Abstract
In this chapter, different encoding strategies for DNA- or PNA-encoded chemical libraries are presented. In general, DNA encoding can be distinguished into a DNA-recorded regimen, which encodes the individual chemical reaction steps, and which is most used in practice today, and a DNA-templated regimen where DNA templates guide the chemical synthesis. Various barcoding strategies have been employed most often for the generation of split-and-pool- or DNA-templated single-pharmacophore DELs, but also for dual-pharmacophore DELs displaying fragments/compounds on both ends of a DNA heteroduplex, both in a static and a dynamic setup. In addition to solution-phase protocols, encoded one-bead-one-compound libraries and solid phase-initiated DNA-encoded library synthesis strategies are reported.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- DNA-encoded libraries
- DNA-routing
- DNA-templated synthesis (DTS)
- Double-stranded DEL
- Dual-pharmacophore DEL
- Dynamic combinatorial DELs
- Encoded combinatorial chemistry
- Encoded self-assembled chemical (ESAC) libraries
- Encoded solid-phase chemistry
- Genetic barcode
- Genetic tag
- One-bead-one-compound libraries
- PNA-encoded libraries
- Single-pharmacophore DEL
- Single-stranded DEL
- Splint ligation
- T4 ligation
- yoctoReactor
1 Introduction: Encoding Molecules with Genetic Tags
Encoded chemical library technologies are based on the covalent linkage of chemically synthesized compounds to synthetic oligonucleotides that serve as barcode identifiers for the compound structure (Fig. 1a). This approach to screening library handling is commonly called phenotype–genotype coupling. It has its biological precedent in the phage-display, ribosome-display, or mRNA-display libraries which are widely used for screening for bioactive peptides and proteins. Like their biological relatives, genetically tagged chemically synthesized libraries can be screened for bioactive compounds on targets as complex mixtures, without the need for the large infrastructure needed to screen discrete libraries of compounds (high-throughput screening/HTS).
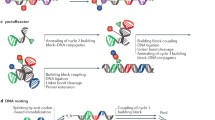
Solution-phase synthesis of DNA-encoded libraries (DEL) by iterative cycles of chemical synthesis and barcode ligation. (a) Schematic presentation of a DNA-encoded molecule. (b) Split-and-pool encoded combinatorial chemistry workflow. (c) Structure and sequence of the “headpiece” DNA oligonucleotide. (d) Generic scheme of the synthesis of a DNA-encoded library initiated with the short hairpin duplex “headpiece” DNA
A seminal publication by Richard A. Lerner and Sydney Brenner in 1992 described for the first time the concept to synthesize molecular chimeras consisting of a genetic tag that was covalently connected to a chemically synthesized molecule [1]. In those early times of encoded chemistry, libraries were synthesized on beads that contained two linkers, one allowed for chemical coupling of amino acid building blocks by peptide coupling, the other for chemical coupling of DNA nucleotides by phosphoramidite nucleic acid chemistry [2, 3]. Coupling of amino acid building blocks and of DNA nucleotide building blocks was performed iteratively, and in combinatorial manner, so that each cycle of synthesis and encoding led to an exponential growth of the library, yet the different chemistries need for peptide synthesis and for phosphoramidite synthesis posed serious constraints to the initial setup. Since the infant days of encoded combinatorial chemistry, the field of encoded library technologies has branched out, and today several barcoding strategies are available for the synthesis of encoded libraries, which will be discussed in this chapter. They can broadly be divided into DNA-recorded techniques where the genetic tag serves to record the synthetic steps and techniques that use the DNA barcode for templating chemical reactions [4]. The class of DNA-recorded libraries comprises split-and-pool encoded solution-phase combinatorial synthesis [5,6,7], solid phase-initiated DNA-encoded combinatorial chemistry, and DNA-encoded solid-phase chemistry [8], as well as hybrid DNA/PNA-encoded chemistry [9]. Further techniques that are based on recording reactions with DNA are dual-pharmacophore technologies and DNA-encoded dynamic combinatorial chemistry which enable both fragment screening and screening of complex molecules [10]. DNA-templated approaches comprise DNA-templated DEL synthesis which were mainly used for the construction of macrocyclic structures, and the yoctoReactor (TM) design, both using the DNA templates to direct the oligonucleotide-linked reagents into proximity to the reaction site within complex mixtures [11, 12]. Alternatively, DNA/PNA hybrid duplex structures may be used in templated reaction strategies [9]. Notably, also the DNA-routing approach uses immobilized DNA strands to direct DNA templates to individual reaction vessels [13]. The use of single-stranded designs further enables the pairing with additional modular functionalities which can be exploited for advanced DEL selection strategies [1], such as photocrosslinking [2], affinity maturation of existing ligands [3], or the use of covalent or reversible-covalent warheads [4]. Such strategies may facilitate the identification of valuable compounds that bind to a biological target and serve as a starting point for drug development programs.
2 Split-and-Pool Encoded Solution-Phase Combinatorial Chemistry
Split-and-pool encoded solution-phase combinatorial chemistry, which today is the most used approach to construct DNA-encoded libraries, was first described in a publication in 1995 that described the combinatorial enzymatic ligation of DNA codes (Fig. 1) [14]. Seminal publications from the Neri group (ETH Zurich), and from a team headed by Barry Morgan, then at Praecis, described the first split-and-pool encoded library approaches [15, 16]. Morgan et al. designed a double-stranded DNA to encode compounds [15]. The overall architecture of their barcode is shown in Fig. 1a. It consisted of the encoded compound that was connected via a polyethylene glycol chain to both terminal nucleotides of the duplex DNA, forming a hairpin structure. Terminal primer regions of the DNA construct allowed for amplification of the genetic information by polymerase chain reaction (PCR). Suitable primer sequences have been published by different research groups [15, 16]. Alternatively, validated primers from genomics research, SELEX, or phage-display may be used, if researchers entering the DEL field wish to use other primer sequences. The number of internal degenerate 8–10 mer regions that contained the compound barcode mirrors the number of chemical reaction steps, and the number of barcodes per reaction step mirrors the number of chemical building blocks. Barcode sequences may be designed using freely available statistics software such as R, in order to avoid long homopurine or homopyrimidine sequences that may lead to sequencing artifacts and to define the Hamilton distance of barcodes. The Hamilton distance indicates the minimal number of different nucleosides between any two barcodes of a coding region. It is advisable to set the Hamilton distance to at least two for unambiguous compound identification by sequencing. The design of smaller libraries and/or longer coding regions would allow for a higher Hamilton distance. In the coding scheme published by Morgan et al. [16] any barcode sequences were connected by two-base regions that are needed for enzymatic barcode ligation with T4 ligase.
The workflow of encoded library synthesis was initiated with a short hairpin duplex DNA, called the “headpiece” that contained a polyethylene linker connecting the two arms of the hairpin (Fig. 1c). This linker carries a functional group for attaching the encoded molecule. The hairpin consisted of a duplex DNA sequence of six nucleotides and a two-base overhang that allowed for ligating a set of duplex (ds)DNA oligonucleotides with T4 ligase. This set of dsDNA oligonucleotides contained forward primer sequence, barcodes for the first set of chemical building blocks, and a two-base overhang for the second sticky-end ligation. Following barcode ligation, chemical building blocks were coupled to the linker part of the DNA constructs. As the linker moiety most often contained a primary amino group as a functional handle, any building blocks need to be substituted with a carboxylic acid, and at least one further functional group for subsequent encoded compound synthesis steps. In order to diversify the reagent space in this first cycle of library synthesis, alternative alkyl linkers were developed that contained a leaving group for substitution by nucleophiles [17]. Furthermore, chloroacetic acid or allyl-protected glycine was coupled to the linker, which allowed for synthesis of secondary amines from abundantly available primary amines and aldehydes, respectively [18, 19]. This process of encoded compound synthesis is called a cycle of encoded chemistry and concluded with a purification and pooling step (Fig. 1b). The last step is usually carried out by precipitation of the DNA conjugate from ethanolic solution. The cycles of synthesis and encoding are iterated as defined by library design. In the publication by Clark et al. [16], three and four cycles of encoded library synthesis have been described that yielded 7,000,000 and 800,000,000 encoded molecules, respectively (Fig. 1d). Libraries undergoing more combinatorial cycles have been synthesized to reach higher compound numbers. A library of encoded macrocycles was synthesized through six cycles of encoded peptide chemistry [20]. Today, libraries that are designed by three or even only two cycles [19] are increasingly being designed, likely, because higher numbers of synthesis steps tend to give rise to molecules with higher molecular weight and functional group compositions that may prove more challenging to develop toward drugs.
An alternative barcoding strategy developed by the Neri group used a long single-stranded (ss)DNA that contained a 5′-terminal linker for the encoded compound, the forward DNA primer, a compound barcode and region that was partially complementary to a set of ssDNA oligonucleotides containing the second barcode [16]. After coupling of compounds to the DNA oligonucleotides, all conjugates were pooled and split for the second cycle of encoded synthesis. Unlike the hairpin strategy that used T4 ligase and sticky-end ligation to concatenate barcode oligonucleotides prior to compound synthesis, the Neri group first performed compound synthesis, and then annealed a set of ssDNA barcode oligonucleotides to the first set of barcode ssDNA for primer elongation by Klenow fill-in (Fig. 2a-1). This strategy can be used to synthesize two-cycle libraries. A third cycle of library synthesis to reach a million-sized encoded library was demonstrated following digest of the construct with a restriction endonuclease and sticky-end ligation with T4 ligase of a third DNA barcode [21]. A more versatile encoding strategy for concatenating (theoretically) any number of ssDNA barcodes was developed by the Scheuermann and Neri groups and used 20-mer splint oligonucleotides that were partially complementary to barcode oligonucleotides. Here, the internal compound barcodes were accordingly flanked by longer 10-mer sequences that were annealed to the splint ssDNA [22, 23] (Fig. 2a-2).
All solution-phase encoded library strategies require chemical building blocks and chemical reactions which are adapted to the process. The building blocks need to be functionalized to enable the encoded compound process, and there also needs to be a diverse set of them available to enable large split sizes, e.g. numbering hundreds of building blocks. The less cycles are used for DEL design, the more important is the ready availability of a class of building blocks. Library chemistry must be compatible with aqueous solvents and with the DNA chemical reactivity. Furthermore, methodology needs to be uniformly high-yielding and to perform robustly on a broad variety of substrates. The available reactions for library design and DNA damage reactions that need to be avoided are discussed in greater detail in chapter “Advancements in DEL-Compatible Chemical Reactions” of this volume.
In order to expand the scope of DEL design, the first cycles of library synthesis may optionally be carried out on DNA sequences covalently connected to a solid-phase matrix, for instance, on controlled pore glass (CPG) (Fig. 2b). This material is a standard solid phase for automated chemical oligonucleotide synthesis and allows for working under strictly dry conditions. Features of this approach are the option to perform reactions under strictly dry conditions, DNA nucleobase protection that prevents chemical deamination, and the possibility to remove reagent excess, and especially potentially harmful contaminants such as metal ions, under stringent washing conditions. Following on-DNA compound synthesis, the DNA oligonucleotides are ligated to further DNA barcodes. The DNA oligonucleotides may be ssDNAs that contain a partially primer sequence [24], primer and barcode [25, 26], short headpiece adapters such as the hexathymidine (hexT; Fig. 2b-1) headpiece [27, 28], or barcodes that are optionally chemically stabilized (csDNA; Fig. 2b-2) for greater reaction tolerance [29]. The latter two barcoding strategies used 4-mer overhangs to concatenate barcodes [30]. Both the hexT headpiece and chemically stabilized csDNA that is composed of pyrimidine nucleobases and 7-deazaadenine have been shown to tolerate reaction conditions that are hardly compatible with native DNA such as low pH and transition metal catalysis under forcing conditions. Thus, they expand the accessible chemical reaction space for solution-phase DEL design.
3 Split-and-Pool Encoded Solid-Phase Combinatorial Chemistry
The Kodadek and Paegel groups developed DNA-barcoding strategies for one-bead-one-compound (OBOC) synthesis (Fig. 3a) [31, 32]. Their DNA-encoded solid-phase synthesis (DESPS) approach employed a functionalized linker on Tentagel macrobeads (Fig. 3b). The bifunctional linker was functionalized with an azide to install a 6-mer dsDNA headpiece “HDNA” whose two strands were connected via an azide-substituted polyethylene glycol chain via copper-catalyzed azide alkyne cycloaddition (CuAAC). Like the headpiece solution-phase DEL strategy, the DESPS headpiece contained an overhang for ligation of the forward primer and first barcode (Fig. 3a). Following enzymatic ligation of the first dsDNA, the solvent could be exchanged by simple filtration, and a first encoded compound synthesis step was performed on the second arm of the bifunctional linker. This arm contained an amine as handle for encoded compound synthesis, and additionally a chromophore and an arginine residue to facilitate compound synthesis quality control by chromatographic and mass spectrometric analysis. Like the headpiece DEL strategy, encoded OBOC libraries were synthesized by iterative split-pool combinatorial cycles of compound synthesis and enzymatic ligation of DNA barcodes, here via three-base overhangs, resulting in oligomer compounds and DNA barcodes flanked by PCR primer sites. The reaction scope for this technology takes advantage of the option to exchange aqueous solvents by organic solvents and encompasses carbonyl chemistry, as well as cross-coupling reactions, aldol reactions, and a suite of aldehyde-diversifying reactions [8]. Encoded OBOC libraries may be screened by affinity-based selection screens [32]. In the future, they may offer the option to perform functional and even phenotypic screens, after cleavage of the encoded molecules from the DNA-encoded beads [33].
4 PNA-Encoded Chemistry
Peptide-nucleic acid (PNA)-encoding strategies have been developed by the Winssinger group as an alternative to barcoding of compound synthesis with DNA [9]. Peptide-nucleic acids (PNAs) are artificially synthesized oligomers, having a backbone composed of repeating N-(2-aminoethyl)glycine units linked by peptide bonds, to which the various purine and pyrimidine bases are attached by a methylene bridge and a carbonyl group. PNA oligonucleotides specifically and strongly hybridize to complementary DNA or RNA sequences, forming duplexes that are resistant to degradation by either nucleases or proteases (Fig. 4a) [34].
Contrarily to unprotected DNA, PNA as coding oligomer does not easily undergo depurination, lacking the ribose sugar backbone and has a much higher thermal and pH-stability, enabling the use of a much broader spectrum of chemical methods for library preparation. Molecular diversity in PNA-encoded libraries can be achieved via a broad array of chemistries, including reactions catalyzed by transition metals, transformations yielding heterocyclic scaffolds, cyclization, and protic acid-promoted reactions [35, 36]. Currently, a considerable amount of peptide- and non-peptide-based PNA-encoded libraries have been reported, proving the broad applicability of the chemistry and the robustness of the encoding strategy [9].
However, the fundamental problem with PNA-encoding is that it does not benefit from the efficiency of enzymatic barcode ligation nor is the code amplifiable by PCR or PCR-like techniques. The simplest format for the identification and quantification of selection outcomes from PNA-encoded libraries generally includes direct hybridization of the selected compounds, bearing a fluorophore on each PNA tag, to a DNA microarray (Fig. 4b) [37, 38]. Nevertheless, the success of this screening method strictly depends on the concentration of PNA tags, which should be sufficient for microarray-based detection [39].
To circumvent this limitation, novel screening strategies have been developed which offer the possibility to perform indirect DNA amplification of PNA tags prior to microarray analysis (Fig. 4c) [40, 41].
A merger of DNA- and PNA-encoding has been achieved with the development of a mating strategy that uses DNA template strands to direct the combinatorial ligation of PNA-encoded peptides. The resulting DNA/PNA-encoded peptides can be deconvoluted via amplification and sequencing of the DNA template strands [42].
5 Dual-Pharmacophore Encoding Strategies
In contrast to the DEL construction schemes described above featuring the display of just one chemical molecule on DNA (single-pharmacophore setup), dual-pharmacophore DELs feature the display of chemical entities (fragments and drug-like molecules) on the extremity of both the 5′ and 3′ end of DNA heteroduplexes. This approach had been pioneered by the Neri/Scheuermann group at ETH Zurich who reported the first such Encoded Self-Assembling Chemical (ESAC) library in 2004 [43]. Two sets of partially complementary sub-libraries, to which chemical compounds were conjugated at the 5′ and the 3′ terminus, respectively, were allowed to combinatorially assemble into stable DNA heteroduplexes (Fig. 5a). These fragment-like sub-libraries when assembled to a DNA heteroduplex would allow for a simultaneous screening of pairs of fragments for target binding, profiting from the chelate effect [44]. Initially, after PCR amplification ESAC libraries were read by hybridization to complementary oligonucleotides on microarrays, yet this read-out was performed individually for both sub-libraries, missing identification of pairs of synergistic binding moieties, initially limiting the potential of ESAC libraries to the lead expansion of known ligands [43]. The full potential of dual-display ESAC technology could only be uncovered using a novel setup that was compatible with the upcoming next-generation DNA sequencing. To this aim, a novel encoding methodology for enabling the transfer of the coding sequence from one strand to the other was proposed, based on the use of abasic DNA segments in one of the two sets of partially complementary oligonucleotides (Fig. 5a) [45]. The coding information of the 3′-strand is transcribed to the shorter 5′-strand by a DNA-polymerase assisted fill-in reaction (Fig. 5a). Together with the development of next-generation DNA sequencing, this method expanded the use of ESAC technology beyond affinity maturation and enabled the de novo discovery of pairs of synergistic binders [45]. A library consisting of 111′100 heteroduplexes obtained by the self-assembly of two mutually complementary sub-libraries of 550 × 202 fragments provided a single pair of fragments which was strongly enriched against alpha-1-acid glycoprotein (AGP), while the individual fragments did not show significant enrichment in the selection and did not display a measurable Kd. Furthermore, in ESAC selections against carbonic anhydrase IX, a validated tumor-associated antigen, a highly enriched pair of acetazolamide (a known 20 nM CAIX ligand) and a phenolic compound could be identified with 40-fold improvement in affinity compared with acetazolamide alone, which is currently in clinical development. In order to further promote the applicability of dual-pharmacophore DEL technology, novel encoding methodologies have recently been postulated: “large-encoding design” (LED) (Fig. 5b) features the assembly and encoding of multi-building block sub-libraries, which allows for the construction and screening of DELs of unprecedented sizes and designs. This technology features sub-libraries with a stable hybridization domain and non-hybridizing encoding parts in a Y-shape manner. After combinatorial assembly and selection on a target of interest, the codes of the selected heteroduplexes can be PCR-amplified using a junction primer and a terminal primer and further ligated to a single PCR product containing all coding information and which can then be interrogated by next-generation DNA sequencing [46]. The selection of dual-pharmacophore DELs is generally performed on purified and immobilized proteins, but recent work on the screening of dual-pharmacophore DELs against carbonic anhydrase IX (CAIX) expressing tumor cells revealed the potential of on-cell selection protocols profiting from a substantial increase of ligand recovery and selectivity through the bivalent display of ligands [47].
Encoding strategies for dual-display DELs. (a) In encoded self-assembling chemical libraries (ESACs), two sub-libraries encoded with partially complementary DNA strands are constructed separately. Despite the variable sequences coding for the first fragment, hybridization of the two sub-libraries is possible owing to the introduction of an abasic spacer region. The extension of the shorter strand by a DNA-polymerase transfers the second code onto the first DNA strand. (b) A large-encoding design enables the construction of ESACs displaying more than three fragments. Two individually synthesized sub-libraries are stably annealed through a short hybridization region. After a fill-in reaction aided by annealing of a junction primer (JP) and terminal primer (T3P) and ligation, a single strand containing all coding sequences is generated. (c) A circular encoding construct can be generated through stable hybridization of two sub-libraries. Two relay primers assist the fill-in reaction followed by ligation, yielding a single strand containing all coding sequences
A formidable challenge associated with the discovery of bidentate ligands from dual-display DELs relies on the identification of an optimal linker which connects the selected fragments/molecules. In most cases, several linkers differing in length, geometry, and rigidity may have to be tested, to identify the optimal solution for the scaffolding of synergistic pharmacophores. The choice of optimal linkers for the coupling of synergistic pharmacophores may be incorporated into future DNA-encoded library design. Recently, a strategy for “dimerizing” non-related single-stranded split-and-pool DELs into dual-pharmacophore DELs which allows for a modulation of fragment spacing and orientation has been devised as a circular construct (Fig. 5c), which may further broaden the scope of dual-pharmacophore DEL technology [10].
6 Conversion from Double-Stranded to Single-Stranded Library Format
A single-stranded library format is required for many of the recently reported advanced selection strategies. Apart from the deliberate single-stranded DEL synthesis two approaches have recently been proposed to convert double-stranded DELs into a single-stranded format. Selective digestion of one strand by exonuclease [48] or the use of a reversible-covalent DNA-headpiece designed to enable reversible interstrand photocrosslinking of a 3-cyanovinyl carbazole nucleoside on one strand with a thymine base on the opposite strand. Irradiation with either UV-A or UV-B light results in reversible conversion from the double- to the single-stranded DEL format [49]. Both methods hold the promise to enable transitioning between the robust double-stranded format and the versatile single stranded DEL format.
7 Dynamic Dual-Pharmacophore DEL Strategies
Dynamic combinatorial chemistry enables the identification of compounds binding to target structures with high affinity by connection of smaller fragments in the binding site [50]. In the context of DNA-encoded chemical libraries dynamic approaches rely on the incubation with the target to guide fragment assembly. In contrast to static dual-pharmacophore DNA-encoded libraries where a stable DNA heteroduplex is preformed, dynamic approaches feature a transiently stable DNA duplex upon incubation of the DNA-tagged fragment sub-libraries with the target. Tagging of fragments with short complementary DNA sequences allows for continuous shuffling of such tagged fragments, until pairs of fragments are binding to a target structure. After the equilibrium is reached, fragment pairs are locked to retain the information on binding pairs for decoding. A first pilot report was reported by Hamilton et al. in 2005, where a stable heteroduplex was heated above the melting temperature thus permitting the reshuffling of fragment pairs, which eventually lead to the enrichment of duplexes displaying the binding fragments [51]. About a decade later the dynamic concept in dual-pharmacophore DEL technology was elaborated independently by the groups of Yixin Zhang and Xiaoyu Li. A first dynamic proof-of-principle library was constructed and subjected to heating above the duplex melting temperature after the first round of selections. The resulting exchange of strands was found to lead to an enrichment of high-affinity duplexes [52]. A Y-shaped encoding setup with a very short hybridization region was proposed (Fig. 6a) where the locking of the equilibrium was accomplished by enzymatic ligation [53]. Further methods were established in order to lock the equilibrium state, e.g., by photocrosslinking of the two DNA strands [54], or by using short, complementary anchor DNA sequences [54] (Fig. 6b). More recently, chemical crosslinking by using p-stilbazole- or psoralen-modified DNA strands was proposed and tested [55, 56]. An interesting approach to facilitate ligand development after the identification of binding small molecule pairs, by linking of the small molecules rather than the two DNA strands was recently devised. Fragment conjugation was performed by imine formation of a single-stranded DEL containing a primary amine and a known ligand containing an aldehyde group, resulting in a dynamic imine library. After incubation with the target, the equilibrium could be locked by reduction of the imine [57]. Recently, an interesting PNA-based dynamic combinatorial supramolecular approach has been reported which is based on the use of a constant short PNA tag to direct the combinatorial pairing of fragments [58]. While the concept of dynamic DELs is very promising, it must be mentioned that, until today, no real, large dynamic DEL, numbering hundreds of thousands, if not millions of compounds, has been reported in literature.
In DNA-encoded dynamic combinatorial chemical libraries, the duplexes are unstable and duplexes binding with high affinity are stabilized by incubation with the target. (a) For Y-EDCCL, the equilibrium reached after incubation with the target was locked by ligation of both strands. (b) After incubation with the target, the equilibrium can be locked by crosslinking prompted by UV irradiation. A relay-primer-bypass strategy relying on a terminal primer and a relay primer and subsequent ligation affords a single DNA strand comprising all coding sequences
8 DNA-Templated Synthesis
An alternative strategy for the assembly of DNA-encoded chemical libraries features the use of DNA-templated synthesis (DTS) [24, 59, 60] (Fig. 7a). In this case, pre-defined DNA templates are employed both for library encoding and for directing the library construction. This setup was pioneered by David R. Liu et al. in 2001 when they demonstrated that chemical reactions are promoted by bringing DNA-linked reagents into close proximity through Watson–Crick base-pairing [24]. The effective molarity of such reactions may be very high, thus allowing to conduct reactions which are otherwise considered difficult or impossible to implement with conventional chemistry. The concept was used not only for reaction discovery [61] but also for the construction of DNA-encoded, macrocyclic libraries. Small organic compounds chemically linked to short single-stranded biotinylated oligonucleotides as donors are transferred upon hybridization to suitable complementary DNA sequences on the template, followed by cleavage and avidin-assisted “donor” strand removal [24]. In a stepwise fashion the template library is annealed with a first set of code-complementary oligonucleotides bearing each a different reactant, which is then chemically attached to the template (Fig. 7a). The linker between the short oligonucleotide and the reactant is subsequently cleaved and this process could be repeated two more times leading to a trimeric linear library. Finally, the library may be cyclized and used for selection experiments against target proteins. The first reported DTS-DEL comprised 65 library members made from three sets of four amino acids, followed by cyclization through Wittig olefination (see Fig. 4).
DNA-templated synthesis uses pre-defined DNA template strands to direct library construction. (a) The hybridization of a library of DNA templates with a library of DNA-linked reagents brings reactants into proximity. Once the coupling of the first building block is complete, the hybridization and reaction steps are sequentially repeated for the remaining reagent libraries. (b) A universal template featuring tri-deoxyinosine anticodons enables hybridization with encoded reagents irrespective of their coding DNA sequence. After hybridization, the reagent DNA is ligated to the universal template and the chemical reaction takes place. This process is repeated for the remaining reagent libraries
This proof-of-principle study was successful in selections against carbonic anhydrase. After technical optimization, a similar, yet larger macrocyclic library of 13,824 DNA-encoded compounds was constructed and reported in 2008 [24, 62], yielding hits for Src kinase and VEGFR2 [63], and six macrocycles against insulin degrading enzyme (IDE), which were also co-crystalized [64, 65].
In 2018, Usanov, Liu, and coworkers published a second-generation DNA-templated library of 256,000 macrocyclic members [66]. To do so, the DNA templates were computationally optimized regarding the orthogonality of each hybridization sequence and template assembly was improved through a polymerase-mediated strategy. In addition, macrocyclic Kihlberg rules [67] were applied to the design. With this DEL, selections were performed against IDE. One macrocyclic hit showed high potency with an IC50 of 40 nM [66]. The company Ensemble Therapeutics, co-founded by David R. Liu, used DTS for the construction of macrocyclic DELs in an industrial setting. In 2015, the company reported a DNA-templated macrocyclic DEL in collaboration with Bristol Meyer Squibb, consisting of five sets of building blocks which were eventually cyclized by a copper-catalyzed azide/alkyne cycloaddition (CuAAC) reaction [67]. Four libraries of 40,000 members were screened against XIAP BIR2 and BIR3 domains [68] and inhibitors with the ability to displace bound pro-apoptotic caspases were identified [69]. The positive aspects of DTS for DEL construction certainly encompass high chemical yields, access to unusual chemical reactions, one-pot reactions with reagent oligonucleotide conjugates, as well as specific purification methods [59]. On the other hand, template library generation, code-specificity for a larger set of building blocks, as well as the creation of larger sets of reagent oligonucleotide conjugates, to reach split sizes of, e.g., a few hundred building blocks can prove challenging. In 2013, Li and coworkers reported a smart solution for omitting the tedious template library generation by proposing a “universal template” consisting of a DNA hairpin containing tri-deoxyinosine anticodons which allow for the indiscriminate base-pairing and subsequent ligation with 3-base encoded reagent oligonucleotide conjugates (Fig. 7b). The feasibility of this DTS approach could be demonstrated with proof-of-principle DELs of 64*64*28 3-base codons [70].
9 DNA-Routing
The group of P. Harbury described “DNA-routing” [71] as a different DNA-template based strategy for library construction. This approach enables combinatorial chemistry by iterative sequence-specific immobilization-reaction and purification–elution step. A library of single-stranded DNA templates comprising the codes for the routing is subjected to resin columns with the immobilized complementary DNA code sequences and subsequently chemically modified (Fig. 8). After repooling, this split-and-pool step is then repeated for all diversity elements. In 2004, Halpin and Harbury described the construction of a combinatorial peptide library using a 340-mer oligonucleotide combinatorial template library consisting of alternating 20 base coding and 20 base constant regions obtained by PCR assembly of overlapping complementary 40-mer oligonucleotides [72]. To serve the purpose of routing on complementary DNA resin the double-stranded template library then was converted into single-stranded DNA by reverse-transcription and base-mediated hydrolysis of the RNA strand.
A library of single-stranded DNA templates serves as the starting point for the construction of DELs by DNA-routing. DNA templates are separated by hybridization with anticodons immobilized on resin columns before being chemically modified. After pooling of all eluates, further rounds of sequence-specific separation and reaction can be carried out
The routing anticodon resin is followed by a chemistry step performed on weak anion-exchange columns, varying high and low salt conditions [73]. The combined eluates after the reaction step can subsequently undergo further split-and-pool cycles. The routing can be performed sequence-specifically and efficiently with overall yield of 0.85 per hybridization round. Using this DNA-routing strategy an N-acylated pentapeptide library with a complexity of 106 was constructed [71], using 10 different amino acid building blocks for the first positions and nine carboxylic acids for the N-acylation step. The library included acylated leucine-enkephalin pentapeptides as a control and was subjected to affinity-based selections against the monoclonal antibody 3-E7 [74], a leucine-enkephalin binding antibody with 7.1 nM affinity.
The routing system was further optimized for peptide coupling yielding >90% conversion per reaction step and an 8-mer library of 100 Mio compounds was created and selected against the N-terminal SH3 domain of the proto-oncogene Crk [13]. Evolving over six generations, the hits converged to a small number of peptides with micromolar affinity. Further refinement allowed for a highly parallel performance in 384-well format [75] and the routing system, acting in analogy as an expanded genetic code, was tested for chemical evolution on protein kinase A (PKA) [76]. While the sole use of peptide bond chemistry and the sophisticated code design may pose restraints, the routing system allows for the generation of very large DNA-encoded libraries, numbering millions of compounds.
10 DNA-Directed Chemistry: The “yoctoReactor”
The final DNA-directed approach to be discussed in this chapter is the “yoctoReactor” or “yR” technology. The hallmark of the yR is a DNA sequence design that leads to self-assembly of two-four oligonucleotides which form a cavity with the name-giving volume of a yoctoliter, “yocto” being the lowest prefix in the SI system [12, 77]. Each of these DNA oligomers carries a functionalized chemical building block via a linker positioned within the sequence, and it is composed of a partial sequence for annealing to give the three- or four-way junction (the yR), and a partial sequence that encodes its building block (Fig. 9). The first set of encoded building blocks, i.e. the equivalent of the cycle 1 building blocks in encoded combinatorial chemistry, is coupled to its DNA oligomers via a stable linker, while the cycle 2 and 3 building blocks are coupled to their barcodes via a scissile linker. Self-assembly of the DNA oligomers to the yR directs the chemical building blocks into proximity for a chemical reaction that couples them to the target molecule. Compound synthesis thus leads in effect to covalent coupling of the DNA barcodes. These constructs can be purified from any non-reacted starting materials by denaturing polyacrylamide gel electrophoresis (PAGE). Thus, the purification step of the anneal-react-purify sequence provides high fidelity in library synthesis, as only the successfully synthesized molecules will enter the encoded library. This fidelity constitutes a major advantage versus DNA-recorded chemistry. On the other hand, the rules for DNA-compatible chemistry apply to the yR, too, thus, carbonyl chemistry and nucleophile substation are prevalent in library design; the choice of building blocks for DEL design is limited to those that contain at least two functional groups, precluding tapping into the vast pool of monofunctionalized chemicals; and an additional chemical step is needed to couple building blocks to the barcodes. Following PAGE, the barcodes of the purified products are ligated to concatenate the genetic information of the compounds. Then, the scissile linker is cleaved to provide the final encoded molecules of a reaction step. As a last step, prior to screening, the complementary strand of the barcodes is formed by primer extension. As the yR can be constructed by annealing the DNA oligomers in combinatorial cycles, this technology furnished millions of encoded compounds, too [78].
11 Conclusion
Taken together the various encoding strategies for DELs have enabled the broad field of DEL technology of today. While DNA-templated synthesis and DNA-routing have led to various DELs, split-and-pool-based methods nowadays are mostly used in practice (Table 1).
While for DNA-recorded DELs most care is generally taken for the optimization on the “chemical” side, i.e., by employing high-yielding DNA-compatible reactions to achieve diversity, the quality of the encoding also critically matters for the performance of DELs in selections.
Encoding in single-stranded DNA format may serve important advantages for DEL selection protocols, since such DELs can be paired with either further single-stranded libraries to form dual-display DELs or by pairing them with known leads, covalent warheads, or with photoreactive moieties displayed on the complementary strand, therefore strategies have been devised for both creating single-stranded DELs by ligation and converting a double-stranded DEL into a single-stranded one {Zhao, 2022 #40; Huang, 2022 #41; Gui, 2022 #42} [1, 11, 12]. Sophisticated selection protocols will expand the scope of DEL technology for the identification of small bioactive compounds.
References
Brenner S, Lerner RA (1992) Encoded combinatorial chemistry. Proc Natl Acad Sci U S A 89:5381–5383
Needels MC, Jones DG, Tate EH, Heinkel GL, Kochersperger LM, Dower WJ, Barrett RW, Gallop MA (1993) Generation and screening of an oligonucleotide-encoded synthetic peptide library. Proc Natl Acad Sci U S A 90:10700–10704
Nielsen J, Brenner S, Janda KD (1993) Synthetic methods for the implementation of encoded combinatorial chemistry. J Am Chem Soc 115:9812–9813
Shi B, Zhou Y, Huang Y, Zhang J, Li X (2017) Recent advances on the encoding and selection methods of DNA-encoded chemical library. Bioorg Med Chem Lett 27:361–369
Goodnow Jr RA, Dumelin CE, Keefe AD (2017) DNA-encoded chemistry: enabling the deeper sampling of chemical space. Nat Rev Drug Discov 16:131–147
Gironda-Martínez A, Donckele EJ, Florent Samain F, Neri D (2021) DNA-encoded chemical libraries: a comprehensive review with successful stories and future challenges. ACS Pharmacol Transl Sci 4:1265–1279
Satz AL, Brunschweiger A, Flanagan ME, Gloger A, Hansen NJV, Kuai L, Kunig VBK, Lu X, Madsen D, Marcaurelle LA, Mulrooney C, O’Donovan G, Sakata S, Scheuermann J (2022) DNA-encoded chemical libraries. Nat Rev Methods Primers 2:3
Fitzgerald PR, Paegel B (2021) DNA-encoded chemistry: drug discovery from a few good reactions. Chem Rev 121:7155–7177
Zambaldo C, Barluenga S, Winssinger N (2015) PNA-encoded chemical libraries. Curr Opin Chem Biol 26:8–15
Oehler S, Plais L, Bassi G, Neri D, Scheuermann J (2021) Modular assembly and encoding strategies for dual-display DNA-encoded chemical libraries. Chem Commun 57:12289–12292
Kleiner RA, Dumelin CE, Liu DR (2011) Small-molecule discovery from DNA-encoded chemical libraries. Chem Soc Rev 40:5707–5717
Blakskjaer P, Heitner T, Hansen NJV (2015) Fidelty by design: Yoctoreactor and binder trap enrichment for small-molecule DNA-encoded libraries and drug discovery. Curr Opin Chem Biol 26:62–71
Wrenn SJ, Weisinger RM, Halpin DR, Harbury PB (2007) Synthetic ligands discovered by in vitro selection. J Am Chem Soc 129:13137–13143
Kinoshita Y, Nishigaki K (1995) Enzymatic synthesis of code regions for encoded combinatorial chemistry (ECC). Nucleic Acids Symp Ser 34:201–202
Clark MA, Acharya RA, Arico-Muendel CC, Belyanskaya SL, Benjamin DR, Carlson NR, Centrella PA, Chiu CH, Creaser SP, Cuozzo JW, Davie CP, Ding Y, Franklin GJ, Franzen KD, Gefter ML, Hale SP, Hansen NJV, Israel DI, Jiang J, Kavarana MJ, Kelley MS, Kollmann CS, Li F, Lind K, Mataruse S, Medeiros PF, Messer JA, Myers P, O’Keefe H, Oliff MC, Rise CE, Satz AL, Skinner SR, Svendsen JL, Tang L, van Vloten K, Wagner RW, Yao G, Zhao B, Morgan BA (2009) Design, synthesis and selection of DNA encoded small-molecule libraries. Nat Chem Biol 5:647–654
Mannocci L, Zhang Y, Scheuermann J, Leimbacher M, De Bellis G, Rizzi E, Dumelin C, Melkko S, Neri D (2008) High-throughput sequencing allows the identification of binding molecules isolated from DNA-encoded chemical libraries. Proc Natl Acad Sci U S A 105:17670–17675
Sun Z-M, Yang S-G, Xue L-J, Zhang J, Yang K, Hu Y-J (2022) N-alkyl linkers for DNA-encoded chemical libraries. Chem Asian J 17:e202200016
Franzini RM, Samain F, Abd Elrahman M, Mikutis G, Nauer A, Zimmermann M, Scheuermann J, Hall J, Neri D (2014) Systematic evaluation and optimization of modification reactions of oligonucleotides with amines and carboxylic acids for the synthesis of DNA-encoded chemical libraries. Bioconjug Chem 25:1453–1461
Rianjongdee F, Atkinson SJ, Chung CW, Grandi P, Gray JRJ, Kaushansky LJ, Medeiros P, Messenger C, Phillipou A, Preston A, Prinjha RK, Rioja I, Satz AL, Taylor S, Wall ID, Watson RJ, Yao G, Demont EH (2021) Discovery of a highly selective BET BD2 inhibitor from a DNA-encoded library technology screening hit. J Med Chem 64:10806–10833
Zhu Z, Shaginian A, Grady LC, O’Keeffe T, Shi XE, Davie CP, Simpson GL, Messer JA, Evindar G, Bream RN, Thansandote PP, Prentice NR, Mason AM, Pal S (2018) Design and application of a DNA-encoded macrocyclic peptide library. ACS Chem Biol 13:53–59
Buller F, Steiner M, Frey K, Mircsof D, Scheuermann J, Kalisch M, Bühlmann P, Supuran CT, Neri D (2011) Selection of carbonic anhydrase IX inhibitors from one million DNA-encoded compounds. ACS Chem Biol 6:336–344
Favalli N, Bassi G, Pellegrino C, Millul J, De Luca R, Cazzamalli S, Yang S, Trenner A, Mozaffari NL, Myburgh R, Moroglu M, Conway SJ, Sartori AA, Manz MG, Lerner RA, Vogt PK, Scheuermann J, Neri D (2021) Stereo- and regiodefined DNA-encoded chemical libraries enable efficient tumour-targeting applications. Nat Chem 13:540–548
Bassi G, Favalli N, Vuk M, Catalano M, Martinelli A, Trenner A, Porro A, Yang S, Lean Tham C, Moroglu M, Yue WW, Conway SJ, Vogt PK, Sartori AA, Scheuermann J, Neri D (2021) A single-stranded DNA-encoded chemical library based on a stereoisomeric scaffold enables ligand discovery by modular assembly of building blocks. Adv Sci 7:2001970
Gartner ZJ, Tse BN, Grubina R, Doyon JB, Snyder TM, Liu DR (2004) DNA-templated organic synthesis and selection of a library of macrocycles. Science 305:1601–1605
Franzini RM, Ekblad T, Zhong N, Wichert M, Decurtins W, Nauer A, Zimmermann M, Samain F, Scheuermann J, Brown PJ, Hall J, Gräslund S, Schüler H, Neri D (2015) Identification of structure-activity relationships from screening a structurally compact DNA- encoded chemical library. Angew Chem Int Ed 54:3927–3931
Yuen LH, Dana S, Liu Y, Bloom SI, Thorsell AG, Neri D, Donato AJ, Kireev DB, Schuler H, Franzini RM (2019) A focused DNA-encoded chemical library for the discovery of inhibitors of NAD+-dependent enzymes. J Am Chem Soc 141:5169–5181
Klika Škopić M, Salamon H, Bugain O, Jung K, Gohla A, Doetsch LJ, dos Santos D, Bhat A, Wagner B, Brunschweiger A (2017) Acid- and Au(I)-mediated synthesis of hexathymidine-DNA-heterocycle chimeras, an efficient entry to DNA-encoded libraries inspired by drug structures. Chem Sci 8:3356–3361
Kunig VBK, Potowski M, Akbarzadeh M, Klika Škopić M, dos Santos SD, Arendt L, Dormuth I, Adihou H, Blaž Andlovic B, Karatas H, Shaabani S, Zarganes-Tzitzikas T, Neochoritis CG, Ran Zhang R, Groves M, Guéret SM, Ottmann C, Rahnenführer J, Fried R, Dömling A, Brunschweiger A (2020) TEAD–YAP interaction inhibitors and MDM2 binders from DNA-encoded indole-focused Ugi peptidomimetics. Angew Chem Int Ed 59:20338–20342
Potowski M, Kunig VBK, Eberlein L, Vakalopoulos A, Kast SM, Brunschweiger A (2021) Chemically stabilized DNA barcodes for DNA-encoded chemistry. Angew Chem Int Ed 60:9744–19749
Potapov V, Ong JL, Kucera RB, Langhorst BW, Bilotti K, Pryor JM, Cantor EJ, Canton B, Knight TF, Evans TC, Lohman GJS (2018) Comprehensive profiling of four base overhang ligation fidelity by T4 DNA ligase and application to DNA assembly. ACS Synth Biol 7:2665–2674
MacConnell AB, McEnaney PJ, Cavett VJ, Paegel BM (2015) DNA-encoded solid-phase synthesis: encoding language design and complex oligomer library synthesis. ACS Comb Sci 17:518–534
Mendes KR, Malone ML, Ndungu JM, Suponitsky-Kroyter I, Cavett VJ, McEnaney PJ, MacConnell AB, Doran TM, Ronacher K, Stanley K, Utset O, Walzl G, Paegel BM, Kodadek T (2016) High-throughput identification of DNA-encoded IgG ligands that distinguish active and latent mycobacterium tuberculosis infections. ACS Chem Biol 12:234–243
Hackler AL, Fitzgerald FG, Dang VQ, Satz AL, Paegel BM (2020) Off-DNA DNA-encoded library screening. ACS Combi Sci 22:25–34
Pothukanuri S, Pianowski Z, Winssinger N (2008) Expanding the scope and orthogonality of PNA synthesis. Eur J Org Chem 18:3141–3148
Chouikhi D, Ciobanu M, Zambaldo C, Duplan V, Barluenga S, Winssinger N (2012) Expanding the scope of PNA-encoded synthesis (PES): Mtt-protected PNA fully orthogonal to fmoc chemistry and a broad array of robust diversity-generating reactions. Chemistry 18:12698–12704
Daguer JP, Ciobanu M, Alvarez S, Barluenga S, Winssinger N (2011) DNA-templated combinatorial assembly of small molecule fragments amenable to selection/amplification cycles. Chem Sci 2:625–632
Harris JL, Winssinger N (2005) PNA encoding (PNA=peptide nucleic acid): from solution-based libraries to organized microarrays. Chemistry 11:6792–6801
Debaene F, Da Silva JA, Pianowski Z, Duran FJ, Winssinger N (2007) Expanding the scope of PNA-encoded libraries: divergent synthesis of libraries targeting cysteine, serine and metalloproteases as well as tyrosine phosphatases. Tetrahedron 63:6577–6658
Galloway WR, Spring DR (2011) PNA to DNA to microarray decoding facilitates ligand discovery. Chem Biol 18:1209–1210
Svensen N, Díaz-Mochón JJ, Bradley M (2011) Encoded peptide libraries and the discovery of new cell binding ligands. Chem Commun 47:7638–7640
Svensen N, Díaz-Mochón JJ, Bradley M (2011) Decoding a PNA encoded peptide library by PCR: the discovery of new cell surface receptor ligands. Chem Biol 18:1284–1289
Vummidi BR, Farrera-Soler L, Daguer JP, Dockerill M, Barluenga S, Winssinger N (2022) A mating mechanism to generate diversity for the Darwinian selection of DNA-encoded synthetic molecules. Nat Chem 14:141–152
Melkko S, Scheuermann J, Dumelin CE, Neri D (2004) Encoded self-assembling chemical libraries. Nat Biotechnol 22:568–574
Melkko S, Dumelin CE, Scheuermann J, Neri D (2006) On the magnitude of the chelate effect for the recognition of proteins by pharmacophores scaffolded by self-assembling oligonucleotides. Chem Biol 13:225–231
Wichert M, Krall N, Decurtins W, Franzini RM, Pretto F, Schneider P, Neri D, Scheuermann J (2015) Dual-display of small molecules enables the discovery of ligand pairs and facilitates affinity maturation. Nat Chem 7:241–249
Plais L, Lessing A, Keller M, Martinelli A, Oehler S, Bassi G, Neri D, Scheuermann J (2022) Universal encoding of next generation DNA-encoded chemical libraries. Chem Sci 13:967–974
Oehler S, Catalano M, Scapozza I, Bigatti M, Bassi G, Favalli N, Mortensen MR, Samain F, Scheuermann J, Neri D (2021) Affinity selections of DNA-encoded chemical libraries on carbonic anhydrase IX-expressing tumor cells reveal a dependence on ligand valence. Chem A Eur J 27:8985–8993
Gui Y, Wong CS, Zhao G, Xie C, Hou R, Li Y, Li G, Li X (2022) Converting double-stranded DNA-encoded libraries (DELs) to single-stranded libraries for more versatile selections. ACS Omega 7:11491–11500
Zhao GX, Zhong ST, Zhang G, Li YF, Li YZ (2022) Reversible covalent headpiece enables interconversion between double- and single-stranded DNA-encoded chemical libraries. Angew Chem Int Ed 61:e202115157
Mondal M, Hirsch AK (2015) Dynamic combinatorial chemistry: a tool to facilitate the identification of inhibitors for protein targets. Chem Soc Rev 44:2455–2488
Sprinz KI, Tagore DM, Hamilton AD (2005) Self-assembly of bivalent protein-binding agents based on oligonucleotide-linked organic fragments. Bioorg Med Chem Lett 15:3908–3911
Reddavide FV, Lin W, Lehnert S, Zhang Y (2015) DNA-encoded dynamic combinatorial chemical libraries. Angew Chem Int Ed 54:7924–7928
Reddavide FV, Cui M, Lin W, Fu N, Heiden S, Andrade H, Thompson M, Zhang Y (2019) Second generation DNA-encoded dynamic combinatorial chemical libraries. Chem Commun 55:3753–3756
Li G, Zheng W, Chen Z, Zhou Y, Liu Y, Yang J, Huang Y, Li X (2015) Design, preparation, and selection of DNA-encoded dynamic libraries. Chem Sci 6:7097–7104
Zhou Y, Li C, Peng J, Xie L, Meng L, Li Q, Zhang J, Li XD, Li X, Huang X et al (2018) DNA-encoded dynamic chemical library and its applications in ligand discovery. J Am Chem Soc 140:15859–15867
Zhou Y, Peng J, Shen W, Li X (2020) Psoralen as an interstrand DNA crosslinker in the selection of DNA-encoded dynamic chemical library. Biochem Biophys Res Commun 533:215–222
Deng YQ, Peng JZ, Xiong F, Song YA, Zhou Y, Zhang JF, Lam FS, Xie C, Shen WY, Huang YR et al (2020) Selection of DNA-encoded dynamic chemical libraries for direct inhibitor discovery. Angew Chem Int Ed 59:14965–14972
Farrera-Soler L, Jean-Pierre Daguer J-P, Patrick Raunft P, Sofia Barluenga S, Anne Imberty A, Nicolas Winssinger N (2020) PNA-based dynamic combinatorial libraries (PDCL) and screening of lectins. Bioorg Med Chem 28:115458
Gartner ZJ, Liu DR (2001) The generality of DNA-templated synthesis as a basis for evolving non-natural small molecules. J Am Chem Soc 123:6961–6963
Li X, Liu DR (2004) DNA-templated organic synthesis: Nature's strategy for controlling chemical reactivity applied to synthetic molecules. Angew Chem Int Ed 43:4848–4870
Kanan MW, Rozenman MM, Sakurai K, Snyder TM, Liu DR (2004) Reaction discovery enabled by DNA-templated synthesis and in vitro selection. Nature 431:545–549
Tse BN, Snyder TM, Shen Y, Liu DR (2008) Translation of DNA into a library of 13 000 synthetic small-molecule macrocycles suitable for in vitro selection. J Am Chem Soc 130:15611–15626
Kleiner RE, Dumelin CE, Tiu GC, Sakurai K, Liu DR (2010) In vitro selection of a DNA-templated small-molecule library reveals a class of macrocyclic kinase inhibitors. J Am Chem Soc 132:11779–11791
Maianti JP, McFedries A, Foda ZH, Kleiner RE, Du XQ, Leissring MA, Tang WJ, Charron MJ, Seeliger MA, Saghatelian A et al (2014) Anti-diabetic activity of insulin-degrading enzyme inhibitors mediated by multiple hormones. Nature 511:94–98
Maianti JP, Tan GA, Vetere A, Welsh AJ, Wagner BK, Seeliger MA, Liu DR (2019) Substrate-selective inhibitors that reprogram the activity of insulin-degrading enzyme. Nat Chem Biol 15:565–574
Usanov DL, Chan AI, Maianti JP, Liu DR (2018) Second-generation DNA-templated macrocycle libraries for the discovery of bioactive small molecules. Nat Chem 10:704–714
Doak BC, Zheng J, Dobritzsch D, Kihlberg J (2016) How beyond rule of 5 drugs and clinical candidates bind to their targets. J Med Chem 59:2312–2327
Seigal BA, Connors WH, Fraley A, Borzilleri RM, Carter PH, Emanuel SL, Fargnoli J, Kim K, Lei M, Naglich JG et al (2015) The discovery of macrocyclic XIAP antagonists from a DNA-programmed chemistry library, and their optimization to give lead compounds with in vivo antitumor activity. J Med Chem 58:2855–2861
Wilkinson JC, Cepero E, Boise LH, Duckett CS (2004) Upstream regulatory role for XIAP in receptor-mediated apoptosis. Mol Cell Biol 24:7003–7014
Li Y, Zhao P, Zhang M, Zhao X, Li X (2013) Multistep DNA-templated synthesis using a universal template. J Am Chem Soc 135:17727–17730
Halpin DR, Harbury PB (2004) DNA display I. Sequence-encoded routing of DNA populations. PLoS Biol 2(E173)
Halpin DR, Harbury PB (2004) DNA display II. Genetic manipulation of combinatorial chemistry libraries for small-molecule evolution. PLoS Biol 2:E174
Halpin DR, Lee JA, Wrenn SJ, Harbury PB (2004) DNA display III. Solid-phase organic synthesis on unprotected DNA. PLoS Biol 2:E175
Meo T, Gramsch C, Inan R, Hollt V, Weber E, Herz A, Riethmuller G (1983) Monoclonal antibody to the message sequence Tyr-Gly-Gly-Phe of opioid peptides exhibits the specificity requirements of mammalian opioid receptors. Proc Natl Acad Sci U S A 80(13):4084–4088
Weisinger RM, Wrenn SJ, Harbury PB (2012) Highly parallel translation of DNA sequences into small molecules. PLoS One 7(3):e28056
Krusemark CJ, Tilmans NP, Brown PO, Harbury PB (2016) Directed chemical evolution with an outsized genetic code. PLoS One 11(8):e0154765
Hansen MH, Blakskjaer P, Petersen LK, Hansen TH, Højfeldt JW, Gothelf KV, Hansen NJV (2009) A yoctoliter-scale DNA reactor for small-molecule evolution. J Am Chem Soc 131:1322–1327
Petersen LK, Blakskjær P, Chaikuad A, Christensen AB, Dietvorst J, Holmkvist J, Knapp S, Kořínek M, Larsen LK, Pedersen AE, Röhm S, Sløk FA, Hansen NJV (2015) Novel p38α MAP kinase inhibitors identified from yoctoReactor DNA-encoded small molecule library. Med Chem Commun 7:1332–1339
Acknowledgments
The authors would like to thank Lars Kolster Petersen (Vipergen ApS) for helpful discussions.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Ethics declarations
Elena Detta declares no conflict of interest.
Andreas Brunschweiger is co-founder of the company Serengen GmbH that offers DNA-encoded library services.
This chapter is a review and no humans or animals were involved in the preparation of the chapter.
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Lessing, A., Detta, E., Scheuermann, J., Brunschweiger, A. (2022). Barcoding Strategies for the Synthesis of Genetically Encoded Chemical Libraries. In: Brunschweiger, A., Young, D.W. (eds) DNA-Encoded Libraries. Topics in Medicinal Chemistry, vol 40. Springer, Cham. https://doi.org/10.1007/7355_2022_146
Download citation
DOI: https://doi.org/10.1007/7355_2022_146
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-18628-8
Online ISBN: 978-3-031-18629-5
eBook Packages: Chemistry and Materials ScienceChemistry and Material Science (R0)