Abstract
The serine/threonine kinase c-Jun N-terminal kinase (JNK) 3 is implicated in the pathogenesis of various disorders ranging from neurodegenerative diseases to inflammation, metabolic disease, diabetes, liver diseases, and cancer. Although the number of publications reporting on JNK3 inhibitors has been decreasing in the last few years, this enzyme still constitutes an attractive target. Within the last years, significant progress in the design of JNK3 inhibitors displaying good to excellent selectivity versus other protein kinases has been achieved. However, the development of a JNK-isoform-selective JNK3 inhibitor, which may serve as a tool compound in animal studies to further evaluate the role of JNK3 as a therapeutic target, is highly desirable. This chapter summarizes the progress in the development of reversible and irreversible inhibitors of JNK3.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
The c-Jun N-terminal kinases (JNKs) are serine/threonine kinases belonging to the family of mitogen-activated protein (MAP) kinases. First members of this enzyme class were discovered in the early 1990s and originally termed as stress-activated protein kinases (SAPKs). Three different isoforms with molecular weights ranging from 46 to 55 kDa have been classified within this group, namely, JNK1 (also known as SAPK-γ/MAPK8), JNK2 (SAPK-α/MAPK9), and JNK3 (SAPK-β/MAPK10), which are encoded by the three distinct genes jnk1, jnk2, and jnk3, respectively [1, 2].
The JNKs are activated by different forms of cellular stress via the classical MAP kinase signaling cascade by tandem phosphorylation by two upstream kinases, in detail by a MAP kinase kinase kinase and by MAP kinase kinases 4 or 7 (Fig. 1) [4]. Activated JNK can phosphorylate a broad subset of downstream targets including both transcription factors and nonnuclear substrates [3].
Representation of the different substrates of JNK. The figure was realized using the information from Bogoyevitch and Kobe [3]
Phosphorylation of c-Jun, which takes part in the formation of the activation protein (AP)-1, increases the transcriptional activity of this complex. An analogous effect derives from the phosphorylation of the activating transcription factor (ATF)-2, which modifies gene expression through the formation of dimers with members of the Jun family [5]. Besides these two well-characterized pathways, JNKs can also act on additional nuclear targets, thereby modulating their effects on gene transcription (Fig. 1). Furthermore, the members of the JNK family phosphorylate cytosolic and mitochondrial substrates resulting in a modification of their functionality or in the regulation of their stability [3].
Due to diverse downstream targets, the JNKs are involved in various physiological functions. These kinases are implicated in the regulation of cell survival/apoptosis in response to external stimuli. The role of JNKs in cell death has been described as bivalent, since it depends on the entity of the stimulus and on the signal integration with additional pathways.
Whereas JNK1 and JNK2 are present in all cells and tissues of the human body, JNK3 is almost exclusively expressed in the brain and to a lesser extent in the testis and heart. The deregulation of JNK3 is linked to several disorders ranging from neurodegenerative diseases, like Parkinson’s and Alzheimer’s disease, to inflammation, metabolic diseases, diabetes, liver diseases, and cancer [6,7,8].
Nevertheless, the design of isoform-selective JNK inhibitors is very challenging as JNK1, JNK2, and JNK3 share more than 80% sequence identity. In particular, the sequence comprising the ATP-binding site or the surrounding area is highly conserved among the isoforms (Fig. 2). In these regions, the JNK3 displays 98% and 95% similarity to JNK1 and JNK2, respectively.
The JNK3 has become an attractive therapeutic target. Several inhibitors targeting this enzyme have been reported from both pharmaceutical industry and academia in the last two decades. However, there are only few reported examples of JNK3 inhibitors that display selectivity versus the other JNK isoforms. For reviews about inhibitors of the JNKs, see Koch et al. [9] and Gehringer et al. [10]
A limited number of small molecule inhibitors of JNKs have been investigated in clinical trials (Table 1). Most of them are ATP-competitive inhibitors (GL5001, CC-401, and CC-930) showing no intra-JNK selectivity. The structure as well as the mechanism of action of CC-90001 is undisclosed. Nevertheless, no JNK3 inhibitor has been launched into the market yet. The phase II clinical trial studies of CC-401 and CC-930 were terminated because of an unfavorable benefit/risk profile.
An alternative strategy to target the JNKs is represented by the retro-inverso peptide inhibitor XG-102 (formerly referred to as AM-111) developed by Xigen SA [11, 12]. While tanzisertib, bentamapimod, and CC-401 are classical type I inhibitors binding reversibly to the ATP-binding site, the retro-inverso peptide inhibitor XG-102 occupies the docking site of the JIP scaffold protein. XG-102 consists of 30 D-configured amino acids and 1 glycine residue (Fig. 3). Clinical phase III trials for treatment of hearing loss as well as for post-cataract surgery inflammation and pain were recently completed for this peptide inhibitor.
2 Reversible Inhibitors
2.1 Prototypical JNK Inhibitor SP600125
One of the first reported small molecule JNK3 inhibitors is 1,9-pyrazoloanthrone (SP600125), an ATP-competitive pan-JNK inhibitor developed by Celgene (Fig. 4) [13]. SP600125 was one of the first JNK inhibitors displaying selectivity versus the related p38α MAP kinase, which is also a member of the MAP kinase family. Although SP600125 has been described to be lacking selectivity within the kinome [14, 15], it is a widely used reference compound in biological JNK assays [16,17,18]. SP600125 binds to the ATP-binding site of the JNKs and forms two hydrogen bond interactions with the hinge region. The N1-atom accepts a hydrogen bond from the backbone amide group of Met111 (JNK1 numbering), and the NH group at the 2-position acts as a hydrogen bond donor toward the backbone carbonyl group of Glu109 (JNK1 numbering). In addition, SP600125 is surrounded by hydrophobic residues in the adenine-binding region of the enzyme [15].
Structure and biological activities of SP600125. Biological data are taken from Bennet et al. [13]
2.2 Aminopyrimidines
Aminopyrimidine derivatives as potent inhibitors of the JNKs have been reported from different research groups [19,20,21,22,23,24]. In contrast to previous studies, LoGrasso and coworkers focused on brain-penetrant JNK-selective aminopyrimidines with promising cellular potency. Their aminopyrimidine series comprised about 500 compounds and 12 examples, all showing no inhibition toward p38α MAP kinase at a test concentration of 20 μM, were further profiled [24]. Compound 1 represents the most potent inhibitor of this series displaying IC50 values in the low nanomolar range against JNK3 and JNK1 (Fig. 5). Aminopyrimidine 1 showed moderate cellular activity. It inhibits the phosphorylation of c-Jun in INS-1 cells with an IC50 value of 210 nM. However, compound 1 is not able to penetrate into the brain. Although being less potent in the in vitro enzyme assay, compound 2 showed an increased cell penetration profile resulting in a strong inhibition of cellular c-Jun phosphorylation (IC50 = 54 nM) and promising pharmacokinetic properties [24]. Inhibitor 2 is able to cross the blood-brain barrier (brain/plasma ratio = 75%) and possesses good oral bioavailability in rats (F = 45%). However, its plasma protein binding in different species (mouse, rat, dog, monkey, and human) is high (>92%). The metabolic stability of 2 in liver microsomes is reasonable. When dosed at 1 mg/kg (iv) in in vivo studies in rats, aminopyrimidine 2 has a favorable PK profile with a half-life of 2.4 h and a clearance of 20 mL/min/kg. Furthermore, inhibitor 2 lowers the generation of reactive oxygen species in INS-1 cells. However, aminopyrimidine 2 displays a high affinity toward the cytochrome P450 (CYP450) isoforms 2C9, 3A4, and 1A2 (>70% inhibition at a tested concentration of 10 μM).
The X-ray crystal structure of aminopyrimidine 2 bound to JNK3 (PDB code: 3KVX) revealed an unexpected binding mode [24]. Although the 2-aminopyrimidine motif is a typical hinge-binding motif present in numerous kinase inhibitors, inhibitor 2 does not form any hydrogen bond interactions with the enzyme. It binds in the ATP-binding site of JNK3 with a highly planar arrangement of the aromatic rings (Fig. 6). Compared to another X-ray structure published by the same group (PDB code: 1PMN) [26], the glycine-rich loop was collapsed toward the active site by 2.5 Å. The compression of the binding pocket might be the driver of selectivity.
Another example of the aminopyrimidinyl-based JNK inhibitors is compound 3 (SR-3306) (Fig. 5) [25]. Compared to inhibitor 2, pan-JNK inhibitor 3 lacks the fluoro substituent on one of the benzene rings, and the morpholino moiety at the triazole moiety is replaced by a methylpyridine. In terms of JNK3 inhibition in the biochemical activity assay, compound 3 is an equipotent inhibitor as compound 2. However, in the cellular assay, JNK inhibitor 3 showed a fourfold reduced activity (IC50 = 216 nM) than 2. Pyrimidinylamine 3 possesses a moderate selectivity. At a test concentration of 3 μM, compound 3 inhibited 35 out of the tested 347 kinases. Additionally, compound 3 showed a clean human ether-à-go-go-related gene (hERG) (IC50 > 30 μM) as well as CYP profile (IC50 > 50 μM against 1A2, 2A6, 2B6, 2C8, 2C9, 2C19, 2D6, 2E1, and 3A4 isoenzymes). Furthermore, inhibitor 3 is able to cross the blood-brain barrier (plasma/brain ratio: 30–40%).
Inhibitor 3 displayed a neuroprotective effect in different in vitro and in vivo models of Parkinson’s disease [25, 27] and effectively protects against ischemia/reperfusion injury in rats at a test concentration of 5 mg/kg [28].
2.3 2-Alkylsulfanyl-5-(pyridin-4-yl)imidazoles
2-Alkylsulfanyl-4-(4-fluorophenyl)-5-(pyridin-4-yl)imidazoles are a prominent class of kinase inhibitors and can be considered as open analogs of the early lead p38α MAP kinase inhibitor SKF86002 [29] developed by researchers from Smith, Kline & French Laboratories (Fig. 7). In different target hopping approaches, several compounds of this class served as lead structures for the design of (selective) inhibitors of other protein kinases not belonging to the family of MAP kinases, e.g., the epidermal growth factor receptor kinase (EGFR) [30, 31], as well as the protein kinases CK1δ and CK1ε [32].
Using a rational structure-based design approach, a series of different reversible and covalent JNK inhibitors was established [33, 34]. In case of the reversible series [33], pyridinylimidazole-based p38α MAP kinase inhibitor LN950 [35, 36] that also shows moderate affinity toward JNK3 (IC50 = 181 nM) [33] served as lead structure. With the aim to improve the inhibition of JNK3 for this class of compounds and simultaneously erase their initial p38α MAP kinase inhibition, a broad series of derivatives was generated, and structure-activity relationships (SAR) were established. The lead compound was extensively modified keeping the putative hinge-binding motif (2-aminopyridine) constant in all analogs (Fig. 8). Structural modifications included (bio)isosteric replacement of the five-membered core scaffold as well as variations in the imidazole N-substitution pattern. The 4-fluorophenyl ring located in the hydrophobic region I was replaced by other substituted phenyl rings, by cycloalkyl rings, as well as by (branched) alkyl groups. Moreover, different substituents at the pyridine-C2 position interacting with the hydrophobic region II were probed. Modifications in this position included branched aliphatic moieties as well as (substituted) carbocyclic and phenyl rings.
Structural modifications of the lead structure and structures as well as biological data of the most potent reversible JNK inhibitor PIT0102014 of this series. Biological data are taken from Ansideri et al. [33]
As general SARs, aryl moieties at the pyridine-C2 position were better tolerated by JNK3 than by p38α MAP kinase. A major contribution in shifting inhibitory activity from p38α MAP kinase to JNK3 was achieved by modifying the 4-fluorophenyl ring at the imidazole C4-position. Replacement of this moiety by a small methyl group (compound 6) resulted in a complete loss of p38α MAP kinase activity, whereas only a slight decrease in JNK3 affinity was observed.
As the central core, a 2,4,5-substituted imidazole ring is the most favored one in terms of improving both JNK3 inhibitory activity and selectivity over p38α MAP kinase. Alkylation of the imidazole nitrogen atom either vicinal (compound 7) or distal (compound 8) to the pyridinyl substituent resulted in a decrease of JNK3 inhibitory activities.
The most promising inhibitor of this series (compound 6) displays IC50 values in the low triple-digit nanomolar range for JNK1 and JNK3 and shows a slight selectivity against the JNK2 isoform (Fig. 8).
The binding mode of pyridinylimidazole 6 at the target kinase was confirmed by X-ray crystallography (PDB code: 6EKD) (Fig. 9). This example represents the first reported crystal structure of a 2-alkylsulfanyl-5-(pyridin-4-yl)imidazole in complex with JNK3. The inhibitor molecule shows a typical type I teardrop binding mode. The hinge-binding motif (2-aminopyridine) of 6 forms – as expected – two hydrogen bond interactions with the backbone of Met149. The methyl substituent on the imidazole-C4 position is pointing toward the hydrophobic region I. A water-mediated hydrogen bond network exists around the imidazole core. The imidazole-N3 atom (distal from the pyridine ring) is not interacting with the conserved Lys93 side chain via a direct hydrogen bond. This interaction was observed to be mediated by two water molecules. Additionally, the imidazole-N1 atom (adjacent to the pyridine ring) is also involved in a water-mediated hydrogen bond to Asn152.
Pyridinylimidazole 6 was evaluated further pharmacologically. This inhibitor is metabolically stable in human liver microsomes, and it possesses only low to moderate affinity toward four out of five tested pharmacologically relevant cytochrome P450 isoenzymes as well as toward hERG channels [33].
2.4 Aminopyrazole Derivatives
In another study, LoGrasso, Feng, and coworkers reported about a series of aminopyrazole-based JNK3 inhibitors, which show high isoform selectivity over JNK1 and in some cases also versus JNK2 (Fig. 10) [37].
Structural modifications of the lead structure 9 (SR-4326) and structures as well as biological data of inhibitors 10 and 11 (SR-11935) of this series. Biological data are taken from Zheng et al. [37]
Starting from lead compound SR-4326, a large SAR study comprising derivatives with modifications on the urea moiety as well as on the amide moiety was generated.
Replacement of the methylpyridine moiety present in SR-4326 by a pyrrolidinylpyrazole moiety resulted in aminopyrazole 10 showing a subnanomolar IC50 value in the JNK3 activity assay as well as a good intra-JNK selectivity (Fig. 10). JNK3 inhibitor 10 displays a more than 500- and 200-fold selectivity over JNK1 and JNK2, respectively. However, data from a selectivity screening of 10 has not been reported so far.
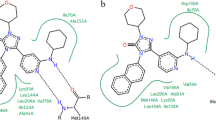
The binding mode of JNK-isoform-selective inhibitor 10 within the ATP-binding site of JNK3 was determined by X-ray experiments (Fig. 11). The R-enantiomer of 10 forms several hydrogen bond interactions with JNK3. The chloro-substituted phenyl ring is located in the hydrophobic region I. The N2 atom of the central pyrazole accepts a hydrogen bond from the NH group of Met149 of the hinge region. A second hydrogen bond toward the backbone of Met149 (carbonyl group) was observed. In this case, the amide NH group serves as a hydrogen bond donor. A water-mediated hydrogen bond interaction is formed between the NH group of the urea group (next to the aniline) and the amino group of the Lys93 side chain. An additional hydrogen bond is established between the NH group of the pyrrolidine ring and Asn89.
Several dual JNK2/3 inhibitors of the aminopyrazole series have also been reported in the same publication [37]. In the biochemical assay, SR-11935 shows a greater than 50-fold isoform selectivity versus JNK1 and potent activity in different cell-based assays in SHSY5Y cells. At a test concentration of 10 μM, SR-11935 displays a good kinome selectivity. However, in this high-throughput screening method, a high affinity for JNK1 was observed. SR-11935 penetrates into the brain (plasma/brain ratio of ∼2:1) showing both good solubility and good DMPK properties. Moreover, this inhibitor displays good microsomal stability, when incubated with human or mice liver microsomes as well as no affinity toward CYP450 isoforms 1A2, 2C9, 2D6, and 3A4. In addition, SR-11935 potently inhibits mitochondrial dysfunction and is noncytotoxic.
3 Covalent Inhibitors
The design of covalent kinase inhibitors has resurged [38,39,40]. In total, a number of six covalent kinase inhibitors were introduced to the market from 2013 to 2018 (Fig. 12). All these inhibitors possess an α,β-unsaturated amide as electrophilic moiety, which targets the side chain of a non-catalytic cysteine residue in the proximity of the ATP-binding site.
As illustrated in the sequence alignment of the JNKs (Fig. 2), all three JNK isoforms possess a cysteine (Cys114 in JNK1/2 numbering; Cys154 in JNK3 numbering) located in the C-lobe adjacent to the hinge region. In this position, this cysteine is unique within the kinome and can be targeted by an inhibitor bearing an electrophilic warhead in a suitable position to form a covalent bond.
3.1 Aminopyrimidine-Based Covalent Inhibitors
The first covalent inhibitors of JNK3 were reported in 2012 by Gray and coworkers [18]. In search for covalent type II inhibitors of the c-Kit and PDGFR kinases, the imatinib-derived pan-JNK inhibitor JNK-IN-1 (Fig. 13) was discovered.
Structures and biological data (after 1 h of incubation) of irreversible JNK inhibitors based on the imatinib scaffold. Structural similarities of imatinib present in 12 (JNK-IN-1) are highlighted in blue color. Biological data are taken from Zhang et al. [18]
A selectivity screening of JNK-IN-1 versus 400 kinases revealed, at a test concentration of 10 μM, besides binding to the classical imatinib targets (Abl, c-Kit and DDR1/2), also a strong binding to all three JNK isoforms. Determination of the biochemical IC50 values against the latter kinases revealed JNK-IN-1 to be a moderate inhibitor of all three JNK isoforms.
Using a structure-based design, a series of analogs was established, and the pan-JNK inhibitor JNK-IN-1 was further optimized regarding its inhibitory activity. Removing the methyl group of the benzene ring at the pyrimidine-C2-amino position and changing the substitution pattern on both benzene rings in the linker bearing the electrophilic warhead resulted in compound JNK-IN-7. This potential covalent inhibitor displayed IC50 values in the low single-digit nanomolar range for JNK1 and JNK2 as well as in the subnanomolar range for JNK3 (1 h incubation). In case of JNK3, JNK-IN-7 shows a 1,000-fold higher activity compared to parent compound JNK-IN-1. JNK-IN-7 also inhibits c-Jun phosphorylation in HeLa and A375 cells with IC50 values of 130 nM and 244 nM, respectively [18].
The X-ray structure of JNK-IN-7 in complex with JNK3 (PDB code: 3V6S) (Fig. 14) reveals that this covalent inhibitor has a different binding mode in the ATP-binding site of JNK3 than imatinib in Abl kinase (PDB code: 1IEP) (Fig. 15) [41].
JNK-IN-7 binds to JNK3 in a type I binding mode with the kinase in the active DFG-in conformation. The covalent link between the electrophilic arylamide and the side chain of Cys154 was also unambiguously revealed. The classical hinge-binding motif (pyrimidinylamine) interacts with the backbone of Met149 via two hydrogen bonds [18].
Imatinib shows a typical type II inhibitor binding mode targeting the inactive DFG-out conformation of Abl kinase. The pyrimidinylamine moiety does not interact via hydrogen bonds with the hinge region. Instead, the nitrogen atom of the 3-pyridinyl moiety accepts a hydrogen bond from the backbone N-H of Met318 (Fig. 15) [41].
In vitro kinase screening against a panel of 442 kinases revealed a very good selectivity for JNK-IN-7. Within this panel, this covalent inhibitor bound or inhibited eight other protein kinases with a KD or IC50 value of 100 nM or lower besides the JNKs.
Reintroduction of the methyl group on the phenyl ring in ortho-position to the pyridine-C2-amino moiety on JNK-IN-7 resulted in JNK-IN-8. This inhibitor displays almost the same inhibitory activity for the JNK3 as JNK-IN-7 yet showing a significantly improved selectivity profile for JNK3 within the JNK family as well as against the rest of the kinome [18].
However, in cell-based assays, JNK-IN-8 shows slightly lower inhibitory activity compared to JNK-IN-7 (IC50 values of 486 nM and 333 nM in Hela and A375 cells, respectively) [18]. In a panel of 442 different protein kinases (also including mutants), JNK-IN-8 only bound to two Kit mutants (V559D and V557D,T650I). In a cellular kinase profiling versus distinct protein kinases, JNK-IN-8 exclusively inhibited JNK1,2,3. Due to its excellent selectivity profile, the covalent pan-JNK-inhibitor JNK-IN-8 was selected by the chemical probes portal as a tool compound to further investigate the role of JNK1/2/3 inhibition (www.chemicalprobesportal.org). However, the same portal recommends using JNK-IN-8 along with validated JNK inhibitors as a positive control.
3.2 Pyridinylimidazole-Based Covalent Inhibitors
The concept of covalently targeting the JNK3 was also transferred to the reversible JNK inhibitors of the 2-alkylsulfanyl-5-(pyridin-4-yl)imidazole series, which are already optimized to interact with different regions in the ATP-binding site of JNK3 (see Sect. 2.3) [34]. For targeting the mentioned cysteine side chain in the JNKs, the electrophilic warhead was attached to the pyridine-C2-amino function via a suitable linker (Fig. 16).
Design of pyridinylimidazole-based irreversible JNK inhibitors as well as structures and biological data (after 50 min (JNK3) or 60 min (p38α) of incubation) of covalent JNK inhibitors 15 (PIT0104026) and 16 (LN2332). Biological data are taken from Muth et al. [34]
An acrylamide was chosen as an electrophilic warhead due to the fact that α,β-unsaturated carbonyl groups are soft electrophiles. On the one side, the acrylamide preferentially reacts with the soft thiol present in the side chain of Cys154. On the other side, acrylamides are less reactive than α,β-unsaturated esters as well as α,β-unsaturated ketones, thereby reducing possible side reactions with thiol groups present in other intracellular proteins.
Potential covalent inhibitors bearing different linkers consisting of two phenyl rings connected by an amide bond were synthesized wherein the linker length and geometry was varied (Fig. 16). The contribution of the covalent bond formation to the inhibitory activity was estimated by comparing the activity of the potential irreversible inhibitors with the one of the corresponding saturated analogs [34].
The linker, consisting of a para-substitution pattern on the phenyl ring connected to the pyridine-C2-amino position as well as a meta-substitution pattern on the second phenyl ring bearing the acryl amide moiety, showed to have the appropriate length and properties to place the reactive electrophile in optimal position to form the covalent bond with the Cys154 side chain. The resulting covalent inhibitor PIT0104026 inhibits JNK3 in the low nanomolar range and possesses an approximately 1,000-fold selectivity versus the p38α MAP kinase. The irreversible binding mode of PIT0104026 was confirmed by mass shift experiments. This covalent inhibitor displays a good selectivity. In a screening versus a panel of 410 kinases, 15 kinases including JNK1,2,3 were inhibited at a test concentration of 1 μM [34].
The inhibitory potency as well as the selectivity of PIT0104026 was further improved by targeting the hydrophobic region I (the so-called selectivity pocket) of the enzyme with a 4-fluorophenyl ring as well as by introducing a methyl substituent on the imidazole nitrogen atom distal to the 4-fluorophenyl moiety. The resulting inhibitor LN2332 shows inhibition of JNK3 in the picomolar range (Fig. 16) [34].
A mass shift corresponding to the molecular weight of LN2332 was detected after its incubation with JNK3 followed by liquid chromatography-mass spectrometry. Since neither the incubation of the saturated counterpart of LN2332 nor the incubation of LN2332 with a JNK3-C154A-mutant resulted in a mass shift, the covalent binding mode of LN2332 with the Cys154 side chain was unambiguously proven.
Further pharmacological profiling identified LN2332 as a promising covalent pan-JNK inhibitor since it remained metabolically stable when exposed to human liver microsomes. LN2332 shows no unpredictable side reactions to other thiol-containing enzymes and has an excellent selectivity profile. In a panel of 410 protein kinases, LN2332 inhibits at a tested concentration of 0.5 μM – besides all three JNK isoforms – only three other protein kinases (Tie-2, MAPKAP2, and CK-1δ). However, cellular data of LN2332 have not been reported so far.
4 Conclusion
Numerous very potent reversible and irreversible small molecule JNK3 inhibitors displaying IC50 values down to the subnanomolar range have been reported within the last years. The binding modes for most of these inhibitors were determined by X-ray experiments. Several examples of recently published JNK3 inhibitors are showing very good to excellent selectivity within the kinome. Most of the discussed inhibitors display no or only low affinity versus non-JNK members of the MAP kinase family.
Moreover, some JNK3 inhibitors displaying intra-JNK selectivity have been reported. Within the class of aminopyrazole-derived reversible inhibitors, several examples of JNK2/3 inhibitors, e.g., SR-11935, showing selectivity versus JNK1 have been reported. Among them, aminopyrazole 10 represents the first reported potent JNK3 inhibitor showing very good JNK-isoform selectivity. However, its kinome-wide selectivity has not been reported.
The targeting of the thiol group present in the non-conserved Cys154, which is located adjacent to the ATP-binding site in JNK3, by electrophilic warheads resulted in the very potent irreversible pan-JNK inhibitors LN2332 and JNK-IN-8. Both compounds show an excellent selectivity within the human kinome. The latter covalent inhibitor, which is also active in cellular assays, is the only available high-quality kinase probe for JNK1/2/3 so far and might be used to further investigate the role of the JNKs in various disorders.
Abbreviations
- AP:
-
Activation protein
- ATF:
-
Activating transcription factor
- CYP450:
-
Cytochrome P450
- EGFR:
-
Epidermal growth factor receptor kinase
- JNK:
-
c-Jun N-terminal kinase
- MAP:
-
Mitogen-activated protein
- SAPK:
-
Stress-activated protein kinases
- SAR:
-
Structure-activity relationships
References
Barr RK, Bogoyevitch MA (2001) The c-Jun N-terminal protein kinase family of mitogen-activated protein kinases (JNK MAPKs). Int J Biochem Cell B 33:1047–1063. https://doi.org/10.1016/S1357-2725(01)00093-0
Bogoyevitch MA (2006) The isoform-specific functions of the c-Jun N-terminal kinases (JNKs): differences revealed by gene targeting. BioEssays 28:923–934. https://doi.org/10.1002/bies.20458
Bogoyevitch MA, Kobe B (2006) Uses for JNK: the many and varied substrates of the c-Jun N-terminal kinases. Microbiol Mol Biol Rev 70:1061. https://doi.org/10.1128/MMBR.00025-06
Davis RJ (2000) Signal transduction by the JNK group of MAP kinases. Cell 103:239–252. https://doi.org/10.1016/S0092-8674(00)00116-1
Hess J, Angel P, Schorpp-Kistner M (2004) AP-1 subunits: quarrel and harmony among siblings. J Cell Sci 117:5965–5973. https://doi.org/10.1242/jcs.01589
Mehan S, Meena H, Sharma D, Sankhla R (2011) JNK: a stress-activated protein kinase therapeutic strategies and involvement in Alzheimer’s and various neurodegenerative abnormalities. J Mol Neurosci 43:376–390. https://doi.org/10.1007/s12031-010-9454-6
Resnick L, Fennell M (2004) Targeting JNK3 for the treatment of neurodegenerative disorders. Drug Discov Today 9:932–939. https://doi.org/10.1016/S1359-6446(04)03251-9
Manning AM, Davis RJ (2003) Targeting JNK for therapeutic benefit: from JuNK to gold? Nat Rev Drug Discov 2:554–565. https://doi.org/10.1038/nrd1132
Koch P, Gehringer M, Laufer SA (2015) Inhibitors of c-Jun N-terminal kinases: an update. J Med Chem 58:72–95. https://doi.org/10.1021/jm501212r
Gehringer M, Muth F, Koch P, Laufer SA (2015) c-Jun N-terminal kinase inhibitors: a patent review (2010–2014). Expert Opin Ther Pat 25:849–872. https://doi.org/10.1517/13543776.2015.1039984
Barkdull GC, Hondarrague Y, Meyer T, Harris JP, Keithley EM (2007) AM-111 reduces hearing loss in a Guinea pig model of acute labyrinthitis. Laryngoscope 117:2174–2182. https://doi.org/10.1097/MLG.0b013e3181461f92
Coleman JKM, Littlesunday C, Jackson R, Meyer T (2007) AM-111 protects against permanent hearing loss from impulse noise trauma. Hearing Res 226:70–78. https://doi.org/10.1016/j.heares.2006.05.006
Bennett BL, Sasaki DT, Murray BW, O'Leary EC, Sakata ST, Xu WM, Leisten JC, Motiwala A, Pierce S, Satoh Y, Bhagwat SS, Manning AM, Anderson DW (2001) SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. P Natl Acad Sci USA 98:13681–13686. https://doi.org/10.1073/pnas.251194298
Fabian MA, Biggs WH, Treiber DK, Atteridge CE, Azimioara MD, Benedetti MG, Carter TA, Ciceri P, Edeen PT, Floyd M, Ford JM, Galvin M, Gerlach JL, Grotzfeld RM, Herrgard S, Insko DE, Insko MA, Lai AG, Lelias JM, Mehta SA, Milanov ZV, Velasco AM, Wodicka LM, Patel HK, Zarrinkar PP, Lockhart DJ (2005) A small molecule-kinase interaction map for clinical kinase inhibitors. Nat Biotechnol 23:329–336. https://doi.org/10.1038/nbt1068
Heo Y-S, Kim SK, Seo CI, Kim YK, Sung B-J, Lee HS, Lee JI, Park S-Y, Kim JH, Hwang KY, Hyun Y-L, Jeon YH, Ro S, Cho JM, Lee TG, Yang C-H (2004) Structural basis for the selective inhibition of JNK1 by the scaffolding protein JIP1 and SP600125. EMBO J 23:2185–2195. https://doi.org/10.1038/sj.emboj.7600212
Goettert M, Luik S, Graeser R, Laufer SA (2011) A direct ELISA assay for quantitative determination of the inhibitory potency of small molecules inhibitors for JNK3. J Pharm Biomed Anal 55:236–240. https://doi.org/10.1016/j.jpba.2011.01.014
Feng YB, Chambers JW, Iqbal S, Koenig M, Park H, Cherry L, Hernandez P, Figuera-Losada M, LoGrasso PV (2013) A small molecule bidentate-binding dual inhibitor probe of the LRRK2 and JNK kinases. ACS Chem Biol 8:1747–1754. https://doi.org/10.1021/cb3006165
Zhang T, Inesta-Vaquera F, Niepel M, Zhang JM, Ficarro SB, Machleidt T, Xie T, Marto JA, Kim N, Sim T, Laughlin JD, Park H, LoGrasso PV, Patricelli M, Nomanbhoy TK, Sorger PK, Alessi DR, Gray NS (2012) Discovery of potent and selective covalent inhibitors of JNK. Chem Biol 19:140–154. https://doi.org/10.1016/j.chembiol.2011.11.010
Alam M, Beevers RE, Ceska T, Davenport RJ, Dickson KM, Fortunato M, Gowers L, Haughan AF, James LA, Jones MW, Kinsella N, Lowe C, Meissner JWG, Nicolas AL, Perry BG, Phillips DJ, Pitt WR, Platt A, Ratcliffe AJ, Sharpe A, Tait LJ (2007) Synthesis and SAR of aminopyrimidines as novel c-Jun N-terminal kinase (JNK) inhibitors. Bioorg Med Chem Lett 17:3463–3467. https://doi.org/10.1016/j.bmcl.2009.03.023
Humphries PS, Lafontaine JA, Agree CS, Alexander D, Chen P, Do QQT, Li LLY, Lunney EA, Rajapakse RJ, Siegel K, Timofeevski SL, Wang TL, Wilhite DM (2009) Synthesis and SAR of 4-substituted-2-aminopyrimidines as novel c-Jun N-terminal kinase (JNK) inhibitors. Bioorg Med Chem Lett 19:2099–2102. https://doi.org/10.1016/j.bmcl.2009.03.023
Song XY, Chen WM, Lin L, Ruiz CH, Cameron MD, Duckett DR, Kamenecka TM (2011) Synthesis and SAR of 2-phenoxypyridines as novel c-Jun N-terminal kinase inhibitors. Bioorg Med Chem Lett 21:7072–7075. https://doi.org/10.1016/j.bmcl.2011.09.090
Song X, He Y, Koenig M, Shin Y, Noël R, Chen W, Ling YY, Feurstein D, Lin L, Ruiz CH, Cameron MD, Duckett DR, Kamenecka TM (2012) Synthesis and SAR of 2,4-diaminopyrimidines as potent c-Jun N-terminal kinase inhibitors. Med Chem Commun 3:238–243. https://doi.org/10.1039/C1MD00219H
Palmer WS, Alam M, Arzeno HB, Chang KC, Dunn JP, Goldstein DM, Gong LY, Goyal B, Hermann JC, Hogg JH, Hsieh G, Jahangir A, Janson C, Jin S, Kammlott RU, Kuglstatter A, Lukacs C, Michoud C, Niu LH, Reuter DC, Shao A, Silva T, Trejo-Martin TA, Stein K, Tan YC, Tivitmahaisoon P, Tran P, Wagner P, Weller P, Wu SY (2013) Development of amino-pyrimidine inhibitors of c-Jun N-terminal kinase (JNK): kinase profiling guided optimization of a 1,2,3-benzotriazole lead. Bioorg Med Chem Lett 23:1486–1492. https://doi.org/10.1016/j.bmcl.2012.12.047
Kamenecka T, Jiang R, Song XY, Duckett D, Chen WM, Ling YY, Habel J, Laughlin JD, Chambers J, Figuera-Losada M, Cameron MD, Lin L, Ruiz CH, LoGrasso PV (2010) Synthesis, biological evaluation, X-ray structure, and pharmacokinetics of aminopyrimidine c-Jun-N-terminal kinase (JNK) inhibitors. J Med Chem 53:419–431. https://doi.org/10.1021/jm901351f
Chambers JW, Pachori A, Howard S, Ganno M, Hansen D, Kamenecka T, Song XY, Duckett D, Chen WM, Ling YY, Cherry L, Cameron MD, Lin L, Ruiz CH, LoGrasso P (2011) Small molecule c-Jun-N-terminal kinase inhibitors protect dopaminergic neurons in a model of Parkinson’s disease. ACS Chem Neurosci 2:198–206. https://doi.org/10.1021/cn100109k
Scapin G, Patel SB, Lisnock J, Becker JW, LoGrasso PV (2003) The structure of JNK3 in complex with small molecule inhibitors: structural basis for potency and selectivity. Chem Biol 10:705–712. https://doi.org/10.1016/S1074-5521(03)00159-5
Crocker CE, Khan S, Cameron MD, Robertson HA, Robertson GS, LoGrasso P (2011) JNK inhibition protects dopamine neurons and provides behavioral improvement in a rat 6-hydroxydopamine model of Parkinson’s disease. ACS Chem Neurosci 2:207–212. https://doi.org/10.1021/cn1001107
Chambers JW, Pachori A, Howard S, Iqbal S, LoGrasso PV (2013) Inhibition of JNK mitochondrial localization and signaling is protective against ischemia/reperfusion injury in rats. J Biol Chem 288:4000–4011. https://doi.org/10.1074/jbc.M112.406777
Griswold DE, Marshall PJ, Webb EF, Godfrey R, Newton J, DiMartino MJ, Sarau HM, Gleason JG, Poste G, Hanna N (1987) SK&F 86002: a structurally novel anti-inflammatory agent that inhibits lipoxygenase- and cyclooxygenase-mediated metabolism of arachidonic acid. Biochem Pharmacol 36:3463–3470. https://doi.org/10.1016/0006-2952(87)90327-3
Günther M, Juchum M, Kelter G, Fiebig H, Laufer S (2016) Lung cancer: EGFR inhibitors with low nanomolar activity against a therapy-resistant L858R/T790M/C797S mutant. Angew Chem Int Ed 55:10890–10894. https://doi.org/10.1002/anie.201603736
Günther M, Lategahn J, Juchum M, Döring E, Keul M, Engel J, Tumbrink HL, Rauh D, Laufer S (2017) Trisubstituted pyridinylimidazoles as potent inhibitors of the clinically resistant L858R/T790M/C797S EGFR mutant: targeting of both hydrophobic regions and the phosphate binding site. J Med Chem 60:5613–5637. https://doi.org/10.1021/acs.jmedchem.7b00316
Halekotte J, Witt L, Ianes C, Krüger M, Bührmann M, Rauh D, Pichlo C, Brunstein C, Luxenburger A, Baumann U, Knippschild U, Bischof J, Peifer C (2017) Optimized 4,5-diarylimidazoles as potent/selective inhibitors of protein kinase CK1δ and their structural relation to p38α MAPK. Molecules 22:522. https://doi.org/10.3390/molecules22040522
Ansideri F, Macedo JT, Eitel M, El-Gokha A, Zinad DS, Scarpellini C, Kudolo M, Schollmeyer D, Boeckler FM, Blaum BS, Laufer SA, Koch P (2018) Structural optimization of a Pyridinylimidazole scaffold: shifting the selectivity from p38α mitogen-activated protein kinase to c-Jun N-terminal kinase 3. ACS Omega 3:7809–7831. https://doi.org/10.1021/acsomega.8b00668
Muth F, El-Gokha A, Ansideri F, Eitel M, Döring E, Sievers-Engler A, Lange A, Boeckler FM, Lämmerhofer M, Koch P, Laufer SA (2017) Tri- and tetrasubstituted pyridinylimidazoles as covalent inhibitors of c-Jun N-terminal kinase 3. J Med Chem 60:594–607. https://doi.org/10.1021/acs.jmedchem.6b01180
Koch P, Bäuerlein C, Jank H, Laufer S (2008) Activated Protein (MAP) kinase: synthesis and biological testing of 2-Alkylsulfanyl-, 4(5)-aryl-, 5(4)-Heteroaryl-substituted Imidazoles. J Med Chem 51:5630–5640. https://doi.org/10.1021/jm800373t
El-Gokha A, Laufer SA, Koch P (2015) An optimized and versatile synthesis to pyridinylimidazole-type p38α mitogen activated protein kinase inhibitors. Org Biomol Chem 13:10699–10704. https://doi.org/10.1039/c5ob01505g
Zheng K, Iqbal S, Hernandez P, Park HJ, LoGrasso PV, Feng Y (2014) Design and synthesis of highly potent and isoform selective JNK3 inhibitors: SAR studies on aminopyrazole derivatives. J Med Chem 57:10013–10030. https://doi.org/10.1021/jm501256y
Singh J, Petter RC, Baillie TA, Whitty A (2011) The resurgence of covalent drugs. Nat Rev Drug Discov 10:307–317. https://doi.org/10.1038/nrd3410
Liu QS, Sabnis Y, Zhao Z, Zhang TH, Buhrlage SJ, Jones LH, Gray NS (2013) Developing irreversible inhibitors of the protein kinase cysteinome. Chem Biol 20:146–159. https://doi.org/10.1016/j.chembiol.2012.12.006
Chaikuad A, Koch P, Laufer SA, Knapp S (2018) The cysteinome of protein kinases as a target in drug development. Angew Chem Int Ed 57:4372–4385. https://doi.org/10.1002/anie.201707875
Nagar B, Bornmann WG, Pellicena P, Schindler T, Veach DR, Miller WT, Clarkson B, Kuriyan J (2002) Crystal structures of the kinase domain of c-Abl in complex with the small molecule inhibitors PD173955 and imatinib (STI-571). Cancer Res 62:4236–4243
Acknowledgments
Many thanks to Dr. Francesco Ansideri and Stanislav Andreev for helpful discussions. Dr. Ansideri is also acknowledged for providing Fig. 1. Kristina Schmidt is gratefully acknowledged for proofreading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Ethics declarations
Conflict of Interest: Author declares that he has no conflict of interest.
Ethical approval: This article does not contain any studies with human participants performed by the author.
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Koch, P. (2020). Inhibitors of c-Jun N-Terminal Kinase 3. In: Laufer, S. (eds) Proteinkinase Inhibitors. Topics in Medicinal Chemistry, vol 36. Springer, Cham. https://doi.org/10.1007/7355_2020_98
Download citation
DOI: https://doi.org/10.1007/7355_2020_98
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-68179-1
Online ISBN: 978-3-030-68180-7
eBook Packages: Chemistry and Materials ScienceChemistry and Material Science (R0)