Abstract
Spider silk is an archetypal biopolymer material with extreme tensile properties arising from its complex hierarchical assembly. While recent advances in sequencing have yielded abundant insights, relatively little is known concerning post-translational modifications (PTMs) in spider silk. Here, we probe the PTM landscape of dragline silk from the Jorō spider (Trichonephila clavata) using a combination of mass spectroscopy and solid-state nuclear magnetic resonance (NMR). The results reveal a wide array of potential modifications, including hydroxyproline, phosphorylation, and dityrosine cross-links, encompassing the different spidroin constituents. Notably, the MaSp3 repetitive region displayed numerous PTMs, whereas MaSp1 and MaSp2 variants showed distinct phosphorylation patterns in its terminal domains. The N-terminal domain (NTD) phosphorylation sites were found predominantly at the dimer interface, suggesting a modulatory function with respect to its pH-driven dimerization function, a hypothesis supported by studies using phosphomimetic NTD mutants. Possible roles of phosphoserine in limiting β-sheet formation, and hydroxyproline in disrupting β-turns are also discussed.
Impact statement
Spider silk is an archetypal biomaterial that can outperform our most sophisticated artificial fibers. The secret to its mechanical properties lies in its complex hierarchical structure—encompassing the nano- to macroscales—that forms through a process of molecular self-assembly of the constituent spidroin proteins. While recent advances in "biomateriomics” have given us tremendous insights into the sequence–function relationships that determine spider silk behavior, the picture is still far from complete. One area that has received little attention is posttranslational modifications (PTMs). PTMs are ubiquitous biological phenomena that are crucial for providing dynamic control of the proteome, and effectively expand the structural and functional design space of proteins beyond that provided by the canonical amino acids. Here, we undertook a comprehensive analysis of PTMs from spider dragline silk fiber, which revealed numerous potential sites for a wide array of modifications. The results provide a fascinating window into additional layers of complexity underlying the mechanical behavior of spider silk, and suggest further avenues for creating novel, dynamically tunable, bioinspired materials.
Graphical abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The extraordinary mechanical performance, biocompatibility, and biodegradability of spider silk have made it an important forerunner in the race to design novel materials inspired by the natural world.1 Spiders can produce up to eight structurally and mechanically distinct silks that are produced by different silk glands within their abdomen, which exhibit a variety of properties depending on their respective usage, from prey capture to protective egg sacs formation.2 Major ampullate (MA) silk (aka dragline silk) is the foremost studied type of spider silk due to its accessibility and superior mechanical performance.
MA silk is composed of multiple high-molecular weight proteins (250–350 kDa) known as the MA spidroins (MaSps).3,4 These MaSp proteins exhibit an extensive repetitive region flanked by relatively short, conserved N- and C-terminal domains (NTD and CTD, respectively). The NTD and CTD are thought to facilitate self-assembly functions (i.e., the controlled transformation of soluble spidroins into hierarchically organized insoluble fibers in response to diverse physicochemical changes in the spider’s fiber-spinning apparatus).5 Such functions include maintenance of solubility, formation of prefibrillar complexes, pH-driven dimerization, and modulation of liquid–liquid phase separation (LLPS).6,7,8 The repetitive region, on the other hand, consists of extended runs of short amino acid motifs and is thought to be largely responsible for the material properties of the mature silk fiber.9,10 The arrangement of motifs in the repetitive sequences make up two structurally distinct components within silk fiber: first, the crystalline component, formed by polyalanine ((A)n) runs, which form β-sheets that stack into nanocrystalline blocks and is thought to impart silk with its strength. Second, a surrounding amorphous component consisting primarily of the glycine-rich sequences from the repetitive domain is thought to impart silk with its extensibilty.10,11,12,13,14 The interplay between these regions is thought to play a major role in ensuring the impressive mechanical performance of dragline silk.15,16 Variations in the abundance of the different MaSp proteins and their constituent motifs, resulting from changes in expression, can explain some but not all variation observed within MA silks’ mechanical properties.11,17,18,19,20
Although techniques such as nuclear magnetic resonance (NMR) spectroscopy and x-ray crystallography have elucidated the structure of the NTD and CTD, as well as the main secondary structures within the repetitive region, a complete picture of the hierarchical structure of the repetitive region is yet to be resolved.14,21,22,23 This is likely due to the difficulty of trying to simultaneously analyze the structure of multiple highly repetitive proteins that comprise a single strand of natural MA silk. With increasing knowledge of the silk genome, we have been able to draw increasingly detailed correlations between silks structure and amino acid sequence, and these insights have been successfully applied toward the production of artificial spider silks with native-like properties.8,24,25,26,27 However, one largely overlooked area is the silk proteome and the role of posttranslational modifications (PTMs).
PTMs are pivotal chemical alterations that can modulate the structure, localization, and function of proteins after their synthesis, and in effect expand proteomic diversity beyond the space afforded by the regular 20 amino acids. These modifications are ubiquitous and encompass a diverse array of covalent changes, such as phosphorylation, glycosylation, acetylation, methylation, ubiquitination, and more.28,29 Phosphorylation, for instance, is recognized as fundamental in regulating a vast array of dynamic biochemical processes such as enzyme activity and cellular signaling cascades.30
PTMs also play crucial roles in modulating the conformation of structural proteins in multicellular organisms, which can often form extended fibers through hierarchical self-assembly. Collagen is the archetypal example, where the conversion of proline (Pro) residues into hydroxyproline (Hyp) is vital to achieving a triple-helical fibril architecture, while oxidation of lysine (Lys) enables covalent cross-linking between adjacent helical bundles.31,32,33 In elastin, lysine PTMs facilitate molecular cross-linking, while Hyp is also reported.32,34 In insects, the rubberlike polymer networks of resilin are cross-linked via dityrosine (diTyr) cross-links, another type of PTM.35
It is also becoming clear that PTMs play important roles in modulating the structure, function, and assembly of externally acting protein biopolymers produced by various organisms.36,37 The underwater attachments (byssus) produced by mussels, for instance, are hierarchically organized, multicomponent assemblies that rely extensively on modified phenylalanine (Phe), known as DOPA, for substrate binding.37,38 Recently, the protein constituents of the slime of velvet worms were found to be heavily phosphorylated, while disulfide bonding and Hyp were also observed.39,40 In insect silks and related fibers, high levels of phosphorylation were detected in the Bombyx mori fibroin heavy chain, while the presence of modified Lys and Tyr suggests cross-linking functions.41,42 In contrast, the underwater silk produced by the caddisfly larva contain β-sheet crystalline structures made from phosphorylated serine (pSer) repeats, as well as diTyr cross-links.43,44
Turning to spider silk, increasing evidence suggests a widespread occurrence of PTMs in the different silk types. The aggregate spidroins (AgSp) constituting the sticky glue of spider webs are both glycosylated and phosphorylated, particularly via serine (Ser) and threonine (Thr) residues.45,46,47 In flagelliform spidroin, the main component of flexible capture threads, Hyp was identified within the GPGGX motifs found throughout its repetitive sequence.48 PTMs have also been previously reported in MA silk. Using solid-state NMR, Craig et al.49 noted the occurrence of Hyp in the dragline silk from Argiope, presumably from Pro-rich MaSp2, hypothesizing that the additional –OH groups could stabilize the GPGXX β-turn conformations50 through additional hydrogen bonding interactions. Mass spectrometry methods have also been used to probe the presence of PTMs in dragline silk of nephilid spiders.51,52,53,54 Analysis of MaSp1 has detected pSer and pTyr in the repetitive regions, and pSer and pThr in the N-terminal domain, with apparent variations between species. For MaSp2, pSer was found in repetitive region, within or directly flanking the polyalanine (Ala) runs, along with pTyr, as well as in C-terminal domain; however, Hyp was not reported. Apparent discrepancies between the different data sets may reflect real variations in PTM profiles across different species/individuals or might be attributed to limitations inherent in the techniques or sequences used for analysis. In any case, so far there has been limited consensus regarding the extent and identity of PTMs associated with spider silk, and the overall structural and functional significance of these modifications have been greatly underappreciated.
The present study aims to elucidate the comprehensive PTM profile of spider major ampullate silk from the Jorō spider, Trichonephila clavata. This was achieved through a multidimensional approach employing tandem mass spectrometry as well as solid-state NMR characterization techniques to identify PTMs and gain insight on their structural consequence. For full characterization, we took advantage of the most up-to-date sequence data from these species, encompassing the full-length sequences for MaSp1, MaSp2, and MaSp3, including subvariants, as well as the various nonspidroin proteins (termed SpiCE) that were recently identified as major components of dragline silk from this species.55 To facilitate identification of phosphorylation sites via MS, phosphopeptide enrichment techniques were carried out, based on HAMMOC and immunoprecipitation, and diet-based isotopic labeling of silk samples was used to significantly enhance signals for NMR analysis. Overall, a diverse array of PTMs were identified, encompassing different protein components, which together suggest unexplored layers of complexity in the sequence and conformational space relevant to the formation of dragline silk.
Results
A comprehensive map of PTMs in MA silk
Identification of PTMs from the dragline silk samples was implemented by comparing the LC–MS/MS data against a reference database of protein sequences generated from the draft genome of T. clavata, as recently reported.55 This reference data set contained the full-length amino acid sequences of the large major ampullate spidroins proteins (MaSp1, MaSp2, MaSp3), including their subvariants; also contained are the associated smaller molecular weight nonspidroin polypeptides abundantly expressed in the dragline fiber, the so-called SpiCE proteins.
Our analysis identified a diverse array of PTMs at different sites, encompassing the whole set of MaSp sequences. A general overview is presented in Figure 1, with a full list of the observed fragments and relevant annotations provided in Supplementary Data File 1, while representative MS/MS spectra of identified peptide ions are shown in Figure S1. Initial analysis of the unenriched dragline silk samples revealed the presence of hydroxyproline (Hyp) across all the major MaSp variants and several phosphoserine (pSer) sites. Subsequent enrichment for phosphopeptides revealed numerous additional phosphorylation sites, including phosphothreonine (pThr), phosphotyrosine (pTyr), and pSer encompassing all the MaSp subtypes. Taken together, 396 potential PTM sites were detected across the eight MaSp variant sequences, along with 23 sites in the smaller SpiCE proteins (Figure 1).
A comprehensive map of observed posttranslational modifications (PTMs) in spider major ampullate silk. Shown are the observed PTM positions with respect to the full-length sequences of the predominant MaSp and SpiCE components of dragline silk from T. clavata. A detailed breakdown of the fragments used to generate the map can be found in the Supplementary material. The numbered residues correspond to phosphorylation of Ser (S, aqua), Thr (T, purple), or Tyr (Y, orange) and hydroxylation of Pro (P, blue). Each protein is drawn to scale, with the scale bar corresponding to 300 residues. NTD, N-terminal domain; CTD, C-terminal domain.
Some general observations are noted (Figure 1). For both MaSp1A and MaSp2, phosphorylation via Ser and Thr were observed at the terminal domains, with conserved sites at the NTD, whereas at the CTD different modification sites are found between MaSp1 and MaSp2 (see following sections). Conversely, phosphorylation events were not detected for the MaSp3 terminal domains. In the repetitive regions, interestingly, various pTyr sites were found interspersed throughout the MaSp1A sequence, while such modifications were not detected in MaSp2. On the other hand, Hyp sites were found associated with the proline-rich MaSp2 repetitive sequence; such sites were localized proximal to the NTD or CTD, with the analysis failing to detect Hyp in the central part of the repetitive region. Overall, however, the highest number of PTM sites was found associated with MaSp3, with multiple instances of pSer, pTyr, and Hyp within the repetitive region. There is a caveat, however, in that the repetitive nature of the sequences can at times preclude the precise assignment of PTM sites, which is especially relevant to MaSp3, where long-range sequence conservation is the most pronounced; for instance, the pattern GYGPGGASGAAAAAAAADGGRGGLY occurs 16 times within the repetitive region of MaSp3B1, thus leading to ambiguity in the placement of fragments within it relative to the full sequence.
The PTMs were found associated with specific contexts in the MaSp repetitive regions. Hyp found in MaSp2 and MaSp3 occurred largely in the context of GPGXX to make a GOGXX motif (where O is Hyp), as previously suggested.49 Moreover, pTyr was observed throughout the repetitive region of MaSp1 and MaSp3 in the context of GGpY motif, and additionally in MaSp3 as part of a GpYGPG motif; however, as noted, we failed to observe it in MaSp2, which contrasts with the previous results from T. clavipes.19 On the other hand, pSer was generally found proximal to the poly-Ala regions in MaSp1 (GpSAAAAAAG) and MaSp3 (pSAAAAAAD and pSGAAAAAAD), and to a lesser extent in LGpSQG, a commonly observed motif in the glycine-rich region of MaSp1.
Regarding the SpiCE proteins, PTMs were identified in two of the sequences, with SpiCE-NMa1 and SpiCE-NMa3 bearing 16 and 6 phosphorylation/hydroxylation sites, respectively, with SPiCE-NMa1, considering its short sequence (249 residues), representing the highest PTM density for all the sequences surveyed.
Structural impact of PTMs on the repetitive region
We conducted solid-state NMR experiments on isotopically enriched MA silk to examine the conformation and structural impact of PTMs on dragline silk, as well as to corroborate the findings from LC–MS/MS. One-dimensional 13C CP-MAS experiments indicate the 13C labeling treatment of the silk was successful (Figure S2), with significant labeling of Tyr and Ser residues as well as Gly, Ala, Pro, Gln, Glu, which can result from redistribution of the 13C through metabolic pathways involved in amino acid synthesis. The labeled spectra also reveal the presence of hydroxylated proline (Hyp) (~71.4 ppm).
Initial monitoring of phosphorylation was carried out through 31P CP-MAS on the silk sample, along with an O-phospho-L-serine standard to identify the phosphorus species associated with phosphorylation (Figure 2a). The silk displayed a major peak at 0.13 ppm associated with pSer and pTyr, similar to that observed in T. clavipes silk.53 An additional smaller downfield peak at 18.1 ppm was also observed, which based on a previous study, could be ascribed to a strained five-membered phosphate ring.53
31P CP-MAS, 13C–13C DARR, and 1H–13C HETCOR analysis of T. clavata major ampullate silk. (a) 31P CP-MAS spectra of T. clavata silk (black) and O-Phosphos-L-Serine (green) displaying main phosphate peaks associated with protein phosphorylation as well as a strained cyclic phosphate peak.53 (b) 13C–13C DARR spectra of labeled T. clavata silk displaying the major observable posttranslational modifications and their cross peaks, as well as the main peaks of other known and observable amino acids in the silk. DiTyr is indicated in red, pTyr in yellow, pSer in green, and Hyp in blue; ssb = spinning side band. The area between 82 and 98 ppm of the spectra has been omitted. (c) 1H–13C HETCOR of labeled T. clavata silk displaying the peak assignments of main observable amino acids and associated cross peaks.
In addition, 2D 13C–13C DARR and 1H–13C HETCOR experiments were performed to probe the characteristic chemical shifts associated with the different PTMs. These revealed the presence of Hyp, pSer, pTyr (but not pThr), and additionally diTyr signals were observed (Figures 2b, S3–S8, Table I). The peaks were assigned based on previous literature and on the Biological Magnetic Resonance Data Bank (BMRB).49,56,57
Hyp was identified by the 70.8 ppm Cγ resonance and an associated Cα cross peak (62.1 ppm). The expected Cδ peak (55.2 ppm) was found (Figure 2a, Table I) which, however, overlaps with a potential spinning side band that may partially contribute to the signal (Figure S3). For the unmodified Pro residue, the associated upfield Cα shift (~60 ppm) is consistent with the proposed Type II β-turn conformation, as previously reported for spider dragline silk.50,58 In the case of Hyp, however, the measured Cα shift corresponds to those reported for random coil conformations, which suggests that the addition of hydroxyl groups might interfere with the formation of β-turns, as observed in elastin model peptides.59,60,61
Phosphorylation of Ser occurs on the Cβ hydroxyl group and generally results in a 2 ppm downfield shift in the Ser Cβ, which was identified with DARR (Cα = 58.2 ppm, Cβ = 65.3 ppm, Figures 2a and S4), and HETCOR via the pSer Cβ/Hβ peaks (Cβ = 65.3 ppm, Hβ = 4.02 ppm). The DARR pSer Cα/Cβ shifts are consistent with random coil conformation and distinct from the observable β-sheet (Ca = 55.0 ppm, Cβ = 63.6 ppm) and random coil (Ca = 58.0 ppm, Cβ = 63.1 ppm) cross peaks (Figures 2 and S4). The pSer Cα/Cβ shifts match the reported random coil pSer chemical shifts in other proteins,62,63 whereas they are distinct from the shifts reported from pSer involved in β-sheet structures (for instance in caddisfly silk).43 The association of pSer with disordered conformations suggests that the phosphorylation may inhibit the propagation of β-sheet structures when located in proximity to the poly-Ala regions, as is often the case, as described.
The Tyr in the DARR displays interesting Cε/Cζ and Cγ/Cζ cross peaks that exhibit small but distinct shoulders at 119.9 ppm and 125.2 ppm, respectively (Figures 2b, S5, and S6). These are unusual shifts for Tyr not previously documented in silk and are consistent with the downfield shift in TyrCε associated with phosphorylation and the formation of diTyr, respectively.62,64,65 Both the pTyr and diTyr show Cα and Cβ shifts indicative of a 31-helical conformation, which align with the previously documented shifts associated with the GGY motif (Figures 2b, S5 and S6, Table I).56,66 The pTyr 31-helical conformation somewhat contrasts with the previous work that modeled MaSp1 with PTMs which suggested that phosphorylation significantly decreased the abundance of 31-helices in favor for α-helical structures.51
PTMs in the terminal domains
The LC–MS/MS data revealed pSer, pThr, pTyr, and Hyp at various sites in the NTD and CTD of the MaSp proteins (Figures 3, S9). While these structurally ordered domains function in the absence of PTMs, the detection of modifications in the native silk material implies the possibility of regulatory roles, as seen in numerous cases with phosphorylation.30 To better understand their potential impact, we modeled their structural impacts on the NTDs from the different MaSp subtypes. We used AlphaFold2 to generate initial models corresponding to the T. clavata sequences (Figure S10). The predicted models all had close structural homology to previously determined empirical structures, with the characteristic 5-helical bundle architecture. High confidence metrics, including for the more sequentially divergent MaSp3 (~30% sequence identity), are consistent with the proposed conservation of biochemical function of the NTDs across the different spidroin types.67
Distribution of PTMs in the MaSp NTDs. (a) Aligned T. clavata MaSp NTD sequences showing positions of identified PTMs in colored text. Grayed out sequences correspond to the secretory peptide. Shown above is the predicted secondary structure, with the five alpha-helical elements (H1–H5) represented as rectangles. Residues are numbered according to the predominant literature. (b) Positions of PTMs identified via LC–MS/MS are mapped onto the predicted NTD monomer structures of the MaSp1 and MaSp2 variants. A large proportion of the PTMs map onto the dimerization interface spanning the helix 2–helix 3 region, situated close to the crucially important charged residues that form distinct cationic and anionic clusters having a dipolar orientation (top and bottom portions, respectively).
Mapping the PTM sites onto the NTD structures shows that they localize onto the solvent accessible surface, with none deeply buried within the 5-helix fold (Figure 3). The phosphorylation sites were generally conserved between variants, with 12 positions in common between MaSp1 and 2. Interestingly, three of the phosphorylation sites (positions 30, 34, and 50) were conserved between MaSp1 and MaSp2 homologs despite the occurrence of Thr/Ser substitutions at these locations. It is notable that a large proportion of the candidate phosphorylation sites were found at the homodimerization interface, formed by the region spanning helices 2 and 3, and which features conserved clusters of positively and negatively charged residues that produces a distinctive dipolar configuration.6 This observation suggests a possibility for the PTMs to modulate the dimerization process (see next section). A closer examination of the locations and patterns of the phosphorylation sites shows that S42, T43, T47, S75, and S76 would be localized in the vicinity of the anionic cluster (comprising D39, D40, D46, E79, E84), whereas S58, S61, S62, and S64 would localize around the cationic cluster (comprising K54, K60, K65 at the interhelical loop), whereas residues such at S/T50 would occupy a somewhat intermediate region (Figure 3b). Notably, the phosphorylation sites near the positively charged cluster were found either alone or in pairs, with the S61/S62, T50/S64, S58/S61, and T47/S58 pairs being observed together in the same fragment in the MS experiments, whereas at S42, T43, S75, and S76 such pairs were not observed.
Probing the effects of phosphorylation on NTD dimerization
The detection of phosphorylation sites near the charged clusters in the NTD is intriguing because the dipolar nature imposed by such charges on the subunit surface is thought to assist a prearrangement of the NTD monomers prior to dimerization, with the oppositely charged clusters of each NTD subunit attracting one another.68 To examine the possible impact of phosphorylation on NTD dimerization, we employed a phosphomimetic strategy, whereby the charge and added side chain length associated with phosphorylation of Ser/Thr can be approximated by a genetic substitution to Asp.69 Asp substitution is found to be additionally appropriate based on the 31P CP-MAS spectra, which indicates the phosphate groups within the silk are protonated, having -1 charge matching the side chain of Asp. This protonation state is logical given the low pH of the spinning duct immediately prior to fiber assembly.
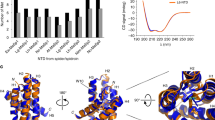
To predict how phosphorylation would impact the electrostatics of the charged clusters, we carried out simulations via APBS on the NTD structures based on some of the observed phosphorylation sites/pairs, either around the positively charged cluster (MaSp1A1 T47D/S58D, S61D/S62D, and MaSp1A2 T50D/S64D) or the negatively charged cluster (MaSp1A2 T43D, MaSp2A1 S50D, and S42D). The results confirm that the simulated phosphorylation sites would indeed induce significant changes to the surface electrostatics, crucial to NTD dimerization, with the effect depending on the location and number of modified sites (Figure 4a). A reduction of the overall positive charge is observed when Asp substitution occurs near the positively charged cluster, and conversely an intensification of the negative charge is predicted when the simulated phosphorylation occurs in proximity to the negatively charged cluster.
N-terminal domain (NTD) phosphomimetic variants’ impact on surface charge distribution and dimerization dynamics. (a) Adaptive Poisson-Boltzmann Solver surface electrostatic simulations of the phosphomimetic NTD variants for MaSp1A1, MaSp1A2, and MaSp2A1 showing the locations of the mutations at and around the dimerization interface and the resultant effects on surface electrostatic potential. (b) pH-driven assays based on tryptophan fluorescence shift were performed for each NTD phosphomimic and compared to wild type. The estimated pK values indicate the midpoint of the dimerization response after curve fitting.
To further assess the impact of phosphorylation on NTD dimer formation, we expressed and purified the above six phosphomimietic variants and their wild-type counterparts (MaSp1A1, MaSp1A2, and MaSp2A1) and tested dimerization behavior by monitoring shifts in intrinsic fluorescence. The mutations produced no apparent ill effects on expression or solubility, and all variants demonstrated pH-dependent dimer formation, although changes in pH responsiveness were observed (Figure 4b). Notably, the introduction of Asp adjacent to the positively charged clusters bearing Arg/Lys resulted in a downward shift in pK such that dimerization was triggered under more acidic conditions compared to the unmodified MaSp1 NTDs, while opposite effects were found with the placement of Asp near the negatively charged clusters (i.e., a shift in dimerization toward higher pH values). The most pronounced effect was found in the MaSp1A1 double-mutant S61D/S62D, with two consecutive substitutions in the loop bridging helix 2 and helix 3 localized next to the main cationic patch on the folded structure, which produced a downward shift in the pK of around −0.37 (Figure 4b). As expected, single substitutions produced smaller perturbations in dimerization response compared to double mutants, as seen in the case of the Asp substitutions near the anionic clusters (MaSp1A2 T43D, MaSp2 S42D, and S50D, with pK shifts of + 0.10 to 0.12). While the actual prevalence of the corresponding NTD modifications within native dragline silk is difficult to assess in the present work, our results demonstrate how specific phosphorylation events can strikingly modulate the pH-dependent assembly of the spidroin building blocks, and might be employed to fine-tune and expand on the range of possible interactions during silk fiber formation process.
Discussion and conclusion
The current methodology, particularly the use of phosphopeptide enrichment techniques, has allowed us to generate the most comprehensive map of PTMs in spider dragline silk, encompassing all the major constituent proteins, including all structural domains within the MaSp proteins. Aided by recent sequencing advances, we could detect previously unknown modifications in the MaSp3 subtypes, particularly multiple pSer, pTyr, and Hyp site within the repetitive region, and also in the small SpiCE proteins. With regard to MaSp1 and MaSp2, the results reveal pSer and pThr as major PTMs within the terminal domains particularly in the NTD near the dimerization interface. Within the repetitive region, pSer was detected flanking the poly-Ala in MaSp1A1 and MaSp3 and within the LGpSQG motif in MaSp1A2. Hyp was also found to be prevalent, predominantly within in MaSp2 and MaSp3 forming as a part of the GPGXX motif, whereas pTyr was found in MaSp1A and MaSp3 as a part of the GGY and GYG motifs, respectively.
In addition, the solid-state NMR results have given us insights into how PTMs might influence silk structure, particularly regarding pSer and Hyp. The pSer was observed predominantly in a random coil conformation; however, many of the phosphorylation sites occur on Ser that would form as a part of the β-sheet forming (A)n runs. Phosphorylation of Ser within the β-sheet forming regions will likely have a disruptive effect, possibly similar to the effect of Asp in a similar context, which acts as a β-sheet terminator on the N-terminal side of β-sheets.70 In dragline silk, pSer could therefore act as a flexible β-sheet terminator within silk, supressing β-strand formation in the N-terminal direction upon phosphorylation (Figure 5a). Similarly, in instances where pSer is found in the interior of (A)n runs (i.e., GAAAApSAAG), the added phosphate group would likely also interfere with β-sheet formation. Interestingly, pSer has also been associated with decreased crystallinity in a recombinant protein system based on MaSp1,71 although the pSer in that case was located within the amorphous region, as opposed to the poly-Ala region.
Predicted impact of posttranslational modifications on the conformation of the repetitive region. (a) Representative models illustrating how pSer located upstream of the poly-Ala region may disrupt the propagation of β-sheets toward the N-terminal direction of the β-strand by increasing the propensity for random coil conformation. (b) Representative models comparing the structures of the GPGXX motif found in the MaSp2 repetitive region. In the unmodified state (top), the motif adopts a regular β-turn conformation featuring an intramolecular H-bond. Hydroxylation to Hyp (bottom) promotes the random coil conformation, which could potentially facilitate H-bonding with neighboring chains.
A similar inhibitory effect on secondary structure appears to be the case with Hyp found in the context of GPGXX motifs (Figure 5b), where the Hyp residues are seen associated with random coil conformation rather than with β-turns, as proposed for the unmodified proline residue. Interestingly, our findings here contrasts with the previous hypothesis that Hyp in MA silk promotes the stability of GPGXX β-turn structures via increased intramolecular H-bonding.49 There is, however, the possibility that the additional hydroxyl group in Hyp could promote intermolecular hydrogen bonding interactions within spider silk, to the detriment of internal β-turn conformations.
In contrast to Hyp and pSer, the pTyr and diTyr modifications did not appear to have any major effect on the backbone conformation. Although the impact of pTyr in spider silk currently remains unknown, it may play a role in influencing the molecular assembly of silk, such as LLPS behavior in which Tyr is thought to play a significant role in disordered proteins like silk.8,72,73 For diTyr, the occurrence of intermolecular cross-links within the amorphous regions of neighboring spidroin chains may have a significant influence on the mechanical properties of the dragline fiber.
In MaSp1 and MaSp2 variants, phosphorylation in the NTD near the dimer interface was seen to influence dimerization behavior by altering the charge distribution on the protein surface, either delaying dimerization with respect to the acidification gradient (due to disruption of the positively charged cluster) or accelerating dimerization when negative charge is added to the negatively charged cluster. Through such effects, phosphorylation may be modulating interactions between NTD monomers, possibly even promoting heterodimer formation through fine control of surface charge potentials. This, however, needs to be examined further to understand the true nature of the phenomenon. This finding ultimately adds a layer of complexity in our understanding of how spiders utilize MaSp proteins, and how they interact, beyond simple differential MaSp expression and spinning effects, which can account for some but not all material property variations observed in natural spider silk.19,74,75
From this study, we uncover a preponderance of PTMs in spider dragline silk involving the various component MaSp proteins and their different structural domains. The characterization presented here gives us the first insights into the structural impact of these PTMs as well as their role as a possible regulatory function particularly in the NTD. At the same time, we acknowledge limitations of the study, particularly the fact that the methods do not allow for a quantitative assessment of the PTM abundance, which hopefully might be addressed in the future using more specialized experimental approaches. This study highlights a significant gap in our understanding of spider silks proteome as a major part of silks broader materiome, and the potential for more sophisticated artificial silks that incorporate the added functionality and complexity associated with the silk proteome.
Materials and methods
Silk collection and 13C labeling
Six adult female T. clavata were collected from around Wakoshi, Saitama, Japan. The spiders were given unique marks on their abdomen with nontoxic paint POSCA markers and housed in an indoor enclosure and allowed to build webs freely for four weeks. These webs were the primary method of feeding the spiders with live crickets every two days. In addition, four of the spiders were also directly fed a solution of 13C 15N uniformly labeled serine (10% w/v) and phenylalanine (9% w/v) mixed with cricket homogenate on alternating days using a micropipette. MA silk was collected on alternating days using a reeling apparatus76 at a rate of 1.28 m/min for 40 to 90 min until the glands were depleted of silk. A total of 28 mg of unlabeled silk fiber, including the silk from the first collection of all six spiders, and 19 mg of labeled silk from weeks three and four was collected from the spiders used in the analysis.
Sample preparation for proteomics
Collected MA silk samples were immersed in 6 M guanidine-HCl (pH 8.5) and homogenized using Biomasher II (Nippi Inc.) and sonication (Bioruptor II, BM Equipment Co., Ltd.) to extract proteins. The protein concentration of each sample was quantified using a Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). Three workflows were used subsequently to prepare samples for proteomics and facilitate identification of PTM sites.
-
1.
Simple digestion: each lysate (50 µg proteins in 50 µL) was reacted with 9.8 mM dithiothreitol (DTT) at 37°C for 30 min followed by 46.7 mM iodoacetamide (IAA) at 37°C for 30 min in the dark. After fivefold dilution with 50 mM ammonium bicarbonate, the samples were digested using 1.1 µg of lysyl endopeptidase (Lys-C) at 37°C for 3 h followed by 1 µg of trypsin at 37°C for 16 h (enzyme/substrate = 1:50, w/w). In addition, the diluted lysates were treated with 1 µg of chymotrypsin at 37°C for 18 h. Each digested sample was acidified by trifluoroacetic acid (TFA) and desalted using a C18 stage tip.77 Samples were dried under reduced pressure and stored at −30°C until nanoLC-MS/MS data collection.
-
2.
Hydroxy acid-modified metal oxide chromatography (HAMMOC):78 each lysate (100 µg proteins in 50 µL) and the mixture (1000 µg proteins in 500 µL) were treated with DTT, IAA, and protease (Lys-C/trypsin or chymotrypsin) followed by desalting using C18 stage tips as with workflow (1.). The elution was transferred to a TiO2 stage tip, and phosphopeptides were enriched by HAMMOC. Briefly, the loaded sample was washed twice by 0.09% TFA/10% lactic acid (LA)/72% acetonitrile (ACN), once by 0.1% TFA/25% LA/60% ACN and twice by 0.1% TFA 80% ACN. The binding peptides were eluted four times by 0.5% piperidine/15% ACN. Each effluent was acidified by TFA and desalted using a C18 stage tip. Samples were dried under reduced pressure and stored at −30°C until nanoLC-MS/MS.
-
3.
Immune precipitation (IP): mixed lysate (960 µg proteins in 480 µL) was subjected to enzymatic digestion followed by HAMMOC as with workflow (2.). The four effluents of HAMMOC were acidified by TFA, combined and desalted using a C18 stage tip. The phosphopeptide sample enriched from 3840 µg of proteins was dried, redissolved in the IP buffer (50 mM Tris–HCl, pH 7.5, 50 mM NaCl), and mixed gently with P-Tyr-1000 beads (#8803, Cell Signaling Technology) at 4°C for 20 h. After snap centrifugation, the supernatant was removed as flow-through and the beads were washed 4 × with IP buffer. Peptide fragments bound to the anti-pTyr antibodies were detached from the beads by mixing with 0.1% TFA twice. The flow-through and binding fractions were desalted using C18 stage tip, dried under reduced pressure, and stored at − 30°C until nanoLC-MS/MS.
NanoLC-MS/MS
Each sample was dissolved in 0.1% formic acid (FA), 2% ACN, and analyzed by nanoLC-MS/MS using a nanoElute ultrahigh-performance LC apparatus and a timsTOF Pro mass spectrometer (Bruker Daltonics). Protein digest sample was injected into a self-packed column (ACQUITY UPLC BEH C18 (Waters), 1.7 µm, 75 µm i.d. × 150 mm or 250 mm length, 3 µm tip i.d.), and separated by linear gradient elution. The mixture of solution A (0.1% FA/water) and B (0.1% FA/ACN) was used as mobile phase at the flow rate of 280 nL/min maintaining at 60°C (solution B concentration: 2–35% for 15 min, 35–80% for 5 min, and 80–80% for 5 min with the 150 mm column; 2–35% for 100 min, 35–80% for 10 min, and 80–80% for 10 min with the 250 mm column). The separated peptides were ionized at 1600 V and analyzed by data-dependent acquisition parallel accumulation–serial fragmentation scan (PASEF).79 The LC–MS raw data files with analytical parameters were deposited in the ProteomeXchange Consortium (Accession Number: PXD039602) via the jPOST partner repository80 (Accession Number: JPST001986).
PTM analysis from LC–MS data
The protein sequences generated from the draft genome data set of T. clavata were used as a reference database.55 LC–MS data files of simple digestion samples were analyzed using PEAKS X + software (version 10.5).81 After de novo sequencing followed by database search (mass tolerance of precursor and fragment ion: 20 ppm and 0.05 Da, enzyme: trypsin or chymotrypsin, maximum missed cleavage: 2, fixed modification: carbamidomethylation at Cys, variable modification: acetylation at protein N-term and oxidation at Met, identified criteria: < 1% FDR at the peptide spectrum match level), PEAKS PTM search82 was performed for further PTM analysis (variable modification: phosphorylation at Ser/Thr/Tyr, hydroxylation at Pro, de novo score: >15%). Meanwhile, LC–MS data files of HAMMOC and IP samples were analyzed using FragPipe software (version 15.0).83 Database search was performed using the following conditions: mass tolerance of precursor and fragment ion: 20 ppm and 0.05 Da, enzyme: trypsin or chymotrypsin, maximum missed cleavage: 2, fixed modification: carbamidomethylation at Cys, variable modification: acetylation at protein N-term, oxidation at Met and phosphorylation at Ser/Thr/Tyr, identified criteria: <1% FDR at the protein level. All of the identified peptide sequences and PTMs were merged into a single data set for analysis.
Solid-state nuclear magnetic resonance spectroscopy
Solid-state NMR experiments were run on a 700 MHz JEOL Resonance ECAII/ECZ Spectrometer with a 3.2-mm HX-MAS probe. The silk material was packed into a 3.2-mm rotor with PCTFE spacers and VESPEL (top) and PEEK (bottom) caps. The rotor was spun up to a final speed of 15 kHz at the magic spinning angle (MAS). The 13C and 31P spectra were recorded using cross-polarization MAS NMR (CP-MAS), and processed using MestReNOVA software,84 with 4× zero filling.
Two-dimensional 13C–13C CP-dipolar-assisted rotational resonance (DARR) experiments85,86 were performed at 15 kHz MAS, with a long DARR mixing time of 1 s. The spectrum was acquired with 64 t1 increments, 112 to 272 transients, and a recycle delay of 3 s that resulted in an experimental time of ~12 to 29 h. The spectral data were processed with 8× zero filling and linear prediction in the indirect dimension.
The 2D 1H-13C heteronuclear correlation (HETCOR) spectra were acquired with 1 ms contact time, 64 t1 increments, 48 transients, and a recycle delay of 3 s that resulted in an experimental time of ~5 h. Spectral data were processed with 4 × zero filling and linear prediction in the indirect dimension.
Preparation of phosphomimetic NTD variants
The NTD phosphomimetic variants were created by substituting phosphorylation sites detected by MS/MS with Asp to create the following mutants for MaSp1A1: T47D/S58D, S61D/S62D; MaSp1A2: T43D, T50D/S58D; MaSp2: S42D, T50D.87 The constructs were purchased from Invitrogen (GeneArt synthesis) in a pMA plasmid, and subcloned into a pET15b vector by restriction digestion and ligation methods. Constructs were transformed into BL21(DE3) cells, grown in LB medium with 100 µg/mL ampicillin at 37°C until OD600 = ~1.0, then expression was induced with 0.4 mM IPTG supplemented with 0.01% glucose and grown overnight at 20°C. Cells were harvested by centrifugation, resuspended in 20 mM Tris–HCl pH 7.5, 0.5 M NaCl, 20 mM imidazole, and lysed via sonication. After centrifugation, the soluble fractions were subjected to purification on a 5 mL His-TRAP FF column (Cytiva), subjected to overnight cleavage of the His-tag using thrombin (Sigma), concentrated using VivaSpin (Sartorius), and further purified size exclusion chromatography using a Superdex 75 Increase 10/300 GL column in 20 mM Tris–HCl pH 7.5, 150 mM NaCl using an ÄKTA Explorer instrument (Cytiva). The purified phosphomimetic mutants were run on SDS-PAGE to evaluate purity, molecular weight, and to ensure complete tag removal (Figure S11).
NTD dimerization assays
The dimerization behavior of the wild-type and phosphomimetic NTD variants was evaluated as a function of pH by monitoring changes in tryptophan fluorescence spectra.68,88 Each 80-μL assay contained 5 μM of NTD against a background of 150 mM NaCl, while the pH was controlled by means of a mixed buffer system containing 20 mM each of sodium acetate, MES, and HEPES, with the pH adjusted between 4.75 and 7.25, in 0.25 increments. The assays were prepared in black 96-well plates (Greiner), and fluorescence spectra were obtained at 25°C using a SpectraMax iD3 plate reader (Molecular Devices) with excitation of 280 nm and emission detected from 300 to 400 nm. Response curves were generated by calculating the ratio of the emission intensity between 338 and 353 nm and plotting the results against pH. The dimerization midpoint (pK) of each data set was estimated from the Ec50 values of the sigmoidal curves generated by fitting the data using QtiPlot (IONDEV SRL).8
Structural prediction of the terminal variants
Monomeric structural models for the NTD variants were generated using the ColabFold version of AlphaFold2.89,90 Primary sequences were input and prediction was run using pdb70 template mode, which for identified and used the following homologous PDB structures (with subunit indicated): 2lth_B, 7but_A, 6tv5_A, 2n3e_A, 2mx9_B, 3lr2_B, 7a0o_A, 5iz2_A, 4fbs_A, 5iz2_B, and 2k3q_A. Multiple sequence alignments (MSA) were generated by Many-against-Many sequence searching (MMseqs2) against the UniRef100 and environmental databanks. AlphaFold2-ptm was run with 48 recycles to achieve the highest accuracy for the models. Resultant model quality was assessed based on the prediction confidence metric (pLDDT).
APBS electrostatic simulations
Adaptive Poisson–Boltzmann Solver (APBS) electrostatic simulations on the wild-type and phosphomimetic NTDs were run within PyMOL using pdb2pqr preparation; the APBS map had 0.5 grid spacing with a ± 5 charge range.91
Data availability
The authors declare that the data supporting the findings of this study are available within the paper and its supporting information. Should any raw data files be needed in another format they are available from the corresponding author upon reasonable request. Source data are provided with this paper.
References
F.G. Omenetto, D.L. Kaplan, Science 329, 528 (2010)
R. Foelix, Biology of Spiders, 3rd edn. (Oxford University Press, Oxford, 2011)
N.A. Ayoub, J.E. Garb, R.M. Tinghitella, M.A. Collin, C.Y. Hayashi, PLoS One 2, e514 (2007)
A.D. Malay, H.C. Craig, J. Chen, N.A. Oktaviani, K. Numata, Biomacromolecules 23, 1827 (2022)
F. Vollrath, D.P. Knight, Nature 410, 541 (2001)
G. Askarieh, M. Hedhammar, K. Nordling, A. Saenz, C. Casals, A. Rising, J. Johansson, S.D. Knight, Nature 465, 236 (2010)
F. Hagn, L. Eisoldt, J.G. Hardy, C. Vendrely, M. Coles, T. Scheibel, H. Kessler, Nature 465, 239 (2010)
A.D. Malay, T. Suzuki, T. Katashima, N. Kono, K. Arakawa, K. Numata, Sci. Adv. 6, 6030 (2020)
J. Gatesy, C. Hayashi, D. Motriuk, J. Woods, R. Lewis, Science 291, 2603 (2001)
J.M. Gosline, P.A. Guerette, C.S. Ortlepp, K.N. Savage, J. Exp. Biol. 202, 3295 (1999)
K. Arakawa, N. Kono, A.D. Malay, A. Tateishi, N. Ifuku, H. Masunaga, R. Sato, K. Tsuchiya, R. Ohtoshi, D. Pedrazzoli, A. Shinohara, Y. Ito, H. Nakamura, A. Tanikawa, Y. Suzuki, T. Ichikawa, S. Fujita, M. Fujiwara, M. Tomita, S.J. Blamires, J.A. Chuah, H. Craig, C.P. Foong, G. Greco, J. Guan, C. Holland, D.L. Kaplan, K. Sudesh, B.B. Mandal, Y. Norma-Rashid, N.A. Oktaviani, R.C. Preda, N.M. Pugno, R. Rajkhowa, X. Wang, K. Yazawa, Z. Zheng, K. Numata, Sci. Adv. 8, eabo6043 (2022)
J.E. Jenkins, G.P. Holland, J.L. Yarger, Soft Matter 8, 1947 (2012)
A. Simmons, E. Ray, L.W. Jelinski, Macromolecules 27, 5235 (1994)
J.D. van Beek, S. Hess, F. Vollrath, B.H. Meier, Proc. Natl. Acad. Sci. U.S.A. 99, 10266 (2002)
K. Numata, H. Masunaga, T. Hikima, S. Sasaki, K. Sekiyama, M. Takata, Soft Matter 11, 6335 (2015)
K. Yazawa, A.D. Malay, H. Masunaga, Y. Norma-Rashid, K. Numata, Commun. Mater. 1, 10 (2020)
T.A. Blackledge, M. Kuntner, M. Marhabaie, T.C. Leeper, I. Agnarsson, Sci. Rep. 2, 833 (2012)
S.J. Blamires, I.C. Chao, I.M. Tso, J. Exp. Biol. 213, 3906 (2010)
S.J. Blamires, C.L. Wu, T.A. Blackledge, I.M. Tso, J. R. Soc. Interface 9, 2479 (2012)
B.O. Swanson, T.A. Blackledge, A.P. Summers, C.Y. Hayashi, Evolution 60, 2539 (2006)
N.A. Oktaviani, A. Matsugami, F. Hayashi, K. Numata, Chem. Commun. (Camb.) 55, 9761 (2019)
N.A. Oktaviani, A. Matsugami, A.D. Malay, F. Hayashi, D.L. Kaplan, K. Numata, Nat. Commun. 9, 2121 (2018)
J.L. Yarger, B.R. Cherry, A. van der Vaart, Nat. Rev. Mater. 3, 18008 (2018). https://doi.org/10.1038/natrevmats.2018.8
J. Chen, A. Tsuchida, A.D. Malay, K. Tsuchiya, H. Masunaga, Y. Tsuji, M. Kuzumoto, K. Urayama, H. Shintaku, K. Numata, Nat. Commun. 15, 527 (2024)
A. Koeppel, C. Holland, ACS Biomater. Sci. Eng. 3, 226 (2017)
A. Rising, J. Johansson, Nat. Chem. Biol. 11, 309 (2015)
T. Arndt, P.R. Laity, J. Johansson, C. Holland, A. Rising, ACS Biomater. Sci. Eng. 7, 462 (2021)
O.N. Jensen, Nat. Rev. Mol. Cell Biol. 7, 391 (2006)
C.T. Walsh, S. Garneau-Tsodikova, G.J. Gatto Jr., Angew. Chem. Int. Ed. 44, 7342 (2005)
P. Cohen, Trends Biochem. Sci. 25, 596 (2000)
B. Brodsky, A.V. Persikov, Adv. Protein Chem. 70, 301 (2005)
D.R. Eyre, M.A. Paz, P.M. Gallop, Annu. Rev. Biochem. 53, 717 (1984)
M.D. Shoulders, R.T. Raines, Annu. Rev. Biochem. 78, 929 (2009)
C.E.H. Schmelzer, M.B.M. Nagel, S. Dziomba, Y. Merkher, S.S. Sivan, A. Heinz, Biochim. Biophys. Acta Gen. Subj. 1860, 2169 (2016)
S.O. Andersen, Biochim. Biophys. Acta Gen. Subj. 93, 213 (1964)
A. Miserez, J. Yu, P. Mohammadi, Chem. Rev. 123, 2049 (2023)
A. Rising, M.J. Harrington, Chem. Rev. 123, 2155 (2022)
H.G. Silverman, F.F. Roberto, Mar. Biotechnol. 9, 661 (2007)
A. Baer, S. Hänsch, G. Mayer, M.J. Harrington, S. Schmidt, Biomacromolecules 19, 4034 (2018)
Y. Lu, B. Sharma, W.L. Soon, X. Shi, T. Zhao, Y.T. Lim, R.M. Sobota, S. Hoon, G. Pilloni, A. Usadi, Adv. Sci. 9, 2201444 (2022)
W.Q. Chen, H. Priewalder, J.P. Pradeep John, G. Lubec, Proteomics 10, 369 (2010)
D.J. Raven, C. Earland, M. Little, Biochim. Biophys. Acta Protein Struct. 251, 96 (1971)
J.B. Addison, N.N. Ashton, W.S. Weber, R.J. Stewart, G.P. Holland, J.L. Yarger, Biomacromolecules 14, 1140 (2013)
C.-S. Wang, N.N. Ashton, R.B. Weiss, R.J. Stewart, Insect Biochem. Mol. Biol. 54, 69 (2014)
N.A. Ayoub, L. DuMez, C. Lazo, M. Luzaran, J. Magoti, S.A. Morris, R.H. Baker, T. Clarke, S.M. Correa-Garhwal, C.Y. Hayashi, Front. Ecol. Evol. 11, 1099481 (2023)
E.K. Tillinghast, M.A. Townley, T.N. Wight, G. Uhlenbruck, E. Janssen, MRS Online Proc. Libr. Arch. 292, 9 (1992)
F. Vollrath, E.K. Tillinghast, Naturwissenschaften 78, 557 (1991)
J. dos Santos-Pinto, H. Arcuri, F. Esteves, M. Palma, G. Lubec, Sci. Rep. 8, 14674 (2018). https://doi.org/10.1038/s41598-018-33068-9
H.C. Craig, S.J. Blamires, M.A. Sani, M.M. Kasumovic, A. Rawal, J.M. Hook, Chem. Commun. (Camb.) 55, 4687 (2019)
J.E. Jenkins, M.S. Creager, E.B. Butler, R.V. Lewis, J.L. Yarger, G.P. Holland, Chem. Commun. 46, 6714 (2010)
J.R. dos Santos-Pinto, H.A. Arcuri, H. Priewalder, H.C. Salles, M.S. Palma, G. Lubec, J. Proteome Res. 14, 3859 (2015)
J.R. dos Santos-Pinto, G. Lamprecht, W.Q. Chen, S. Heo, J.G. Hardy, H. Priewalder, T.R. Scheibel, M.S. Palma, G. Lubec, J. Proteomics 105, 174 (2014)
C.A. Michal, A.H. Simmons, B.G. Chew, D.B. Zax, L.W. Jelinski, Biophys. J. 70, 489 (1996)
J.R. dos Santos-Pinto, H.A. Arcuri, G. Lubec, M.S. Palma, Biochim. Biophys. Acta Proteins Proteom. 1864, 1444 (2016)
N. Kono, H. Nakamura, M. Mori, Y. Yoshida, R. Ohtoshi, A.D. Malay, D.A. Pedrazzoli Moran, M. Tomita, K. Numata, K. Arakawa, Proc. Natl. Acad. Sci. U.S.A. 118(31), e2107065118 (2021). https://doi.org/10.1073/pnas.2107065118
T. Izdebski, P. Akhenblit, J.E. Jenkins, J.L. Yarger, G.P. Holland, Biomacromolecules 11, 168 (2010)
P.R. Romero, N. Kobayashi, J.R. Wedell, K. Baskaran, T. Iwata, M. Yokochi, D. Maziuk, H. Yao, T. Fujiwara, G. Kurusu, E.L. Ulrich, J.C. Hoch, J.L. Markley, in Structural Bioinformatics, Methods and Protocols, vol. 2112, ed. by Z. Gáspári (Humana, New York, 2020). https://doi.org/10.1007/978-1-0716-0270-6_14
X. Shi, J.L. Yarger, G.P. Holland, Chem. Commun. 50, 4856 (2014)
H.R. Kricheldorf, D. Müller, Int. J. Biol. Macromol. 6, 145 (1984)
K. Ohgo, J. Ashida, K.K. Kumashiro, T. Asakura, Macromolecules 38, 6038 (2005)
B. Bochicchio, A. Laurita, A. Heinz, C.E. Schmelzer, A. Pepe, Biomacromolecules 14, 4278 (2013)
A.C. Conibear, K.J. Rosengren, C.F.W. Becker, H. Kaehlig, J. Biomol. NMR 73, 587 (2019)
R. Hendus-Altenburger, C.B. Fernandes, K. Bugge, M.B.A. Kunze, W. Boomsma, B.B. Kragelund, J. Biomol. NMR 73, 713 (2019)
C. Chrissian, C.P.-C. Lin, E. Camacho, A. Casadevall, A.M. Neiman, R.E. Stark, J. Fungi 6, 329 (2020)
D.I. Lee, S. Hwang, J.Y. Choi, I.S. Ahn, C.H. Lee, Process Biochem. 43, 999 (2008)
A.D. Malay, K. Arakawa, K. Numata, PLoS One 12, e0183397 (2017)
J.E. Garb, N.A. Ayoub, C.Y. Hayashi, BMC Evol. Biol. 10, 243 (2010)
S. Schwarze, F.U. Zwettler, C.M. Johnson, H. Neuweiler, Nat. Commun. 4, 2815 (2013)
D. Somale, G. Di Nardo, L. di Blasio, A. Puliafito, M. Vara-Messler, G. Chiaverina, M. Palmiero, V. Monica, G. Gilardi, L. Primo, P.A. Gagliardi, Sci. Rep. 10, 591 (2020)
F. FarzadFard, N. Gharaei, H. Pezeshk, S.A. Marashi, J. Struct. Biol. 161, 101 (2008)
S. Winkler, D. Wilson, D. Kaplan, Biochemistry 39, 12739 (2000)
Y. Lin, S.L. Currie, M.K. Rosen, J. Biol. Chem. 292, 19110 (2017)
D. Stengel, M. Saric, H.R. Johnson, T. Schiller, J. Diehl, K. Chalek, D. Onofrei, T. Scheibel, G.P. Holland, Biomacromolcules 24, 1463 (2023)
S.J. Blamires, C.L. Wu, I.M. Tso, PLoS One 7, e31626 (2012)
C. Boutry, M. Rezac, T.A. Blackledge, PLoS One 6, e22467 (2011)
R.W. Work, P.D. Emerson, J. Arachnol. 10, 1 (1982)
J. Rappsilber, Y. Ishihama, M. Mann, Anal. Chem. 75, 663 (2003)
N. Sugiyama, T. Masuda, K. Shinoda, A. Nakamura, M. Tomita, Y. Ishihama, Mol. Cell. Proteomics 6, 1103 (2007)
F. Meier, A.D. Brunner, S. Koch, H. Koch, M. Lubeck, M. Krause, N. Goedecke, J. Decker, T. Kosinski, M.A. Park, N. Bache, O. Hoerning, J. Cox, O. Rather, M. Mann, Mol. Cell. Proteomics 17, 2534 (2018)
S. Okuda, Y. Watanabe, Y. Moriya, S. Kawano, T. Yamamoto, M. Matsumoto, T. Takami, D. Kobayashi, N. Araki, A.C. Yoshizawa, T. Tabata, N. Sugiyama, S. Goto, Y. Ishihama, Nucleic Acids Res. 45, D1107 (2017)
B. Ma, K. Zhang, C. Hendrie, C. Liang, M. Li, A. Doherty-Kirby, G. Lajoie, Rapid Commun. Mass Spectrom. 17, 2337 (2003)
X. Han, L. He, L. Xin, B. Shan, B. Ma, J. Proteome Res. 10, 2930 (2011)
F. Yu, S.E. Haynes, G.C. Teo, D.M. Avtonomov, D.A. Polasky, A.I. Nesvizhskii, Mol. Cell. Proteomics 19, 1575 (2020)
M.R. Willcott, J. Am. Chem. Soc. 131, 13180 (2009)
K. Takegoshi, S. Nakamura, T. Terao, Chem. Phys. Lett. 344, 631 (2001)
K. Takegoshi, S. Nakamura, T. Terao, J. Chem. Phys. 118, 2325 (2003)
N. Dissmeyer, A. Schnittger, Plant Kinases: Methods Prot. (2011). https://doi.org/10.1007/978-1-61779-264-9_6
K. Jaudzems, G. Askarieh, M. Landreh, K. Nordling, M. Hedhammar, H. Jornvall, A. Rising, S.D. Knight, J. Johansson, J. Mol. Biol. 422, 477 (2012)
J. Jumper, R. Evans, A. Pritzel, T. Green, M. Figurnov, O. Ronneberger, K. Tunyasuvunakool, R. Bates, A. Žídek, A. Potapenko, Nature 596, 583 (2021)
M. Mirdita, K. Schütze, Y. Moriwaki, L. Heo, S. Ovchinnikov, M. Steinegger, Nat. Methods 19, 679 (2022)
E. Jurrus, D. Engel, K. Star, K. Monson, J. Brandi, L.E. Felberg, D.H. Brookes, L. Wilson, J. Chen, K. Liles, Protein Sci. 27, 112 (2018)
Acknowledgments
The authors thank S. Ohnuma and Y. Takai (Keio University) for technical support in proteomic analysis.
Funding
The work was funded by the ImPACT Program of Council for Science, research funds from the Yamagata Prefectural Government and Tsuruoka City, Japan, Grants-in-Aid from the Japan Science and Technology Agency Core Research for Evolutional Science and Technology (JST-CREST; Grant No. JPMJCR2091), the Japan Science and Technology Agency Exploratory Research for Advanced Technology (JST-ERATO; Grant No. JPMJER1602), JST COI-NEXT (Grant No. JPMJPF2114), and MEXT Program: Data Creation and Utilization-Type Material Research and Development Project (Grant No. JPMXP1122714694).
Author information
Authors and Affiliations
Contributions
H.C.C. performed most experiments, analyzed the data, and wrote the manuscript; A.D.M. conceptualized the study and wrote the manuscript; F.H. undertook NMR experiments and performed data analysis; M.M. performed LC–MS/MS and subsequent data analysis; K.A. designed the LC–MS/MS measurements and related data analysis; K.N. conceptualized and supervised the study and also wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Craig, H.C., Malay, A.D., Hayashi, F. et al. Posttranslational modifications in spider silk influence conformation and dimerization dynamics. MRS Bulletin (2024). https://doi.org/10.1557/s43577-024-00771-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1557/s43577-024-00771-0