-387-79251-4_8
Published:
Publisher Name: Springer, Boston, MA
Print ISBN: 978-0-387-79250-7
Online ISBN: 978-0-387-79251-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)