Abstract
Bombyx mori bidensovirus (BmBDV) is a unique bipartite DNA virus that causes chronic disease in silkworms, and its interactions with the host give rise to variations of genes expression. Reverse transcription quantitative real-time PCR (RT-qPCR) has extensive applications in mRNA quantification. To obtain reproducible and meaningful quantification of transcripts, it is crucial to identify the best reference genes in preliminary work. In this work, we analyzed the expression of seven candidate reference genes in silkworm larvae infected or uninfected with BmBDV, and compared their expression stability with three statistical methods (geNorm, NormFinder, and BestKeeper). The results revealed that the RPL3 and 28S rRNA were the stable internal control, and UBQ was the least stable gene. Furthermore, the expression of Bombyx mori serine protease (BmSP142) was assessed using RPL3 as internal. The expression of BmSP142 was significantly different between BmBDV-resistant and BmBDV-susceptible silkworm after virus infection, suggesting that BmSP142 might be associated with the resistance to BmBDV in silkworm.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The silkworm, also called Bombyx mori (B. mori L), a completely metamorphic insect, has four developmental stages in its life cycle: egg, larva, pupa, and adult. Silkworm larvae molt four times during the larval period, which corresponds to five instars of larval development. The silkworm is an important economic insect for silk production and has become a model organism in the study of life science (Chen et al. 2014). Sericulture is a principal source of income for farmers in many developing countries. However, silkworm pathogens cause losses in cocoon production of almost 20% each year (Jiang et al. 2013). Almost 80% of the total cocoon loss is caused by viral diseases (Guo et al. 2016), one of which is Bombyx mori bidensovirus (BmBDV). BmBDV belongs to Bidensovirus in Bidnaviridae, which replicates mainly in silkworm midgut columnar cells and causes the fatal flacherie disease. This virus possesses two single-stranded DNA genome segments in separate capsids and encodes a putative protein-primed DNA polymerase (Hu et al. 2013). Up to date, it is the only bipartite animal virus and is the only species in the Bidnaviridae (Hu et al. 2013). However, the interactions between BmBDV and its host at transcription level remain to be detailed.
Reverse transcription quantitative real-time polymerase chain reaction (RT-qPCR) approaches is the main method used to accurately analyze the transcript abundance of genes involved in viral pathogenesis (Long et al. 2015). Due to its superior speed, sensitivity, and reproducibility, as well as the wide range of commercially available instrumentation and reagents, RT-qPCR has been universally adopted as the choice for transcriptomic analysis (Bustin et al. 2005). To achieve accurate and reliable gene expression analyzed by RT-qPCR, it is essential that one or several reference genes can be used as internal control to normalize experimental variations (Bustin et al. 2005; Huggett et al. 2005). The suitable reference genes used in RT-qPCR should be expressed stably under the research conditions. Housekeeping genes which were considered to be constant expression were commonly used for normalization, such as 3-phosphoric acid glycerol aldehyde dehydrogenase (GAPDH), 28S rRNA and atcin. However, most of these genes displayed variable expression levels under different experimental conditions (Thellin et al. 2009; Lee et al. 2002; Thellin et al. 1999). An erroneously strong specific expression pattern for a given target gene could be obtained, if an inappropriate reference gene was used for normalization (Gutierrez et al. 2008). In silkworm, the candidate reference genes were expressed differentially in different developmental stages and varied in response to experimental conditions (Peng et al. 2012). Thus, it is necessary to verify the expression stability of candidate reference genes before the specific experiment.
Previous studies have used actin A3 (Kong et al. 2011) or GAPDH (Bao et al. 2013) as internal reference genes in BmBDV-infectied silkworm. Nevertheless, whether these reference genes keep stable expression remains to be determined in silkworm infected by BmBDV. In this study, we have selected and validated the most suitable internal control gene(s) for the normalization of RT-qPCR data upon BmBDV infection in silkworms. Using statistical algorithm programs (geNorm, Normfinder, and BestKeeper), we evaluated the expression stability of seven candidate reference genes: 28S rRNA, actin A3, ubiquitin (UBQ), GAPDH, ribosomal protein L3 (RPL3), TATA response element binding protein (TBP), and ɑ-tubulin. RPL3 and 28S rRNA were identified as the best reference genes. The expression of Bombyx mori serine protease (BmSP142) varied significantly during BmBDV infection (Li et al. 2017). The reliability of RPL3 and 28S rRNA were validated by expression analysis of BmSP142.
Materials and methods
Silkworm and virus
The BmBDV susceptible Bombyx mori strains Jingsong and Huaba35 were bred by our laboratory. The near isogenic line HuabaBC7 which is resistant to BmBDV was constructed by our laboratory (Chen et al. 2011). The silkworm larvae were reared on fresh mulberry (Morus alba) leaves at 25 ± 1 °C and 80% ± 15% relative humidity with a 12 h light/12 h dark cycle. Thirty animals were used for each experimental group. Newly exuviated fifth-instar larvae were used for this experiment. BmBDV was propagated in fifth instar susceptible lavae and the virus supernatant was prepared as described previously (Hu et al. 2016).
Midgut collection, RNA extraction, quality controls, and cDNA synthesis
The BmBDV-infected fifth instar larvae were dissected, and their midguts were collected at 0, 12, 24, 36, 48, 72, 84 and 96 h post-infection (hpi). One hundred milligrams of the midgut was ground into powder in liquid nitrogen. Total RNA was extracted using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. The quality and concentration of each of the RNA samples was determined with a Nanodrop ND-1000 spectrophotometer (Thermo Fisher Scientific) and an Agilent 2100 Bioanalyzer. Only those RNA samples having A260 to A280 ratios of 1.9 to 2.1 and A260 to A230 ratios of 2.0 to 2.5, as well as an RNA integrity number greater than 7.0, were used for the analysis. The integrity of the RNA was also checked by agarose gel electrophoresis. Approximately 500 ng of RNA was reverse transcribed using the PrimeScript RT Reagent Kit with gDNA Eraser (TaKaRa, Dalian, China) according to the manufacturer’s instructions, and the product was used as the template in RT-qPCR.
Primer design and qPCR
In this study, The sequences of seven reference genes and the target gene were obtained from NCBI database by BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.
cgi). Primer 5.0 software (PREMIER Biosoft International, Palo Alto, CA, USA) was used to design the primers. Primers were synthesized by Generay Biotech Co, Ltd. (Shanghai, P.R. China). The primers of seven reference genes and the target gene for RT-qPCR were shown in Table 1. The fluorescent RT-qPCR reaction system consisted of 5 μL 2 × SYBR Premix Ex Taq™ II (Takara, Dalian, China), 0.2 μL upstream and downstream primers (10 mM), 0.2 μL ROX Reference Dye II (50 × Conc.), 1 μL template cDNA, and 3.4 μL double-distilled H2O in a total volume of 10 μL. The RT-qPCR conditions were as follows: an initial denaturation at 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s, 55 °C 27 s, and 72 °C for 31 s, and finally, PCR amplification specificity was verified by a dissociation curve (75–94 °C). The RT-qPCR analysis was performed for three biological replicates for each sample, and three technical replicates were analyzed for each biological replicate.
Standard curve
Seven reference gene segments were PCR-amplified, and cloned into the pMD18-T vector (Takara, Dalian, China). The recombinant plasmids were extracted using the E.Z.N.A Plasmid Mini Kit I (Takara, Dalian, China) and sequenced as the initial templates. The plasmid concentrations were converted into copies of the reference genes, and the standard templates were diluted to 108, 107, 106, 105, 104, 103, and 102 copies/mL. Then, the 7300 Fast system (Applied Biosystems, Foster City, CA, USA) was used to draw a standard curve.
Data calculation and analysis of gene stability
Three Excel-based softwares the BestKeeper, NormFinder and geNorm programs were used to measure the expression stability of the seven candidate genes. The geNorm was used to calculate the expression stability (M) based on the raw expression data (http://medgen.ugent.be/~jvdesomp/genorm/), and NormFinder uses raw data as an input in the form of expression values generated using the comparative Ct (cycle threshold) values (http://www.mdl.dk/publicationsnormfinder.htm). While BestKeeper evaluated the expression stability based on the SD (standard deviation) and CV (coefficient of variance) for the target genes in all the samples (http://www.genequantification.de/bestkeeper.html). Statistical comparisons were made using GraphPad Prism 6 (GraphPad Software Inc., San Diego, CA, USA).
Results
Analysis of the expression levels of the reference genes
The expression of seven reference genes were evaluated in BmBDV-infected or non-infected silkworm at different time points by RT-qPCR. The primer specificities were confirmed by the single bands of the expected sizes on 1% agarose gel and the melting curves with a single distinctive peak (Fig. 1). Standard curves of the seven candidate genes are shown in Table 2. The linear correlation coefficient (R2) ranged from 0.997 to 0.999, and the PCR efficiency commonly varied from 90 to 120%. The regression coefficient values were greater than 0.99. The absolute values of the straight-line gradient were controlled within approximately three to four.
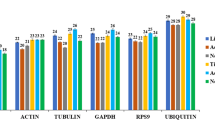
We used the value of the absolute copies to analyze the expression of the control genes at the various time points (Fig. 2). The results showed that the expressions of GAPDH, RPL3, UBQ and 28S rRNA were relatively stable across all the tested samples, while the expressions of actinA3 and ɑ-tubulin exhibited larger variation. TBP was the least expressed gene.
Assessment of the expression stability of the reference genes
We calculated the mean and the standard deviation (SD) of the cycle threshold (Ct) values for all the samples. The number of Ct values was transformed into a quantity via the standard curve based on the PCR efficiency, which allowed the initial copies of the cDNA to be obtained. Three different programs were applied to evaluate the expression stability of the selected reference gene candidates: geNorm, NormFinder, and BestKeeper. To select stably expressed genes, we collected Ct values across all the samples. These raw Ct values were used directly for stability calculations for the BestKeeper analysis, and then they were transformed to relative quantities using the −ΔΔCt method for geNorm, while they transformed to absolute quantities by the standard curves for NormFinder (Sang et al. 2013).
geNorm analysis
geNorm allows the most appropriate mean of the expression of a candidate cDNA to be determined (Jain et al. 2006). The expression stability M for all the reference genes were calculated by the geNorm program based on the average pairwise variation between all the tested reference genes (Bustin et al. 2009). The most stable reference gene has the lowest M value, whereas the least stable one has the highest M value (Bustin et al. 2009). In our analysis, all the reference genes of the BmBDV-infected and uninfected silkworms at different time points had an M value less than the geNorm threshold of 1.5, which is recognized as stability (Fig. 3). The results showed that RPL3 and 28S rRNA had the lowest M values among the seven candidate genes, implying that these two candidate genes are the appropriate reference genes for researching BmBDV infection of silkworms. The data also showed that UBQ and TBP were the least stable control genes.
NormFinder analysis
Another adopted program, NormFinder, which is one of the Visual Basic application tools for Microsoft Excel, is used to calculate the stability value of reference genes based on their intra- and inter-group variations, and it combines both results into a consistent value for each investigated gene (Andersen et al. 2004; Jian et al. 2008). NormFinder not only measures the variation, but it also ranks the potential reference genes by how they differ between studies (Chuaqui et al. 2002). Genes exhibiting lower average expression stability values are regarded as being more stably expressed reference genes (Vandesompele et al. 2002). The ranking generated by this approach (Fig. 4) was approximately similar to that determined by geNorm. Based on our data, NormFinder identified RPL3 and 28S rRNA as the best reference genes, while UBQ and ɑ-tubulin were the least stable control genes for the gene analysis of BmBDV infected-silkworms.
BestKeeper analysis
The BestKeeper program is used to evaluate the expression stability of reference genes using the raw Ct values based on their correlation coefficient (r) to the BestKeeper Index by calculating the SD and coefficient of variance (CV) (Tang et al. 2015). Surprisingly, the BestKeeper analysis revealed that the best correlations were obtained for actin A3 (0.92), ɑ-tubulin (0.90), and RPL3 (0.85) with a P-value of 0.001 across all the samples (Table 3). Because of the high correlation value and low BestKeeper index, actin A3, ɑ-tubulin, and RPL3 were selected as the three likely reference genes. The results varied slightly from the previously mentioned programs, which might result from the distinct statistical algorithms of BestKeeper.
Basing on above analysis and considering that RPL3 and 28S rRNA had the lower SD and CV among these genes, we suggest that RPL3 and 28S rRNA may be the appreciate control genes.
Expression profile of BmSP142 in BmBDV-infected silkworm
The previous studies demonstrated that the expression of BmSP142 varied greatly during BmBDV infection (Li et al. 2017). To verify the reliability of the selected reference genes, we analyzed the expression patterns of BmSP142 by RT-qPCR in BmBDV-infected Huaba35 (BmBDV-sensitive) and HuabaBC7 (BmBDV-resistant) silkworm at different infection times. In consideration of the previous report that TIF-4a (translation initiation factor 4A) was an appropriate reference gene for studies of the effects of BmBDV on silkworms (Guo et al. 2016), the relative expression levels of BmSP142 at different time points across different strains were normalized by RPL3, 28S rRNA and TIF-4A, respectively. Similar expression pattern of BmSP142 was displayed using these there genes as internal control respectively (Fig. 5). The expression levels of BmSP142 were remarkably up-regulated at 24 hpi in resistant silkworm, and significantly higher compared with the sensitive silkworm. Meanwhile, the transcription levels of BmSP142 were markedly up-regulated at 84 hpi in sensitive silkworm, and remarkably higher than that in resistant silkworm.
Discussion
In this study, three approaches were used to investigate seven candidate genes for their potential to be used as reference genes in silkworms after BmBDV infection. RPL3 and 28S rRNA, were found as the most stable genes by geNorm and Normfinder analysis (Figs. 3 and 4). Meanwhile, Bestkeeper has ranked the actin A3, ɑ-tubulin and RPL3 as the three most suitable reference genes, and RPL3 and 28S rRNA had the lower SD and CV among these genes (Table 3). UBQ was consistently identified as the least stable gene by all three algorithms. The analysis of transcript abundant showed that actin A3 and ɑ-tubulin were more variable comparing with the other candidates (Fig. 1). Previous studies reported an interesting property of the absence of the 28S rRNA band usually present in eukaryotic cells when viewed on non-denaturing agarose gels and the dissociation of the 28S rRNA molecule into two 18S rRNA components upon exposure to a brief heat treatment in dipteran species (Balazs and Agosin 1968). Because the expression level of 28S rRNA was significantly higher than that of other genes, 28S rRNA gene may not be ideal for some studies in insect, e.g. in L.cuprina (Bagnall and Kotze 2010). RPL3, which encodes ribosomal protein L3, is expressed stably at different development stages in Rhododendron molle G. Don (Xiao et al. 2016). RPL3 has also been found to be the most stable gene in wing discs of butterfly Heliconius numata at different developmental stages (Prunier et al. 2016). UBQ has also been commonly used as the appropriate internal control for gene expression analysis in different genotype of Vitis viniferarice infected with downy mildew (Monteiro et al. 2013) and in turbot gonad (Robledo et al. 2014). In present study, UBQ was found to be the least stable gene in BmBDV-infected silkworm.
Li et al. (2017) assessed the expression of BmSP142 in BmBDV-infected silkworm by absolute quantification, and their results showed that the expression was significantly up-regulated at 24 hpi in resistant silkworm and up-regulated at 72 hpi in sensitive silkworm. Guo et al. (2016) suggested that the expression level of TIF-4A was stable for BmCPV or BmBDV challenge in silkworms using transcriptome data analysis and RT-qPCR verification. To validate RPL3, 28S rRNA and TIF-4A being the appropriate reference genes in BmBDV-infected silkworm, we evaluated the relative expression levels of BmSP142 in silkworm infected with BmBDV normalized by RPL3, 28S rRNA and TIF-4A, respectively (Fig. 5). Similar expression pattern was observed using these three genes as reference genes respectively. Consistent with the findings of Li et al. (2017), the mRNA levels of BmSP142 changed significantly after virus infection, and the expression patterns of BmSP142 were significantly different between BmBDV-sensitive and BmBDV-resistant silkworm. These results further indicated that RPL3, 28S rRNA and TIF-4A were the reliable reference genes. The expression level of BmSP142 was remarkably up-regulated in resistant silkworm and markedly higher at early stage of infection (24 hpi) than that in sensitive silkworm, suggesting that BmSP142 may play an important role in resistance to BmBDV in silkworm. Genome-wide identification indicate that silkworm possesses 51 serine proteases (SPs) and 92 serine protease homolog genes, which may participate in biological processes such as digestion, development and defence (Ping et al. 2010). BmSP142 was a 35 KDa protein which interacted with nonstructural protein NS1 and NS2 of BmBDV (Bao et al. 2013). Over expression of BmSP142 in BmN cells inhibited the propagation of Bombyx mori nucleopolyhedrovirus (BmNPV), and the replication efficiency of BmBDV and BmNPV treated with purified BmSP142 decreased in silkworm, indicating that BmSP142 may be involved in defence response to virus infection (Li et al. 2017).
Overall, the present study evaluated the expression stability of seven candidate reference genes for RT-qPCR normalization in BmBDV-infected silkworm. RPL3, and 28Sr RNA were identified as the suitable internal control genes under BmBDV challenge. And we further verified that TIF-4A was also a reliable reference gene. The expression patterns of BmSP142 were significantly different in susceptible and resistant silkworm after BmBDV infection, indicating that BmSP142 may be involved in the resistance to virus. This study will be beneficial for investigating the gene expression in silkworm under the BmBDV infection.
Abbreviations
- RT-qPCR:
-
Reverse transcription quantitative real-time polymerase chain reaction
- BmBDV:
-
Bombyx mori bidensovirus
- BmSP142 :
-
Bombyx mori serine protease
- GAPDH :
-
3-phosphoric acid glycerol aldehyde dehydrogenase
- UBQ :
-
ubiquitin
- RPL3 :
-
ribosomal protein L3
- TBP :
-
TATA response element binding protein
- BmNPV:
-
Bombyx mori nucleopolyhedrovirus
- TIF-4A :
-
translation initiation factor 4A
- Ct:
-
cycle threshold
- SD:
-
standard deviation
- CV:
-
coefficient of variance
- hpi:
-
hours post infection
- NCBI:
-
National Center for Biotechnology Information
References
Andersen CL, Jensen JL, Ørntoft TF (2004) Normalization of real-time quantitative reverse transcription-pcr data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64:5245. https://doi.org/10.1158/0008-5472.CAN-04-0496
Bagnall NH, Kotze AC (2010) Evaluation of reference genes for real-time pcr quantification of gene expression in the australian sheep blowfly, lucilia cuprina. Med Vet Entomol 24:176–181. https://doi.org/10.1111/j.1365-2915.2010.00866.x
Balazs I, Agosin M (1968) Isolation and characterization of ribonucleic acid from musca domestica (l.). Comp Biochem Physiol 27:227–237 (PMID: 5758370)
Bao YY, Che LB, Wu WJ et al (2013) Direct interactions between bidensovirus bmdnv-z proteins and midgut proteins from the virus target bombyx mori. FEBS J 280:939–949. https://doi.org/10.1111/febs.12088
Bustin SA, Benes V, Nolan T, Pfaffl MW (2005) Quantitative real-time rt-pcr – a perspective. J Mol Endocrinol 34:597. https://doi.org/10.1677/jme.1.01755
Bustin SA, Benes V, Garson JA et al (2009) The miqe guidelines: minimum information for publication of quantitative real-time pcr experiments. Clin Chem 55:611–622. https://doi.org/10.1373/clinchem.2008.112797
Chen HQ, Yao Q, Bao F et al (2011) Comparative proteome analysis of silkworm in its susceptibility and resistance responses to bombyx mori densonucleosis virus. Intervirology 55:21–28. https://doi.org/10.1159/000322381
Chen KP, Wang JT, Yao Q (2014) Model organism Bombyx mori. Phoenix science press Ltd Nanjing. 4-35pp
Chuaqui RF, Bonner RF, Best CJM et al (2002) Post-analysis follow-up and validation of microarray experiments. Nat Genet 32:509–514. https://doi.org/10.1038/ng1034
Guo H, Liang J, Xia Q (2016) Selection of reference genes for analysis of stress-responsive genes after challenge with viruses and temperature changes in the silkworm bombyx mori. Mol Gen Genomics 291:999–1004. https://doi.org/10.1007/s00438-015-1125-4
Gutierrez L, Mauriat M, Guénin S (2008) The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (rt-pcr) analysis in plants. Plant Biotechnol J 6(6):609–618. https://doi.org/10.1111/j.1467-7652.2008.00346.x
Hu ZY, Li GH, Li GT, Qin Y, Chen KP (2013) Bombyx mori bidensovirus: the type species of the new genus bidensovirus, in the new family bidnaviridae. Chin Sci Bull 58:4528–4532. https://doi.org/10.1007/s00284-014-0613-9
Hu Z, Zhang X, Liu W et al (2016) Genome segments accumulate with different frequencies in bombyx mori bidensovirus. J Basic Microbiol 56(12). https://doi.org/10.1002/jobm.201600120
Huggett J, Dheda K, Bustin S, Zumla A (2005) Real-time rt-pcr normalisation; strategies and considerations. Genes Immun 6:279. https://doi.org/10.1038/sj.gene.6364190
Jain M, Nijhawan A, Tyagi AK, Khurana JP (2006) Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time pcr. Biochem Biophys Res Commun 345:646–651. https://doi.org/10.1016/j.bbrc.2006.04.140
Jian B, Liu B, Bi Y, Hou W, Wu C, Han T (2008) Validation of internal control for gene expression study in soybean by quantitative real-time pcr. BMC Mol Biol 9:1–14. https://doi.org/10.1186/1471-2199-9-59
Jiang L, Zhao P, Cheng T, Sun Q, Peng Z, Dang Y (2013) A transgenic animal with antiviral properties that might inhibit multiple stages of infection. Antivir Res 98(2):171–173. https://doi.org/10.1016/j.antiviral.2013.02.015
Kong J, Hu Z, He Y et al (2011) Expression analysis of bombyx mori, parvo-like virus vd2-orf1 gene encoding a minor structural protein. Biologia 66:684–689. https://doi.org/10.2478/s11756-011-0074-6
Lee PD, Sladek R, Greenwood CMT et al (2002) Control genes and variability: absence of ubiquitous reference transcripts in diverse mammalian expression studies. Genome Res 12:292. https://doi.org/10.1101/gr.217802
Li G, Qian Z, Qiu L et al (2017) Serine protease bm-sp142 was differentially expressed in resistant and susceptible bombyx mori strains, involving in the defence response to viral infection. PLoS One 12:e0175518. https://doi.org/10.1371/journal.pone.0175518
Long X, He B, Gao X et al (2015) Validation of reference genes for quantitative real-time pcr during latex regeneration in rubber tree. Gene 563:190. https://doi.org/10.1016/j.gene.2015.03.026
Monteiro F, Sebastiana M, Pais MS, Figueiredo A (2013) Reference gene selection and validation for the early responses to downy mildew infection in susceptible and resistant Vitis vinifera cultivars. PLoS One 8(9):e72998. https://doi.org/10.1371/journal.pone.0072998
Peng R, Zhai Y, Ding H et al (2012) Analysis of reference gene expression for real-time pcr based on relative quantitation and dual spike-in strategy in the silkworm bombyx mori. Acta Biochim Biophys Sin 44:614–622. https://doi.org/10.1093/abbs/gms040
Ping Z, Wang GH, Dong ZM et al (2010) Genome-wide identification and expression analysis of serine proteases and homologs in the silkworm bombyx mori. BMC Genomics 11:405. https://doi.org/10.1186/1471-2164-11-405
Prunier FP, Chouteau M, Whibley A et al (2016) Selection of valid reference genes for reverse transcription quantitative pcr analysis in heliconius Numata (lepidoptera: nymphalidae). J Insect Sci 16:50. https://doi.org/10.1093/jisesa/iew034
Robledo D, Hernández-Urcera J, Cal RM et al (2014) Analysis of qPCR reference gene stability determination methods and a practical approach for efficiency calculation on a turbot (Scophthalmus maximus) gonad dataset. BMC Genomics 15:648. https://doi.org/10.1186/1471-2164-15-648
Sang J, Han X, Liu M et al (2013) Selection and validation of reference genes for real-time quantitative pcr in hyperaccumulating ecotype of sedum alfredii under different heavy metals stresses. PLoS One 8:e82927. https://doi.org/10.1371/journal.pone.0082927
Tang X, Wang H, Shao C, Shao H (2015) Reference gene selection for qpcr normalization of kosteletzkya virginica under salt stress. Biomed Res Int 2015:823806. https://doi.org/10.1155/2015/823806
Thellin O, Zorzi W, Lakaye B et al (1999) Housekeeping genes as internal standards: use and limits. J Biotechnol 75(2–3):291–295. https://doi.org/10.1016/S0168-1656(99)00163-7
Thellin O, Elmoualij B, Heinen E et al (2009) A decade of improvements in quantification of gene expression and internal standard selection. Biotechnol Adv 27:323–333. https://doi.org/10.1016/j.biotechadv.2009.01.010
Vandesompele J, Preter KD, Pattyn F et al (2002) Accurate normalization of real-time quantitative rt-pcr data by geometric averaging of multiple internal control genes. Genome Biol 3(7). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC126239/
Xiao Z, Sun X, Liu X (2016) Selection of reliable reference genes for gene expression studies onrhododendron molleg. Front Plant Sci 7(e0141853):1547. https://doi.org/10.3389/fpls.2016.01547
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant nos. 31570150 and 31270192), Startup Scientific Research Fund from Jiangsu University for Advanced Professionals (No.11JDG048), and the Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Hu, Z., Deng, Y., Zhang, X. et al. Selection and validation of reference genes for reverse transcription quantitative real-time PCR (RT-qPCR) in silkworm infected with Bombyx mori bidensovirus. Biologia 73, 897–906 (2018). https://doi.org/10.2478/s11756-018-0086-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.2478/s11756-018-0086-6