Abstract
Scale-down models are indispensable and crucial tools for process understanding and continuous process improvement in product life-cycle management. In this study, a scale-down model representing commercial-scale cell culture process of adalimumab biosimilar HS016 was first developed based on constant power per volume (P/V) principle and then qualified by multivariate data analysis (MVDA) and equivalence test method. The trajectories of the bench-scale process lie in the middle of the control range of large-scale process, built by multivariate evolution model based on nutrients, metabolites, and process performance datasets. This indicates that the small-scale process performance is comparable with that of the full-scale process. The final product titer, integrated viable cell density (iVCD), viability, aggregates, acid peak content, total afucosylation level, and high mannose content recognized as key process attributes (KPAs) or critical quality attributes (CQAs) were equivalent across the scales upon comparison using equivalence test method. The qualified scale-down model was then used for process characterization using a definitive screening design (DSD) where five independent variables including pH, shifted temperature, inoculation seeding density, viable cell density (VCD) at first feeding, VCD at temperature shift were evaluated. Three quadratic polynomial models for final product titer, aggregation, and high mannose were then established using the DSD results. The design space was finally developed using a probability-based Monte Carlo simulation method and was verified with the operation setpoint and worst-case condition. The case study presented in this report shows a feasible roadmap for cell culture process characterization.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
The objective of pharmaceutical process development is to establish a robust and well-controlled manufacturing process being capable of consistently delivering a cost-effective and high-quality product in product life-cycle (1). Process characterization is a principal tool to enhance the process understanding and avoid process deviations and batch failure. By means of identifying critical process parameters (CPPs), critical material attributes (CMAs), and critical quality attributes (CQAs) for a given product and process and then exploring the linkage between CPPs/CMAs and CQAs, process characterization applying a quality by design (QbD) principle helps to establish the design space for whole process and leads to a risk-based control strategy for efficient manufacturing (2). For biologics, it is impractical to perform process characterization at full-scale due to high cost, resource limitation, and a huge work undertaking to evaluate hundreds of independent variables. According to ICH guidelines, justified small-scale models being representative of the proposed commercial process can be used to support process design studies (3). Therefore, developing and qualifying a scale-down model is indispensable for process characterization and understanding.
Being different from the scaling-up process, a scale-down model should have a similar or some aspect similar performance of the production-scale process. Therefore, the scale-down models must be qualified after being established to ensure that they have a predictable relationship with full-scale process performance.
Several statistical tools have been applied for scale-down model qualification including risk analysis, Student’s t test, quality range approach, equivalence test, and multivariate data analysis (MVDA) (4,5,6,7,8). The Student’s t test is considered incompetent for this kind of compatibility since the acceptance of the null hypothesis means there is insufficient data to reject it and it does not prove there is no difference, whereas the rejection of the null hypothesis means difference which may be too small to be clinically meaningful (5). Conversely, equivalence test, also known as two one-sided t tests (TOST), tries to reject the null hypothesis of inequivalence to demonstrate that the two sets of datasets are within a meaningful margin (9). Equivalence test, widely used to demonstrate comparability in clinical trials and chemistry, manufacturing, and controls (CMC) studies, has recently been reported to apply into scale-down model qualification (5,6,9,10,11). Nevertheless, the aforementioned methods are limited by individual CQA or CPP comparability and therefore cannot capture the linkage and relationships among variables. Multivariate data analysis (MVDA) method provides a simple approach to compare the complex process by projecting the multidimensional datasets into a few principal components and allow researchers to explore the potential linkage among variables. The MVDA method had been reported using comparability study of cell culture process across scales (7,8,12).
Traditionally, a two-step experimental design has been used for process characterization. First, a low-resolution screening design is explored, and then the response surface design for design space exploration, such as central composite design and Box-Behnken design, is implemented (13). As cell culture process usually lasts for several weeks from seed recovery to cell harvest, high cost of manpower and material resources must be considered. Recently, a novel one-step design method named definitive screening design (DSD) requiring as few as 2 K + 1 experiment runs for K independent variables was proposed by Jones and Nachtsheim (14). In DSD method, the main effects are uncorrelated with two-factor interactions and quadratic effects, and two-factor interactions are not confounded with each other, a key advantage over standard screening design. Therefore, DSD is more efficient than the standard approach in one design for identifying main linear effects, main quadratic effects, and two-factor interactions based on the sparsity-of-effects principle (15). The DSD approach is being increasingly employed in process development and characterization of biologics (16,17,18).
In this study, a scale-down model representing commercial-scale cell culture process of adalimumab biosimilar HS016 was developed first and then qualified for process comparability by a MVDA method combined with an equivalence test method for key process attributes (KPAs) and CQAs comparison across scales. The qualified scale-down model was then used for process characterization studies using a definitive screening design. The design space was finally developed using a probability-based Monte Carlo simulation method.
MATERIALS AND METHODS
Cell Line and Media
A recombinant CHO-S cell line expressing adalimumab biosimilar HS016 supplied by Zhejiang Hisun Pharmaceutical Co. Ltd. (Taizhou, China) was used in this study. A proprietary basal serum-free media and feed solutions were applied for seed expansion and cell culture process.
Cell Culture
A 750-L stainless-steel bioreactor (Sartorius Stedim AG, Goettingen, Germany) with a working volume of 500-L was operated for production-scale manufacturing. Nine batches with a temperature-shift fed-batch process for performance qualification and clinical studies were produced. In-process data and process and product quality attributes were collected for scale-down model qualification. The bench-scale model developing and design space exploration studies were carried out in 3-L glass Applikon bioreactors (Applikon, Schiedam, Netherlands) with a 2-L working volume. The seed was thawed and expanded in consecutive disposal spinner flasks or shake flasks similar to full-scale manufacturing. The culture duration was 11 to 12 days with a bolus feeding at pre-determined viable cell density (VCD). The temperature, dissolved oxygen (DO), and pH, recognized as scale-independent variables, were kept consistent across scales. Constant power per volume (P/V) was selected as scaling down criterion.
Analytical Methods
Daily samples were collected aseptically for viability, VCD, nutrients and metabolites, and product titer as described below. The viability and VCD were measured by Countstar (ALIT Life Science, Shanghai, China), and integral of viable cell density (iVCD) was calculated based on VCD (19). Nutrients and metabolites including glucose, lactate, ammonia, sodium, and potassium were tested using NOVA BioProfile 400 (NOVA Biomedical, Waltham, USA). The HS016 product titer was determined by a HPLC method with Protein A column after clarification. The recombinant antibody in harvested cell culture fluid was captured by a qualified scale-down Protein A chromatography and used for CQAs analysis. A cation exchange (CEX) HPLC method was used for charge variant quantification following carboxypeptidase B (YaxinBio, Shanghai, China) digestion. A size exclusion chromatography (SEC) HPLC method was applied for aggregates analysis. The N-glycosylation profile was characterized by a HPLC method after enzymatic cleavage of glycans by N-glycanase (Prozyme, Hayward, USA) and labeling by InstantPC dye (Prozyme, Hayward, USA). High mannose content including Man5, Man6, Man7, and Man8 was calculated by HPLC method. Total afucosylation level was then obtained by the sum of afucosylation and high mannose content. Agilent 1260 infinity series HPLC system (Agilent Technologies, Inc., Santa Clara, USA) was used for HPLC analysis.
Design of Experiments
The DSD was applied to investigate five independent variables including pH, shifted temperature, inoculation seeding density, VCD at first feeding, and VCD at temperature shift. The independent variables and abbreviations are listed in Table I. The final antibody titer and CQAs including aggregates, acid peak, total afucosylation level, and high mannose content were evaluated as dependent variables. The experimental conditions are listed in Table II.
Statistical Analysis
MVDA combined with the equivalence test was used for scale-down model qualification. SIMCA software (v14.1, Umetrics AB, Sweden) was applied to generate a batch level partial least square (PLS) model based on nine batches of cell culture process datasets at 500-L full-scale. Eight variables were included in the process comparability analysis: product titer, iVCD, viability, glucose, lactate, ammonia, sodium, and sodium/potassium. The six bench-scale datasets were then imported for process comparability assessment. Equivalence test was employed for comparability study using Minitab (v18.1, Minitab Inc., State College, USA). The KPAs or CQAs were considered comparable between scales if the (1-2α)100% two-sided confidence interval (CI) of the mean difference is within an equivalence margin (− δ, δ). The α and δ were set at 0.05 and 3 standard deviations of full-scale data, respectively.
The DSD model fitting was finished by JMP Pro software (v13.0.0, SAS Institute Inc., Cary, USA), and the design space was explored by MODDE Pro (v11.0.1.1878, Umetrics AB, Sweden) using a probability-based Monte Carlo simulation method. The limit of failure probability was set at 1% with 50,000 times iterations. The model error was included, and the simulation resolution was set at 16. The regression equations are shown below:
where Yi is the dependent variable, Xi and Xj represent independent variables, β0 is constant, and βi, βij, and βii are the regression coefficients for linear, interaction, and quadratic terms, respectively.
Design Space Verification
The operation setpoint and worst-case condition in design space were identified and chosen for design space verification as listed in Table III. The studies at each condition were repeated three times. Average relative deviation (ARD) values were calculated as below for verification:
where EV and PV are experimental and predicted values, respectively.
RESULTS AND DISCUSSION
Scale-down Model Development and Qualification
The scaling-up principles and approaches are instructive for scaling down a cell culture process (7). Scale-dependent and scale-independent variables were identified first. Seed density, pH, culture temperature, DO, and feeding VCD recognized as scale-independent variables were kept consistent across scales, whereas scale-dependent variables such as agitation were adjusted for flow field and mass transfer similarity. For a stainless-steel bioreactor process, several strategies based on agitation and aeration criteria have been developed for scale-dependent variable transformation, in which constant power per unit volume (P/V) and volumetric mass transfer coefficient (kLa) are most commonly used (20,21). In our study, constant P/V was selected for agitating determination to mimic the large-scale mass transfer and heat transfer. Datasets from nine cell culture batches of HS016 were collected and taken as the baseline for scale-down model development. Six repeated bench-scale batches were performed, and the datasets were used for comparison. To demonstrate the applicability of the scale-down model, a combination of MVDA and equivalence test method was used for model performance qualification.
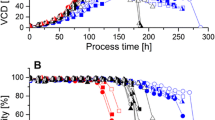
For cell culture process comparison, a PLS batch evolution model, one of the most popular MVDA techniques (8), was created first based on process datasets of nine full-scale HS016 cell culture batches. The model R2 and Q2 were 97.2% and 95.6% showing a good fit and predictability, respectively. The independent variables were projected into four principal components, which are linear combinations of the original variables. The first principal component, representing the direction of greatest variability in the data, explained as most as 92.8% of the total variance. The scores of the first principal component are the weighted combinations of those variables shown in Fig. 1. The batch statistical process control chart displaying the score value of the first principal component over culture day for HS016 full-scale batches is plotted in Fig. 2a. The control range with 3 standard deviations from full-scale means is plotted as red dotted lines. The cell culture process at 500-L scale seems to be well-controlled. The model was then applied to predict performance at 3-L scale imported with the same process variables. Figure 2b shows the predicted score of the first principal component of bench-scale batches process over culture day. The trajectories of the bench-scale process are all in the middle of the control range of large-scale process indicating that the small-scale process performance is comparable with that of the full-scale process.
a Score plot of the first principal component over culture day in the full-scale multivariate model (n = 9). b Predicted score plot of bench-scale data generated from 2-L bioreactors (n = 6). Green dashed lines represent the mean of the first principal component. Red dashed lines represent control range with 3 standard deviations from the means. CS and BS are full-scale and small-scale batches, respectively
Equivalence test was used to compare KPAs at multiple levels including final product titer, iVCD, and viability. The CQAs included aggregates, acid peak, total afucosylation level, and high mannose content from different scales. The most crucial and challenging aspect of equivalence test is establishing a reasonable equivalence margin, representing the maximum difference in means having no practical scientific impact. A subject matter expert definition, specifications, or other decision-making limits can be taken as equivalence margin (22). Recently, the X fold function of the variability of the reference product has been considered as the equivalence margin for its applicability with a limited sample size (11). In our approach, the equivalence margin was set at 3 standard deviation of HS016 full-scale data. The equivalence tests results are shown as Fig. 3. The 90% CI of mean differences of all KPAs and CQAs between two scales divided by equivalence margin fully falls within the normalized margin. Therefore, equivalences are concluded for all the attributes between two scales.
Equivalence test results of final product titer, iVCD, viability, aggregates, acid peak content, total afucosylation level, and high mannose content from different scales. The equivalence margin was set as 3 standard deviations of full-scale data. The black vertical lines marked as − 1 and 1 represent normalized equivalence margin. The black bars represent the 90% CI of the mean difference between scales divided by equivalence margin
The combined result of the MVDA and equivalence test demonstrates that the scale-down cell culture model accurately mimics the full-scale system and is suitable for use in future process characterization studies.
DSD Model
The DSD method was used for experiments design and model development. Five independent variables including pH, shifted temperature, inoculation seeding density, VCD at first feeding, and VCD at temperature shift, recognized as CPPs with a potential high risk or not well-controlled during HS016 cell culture process, were evaluated. The final product titer and four CQAs including aggregates, acid peak, total afucosylation level, and high mannose content were taken as dependent variables. The DSD design and experimental results are listed in Table II.
To identify active main effects and second-order effects, a two-stage modeling method was applied for data analysis with a forward stepwise regression. This was followed by all-subsets regression with heredity restriction. The values of corrected Akaike information criterion (AICc) and Bayesian information criterion (BIC) were calculated and applied for model determination with best prediction capability (23). No acceptable model was found for acid peak indicating that there was no significant relationship between independent variables and acid peak content in the study range. Although a regression model was found for total afucosylation, the normalized value changed from 0.90 to 1.00 (Table III) showing that the independent variables in the study range have little effect on total afucosylation. Therefore, the model for total afucosylation was excluded from following design space exploration.
Three quadratic polynomial models for final product titer, aggregates, and high mannose content were then established, respectively. The regression coefficients and P values are listed in Table IV. The model equations are shown below:
Results showed that the final product titer was between 0.43 and 1.0, and the aggregation and high mannose contents ranged from 0.56 to 1.0 and 0.47 to 1.0 as shown in Table III, respectively. The R2 and adjusted R2 were higher than 96% and 90% for all three models respectively, meaning good fit of the data. The contour plots of models are shown in Figs. 4, 5, and 6. As shown in Fig. 4, the cell culture pH and shifted temperature showed a negative and positive effect on final product titer, respectively. A significant interaction between culture pH and shifted temperature is also found to positively influence the final product titer. The pH and temperature have been reported to impact glucose consumption, base consumption, and lactose production, and consequently change the antibody expression (24). The unwanted impurity aggregates that can cause enhanced immunogenicity or reduced efficacy with a high risk should be controlled during HS016 cell culture (25). The linear and quadratic terms of VCD at first feeding significantly affect the product titer and aggregation as seen in Fig. 4 and Fig. 5. As the VCD at first feeding increased, both product titer and aggregates initially increased but then decreased. Temperature was inversely related to aggregates where higher temperature helps to reduce the aggregation. Glycans known to affect the clinical performance were taken as high-risk CQAs. The afucosylated forms affect the ADCC activity and high mannose forms affect PK profile and ADCC activity (26,27), which are plausible mechanisms of action of adalimumab for Crohn’s disease and ulcerative colitis indication (28). The pH, VCD at first feeding, and VCD at temperature shift were found remarkably effecting on high mannose content, which are all positive effects. Although the shifted temperature had little effect on high mannose content, the quadratic term of the independent variable positively influenced the response, while the interaction between pH and temperature showed a negative effect.
Design Space Development
Design space is defined as a multivariate combination of independent variables that deliver a robust quality product within specification (1). Design space is a key concept in process characterization and quality by design (QbD), and movement within it is not considered as a change and allows for regulatory flexibility. In this study, the final product titer was limited to no less than 0.4 for example, while the contents of aggregates and high mannose were set at not more than 1.0. A Monte Carlo simulation method was then applied for design space exploration. The design space with a 1% probability to fall outside the specification limits is denoted by green color as shown in Fig. 7. The irregular design space is factor-interdependent and hard to describe for industrial manufacturing. Therefore, the largest hypercube design space for operational simplicity was calculated and is plotted as a gray dashed frame in Fig. 7. An operation setpoint, which is a robust or best compromise condition, was also computed and displayed as arrow crossing. The hypercube design space and setpoint values are listed in Table V.
Design space plot with a robust setpoint. The final product titer was limited to not less than 0.4, while the content of aggregates and high mannose content were set at not more than 1.0. The green areas are design space calculated by Monte Carlo simulation with a probability of failure of less than 1%. The largest hypercube design space is plotted as a gray dashed frame. The optimal robust setpoint is displayed as arrow crossing
Design Space Verification
The worst-case condition in hypercube design space, which generates the worst performance, was identified with the lowest final product titer and the highest content of aggregates and high mannose. The verification experiments were carried out with the operation setpoint and worst-case condition. The conditions, prediction results, and experimental results are listed in Table III. The calculated ARD values for responses are lower than 5%, indicating that the target specification can be achieved by conditions within the design space even at the worst-case scenario.
CONCLUSION
A qualified scale-down model is essential and valuable in conducting late-stage process characterization and enhancing process understanding. This study demonstrates the successful establishment of a scale-down mammalian cell culture model for HS016 and its qualification by MVDA and equivalence test methods. MVDA approach provides an effective tool for global process comparability assessment with multidimensional culture process datasets, whereas the equivalence test focuses more on high-risk CQAs comparison between scales. The combined method proposed in this study is effective for scale-down model comparison and can be applied for other complex dataset comparison like process change, technology transfer, and process scale-up.
Based on the qualified scale-down model, five independent variables were selected and investigated using a DSD approach. The active main effects, quadratic effects, and two-factor interaction were identified effectively using only 13 runs. The design space was calculated using a Monte Carlo simulation method and was verified with the operation setpoint and worst-case condition. The verified design space can be extended to full-scale process control and will be further validated for model suitability at full-scale operation setpoint. The results of this work indicate that the DSD approach is powerful for process characterization and early-stage process development.
The case study presented in this report shows a feasible roadmap to characterize HS016 cell culture process based QbD principle and is instructional for other antibody product study.
References
ICH Q8 (R2). Pharmaceutical development. 2009. http://www.ich.org/products/guidelines/quality /article/quality-guidelines.html. Accessed 23 Feb 2019.
Hakemeyer C, McKnight N, St John R, Meier S, Trexler-Schmidt M, Kelley B, et al. Process characterization and design space definition. Biologicals. 2016;44(5):306–18. https://doi.org/10.1016/j.biologicals.2016.06.004.
ICH Q11. Pharmaceutical quality system. 2012. http://www.ich.org/products/guidelines/ quality/article/quality-guidelines.html. Accessed 23 Feb 2019.
Amadeo I, Mauro L, Orti E, Forno G. Establishment of a design space for biopharmaceutical purification processes using DoE. Methods Mol Biol. 2014;1129:11–27. https://doi.org/10.1007/978-1-62703-977-2_2.
Li F, Hashimura Y, Pendleton R, Harms J, Collins E, Lee B. A systematic approach for scale-down model development and characterization of commercial cell culture processes. Biotechnol Prog. 2006;22(3):696–703. https://doi.org/10.1021/bp0504041.
DiCesare C, Yu M, Yin J, Zhou W, Hwang C, Tengtrakool J, et al. Development, qualification, and application of a bioreactor scale-down process: modeling large-scale microcarrier perfusion cell culture. BioProcess Int. 2016;14(1):18–29.
Tescione L, Lambropoulos J, Paranandi MR, Makagiansar H, Ryll T. Application of bioreactor design principles and multivariate analysis for development of cell culture scale down models. Biotechnol Bioeng. 2015;112(1):84–97. https://doi.org/10.1002/bit.25330.
Tsang VL, Wang AX, Yusuf-Makagiansar H, Ryll T. Development of a scale down cell culture model using multivariate analysis as a qualification tool. Biotechnol Prog. 2014;30(1):152–60. https://doi.org/10.1002/btpr.1819.
Schuirmann DJ. A comparison of the two one-sided tests procedure and the power approach for assessing the equivalence of average bioavailability. J Pharmacokinet Biopharm. 1987;15(6):657–80. https://doi.org/10.1007/BF01068419.
US FDA. Statistical approaches to establishing bioequivalence. 2001. https://www.fda.gov/ downloads/drugs/guidances/ucm070244.pdf. Accessed 23 Feb 2019.
Tsong Y, Dong X, Shen M. Development of statistical methods for analytical similarity assessment. J Biopharm Stat. 2017;27(2):197–205. https://doi.org/10.1080/10543406.2016.1272606.
Rathore AS, Pathak M, Jain R, Jadaun GPS. Monitoring quality of biotherapeutic products using multivariate data analysis. AAPS J. 2016;18(4):793–800. https://doi.org/10.1208/s12248-016-9908-z.
Horvath B, Mun M, Laird MW. Characterization of a monoclonal antibody cell culture production process using a quality by design approach. Mol Biotechnol. 2010;45(3):203–6. https://doi.org/10.1007/s12033-010-9267-4.
Jones B, Nachtsheim CJ. A class of three-level designs for definitive screening in the presence of second-order effects. J Qual Technol. 2011;43(1):1–15. https://doi.org/10.1080/00224065.2011.11917841.
Jones B, Nachtsheim CJ. Definitive screening designs with added two-level categorical factors. J Qual Technol. 2013;45(2):121–9. https://doi.org/10.1080/00224065.2013.11917921.
Tai M, Ly A, Leung I, Nayar G. Efficient high-throughput biological process characterization: definitive screening design with the ambr250 bioreactor system. Biotechnol Prog. 2015;31(5):1388–95. https://doi.org/10.1002/btpr.2142.
Erler A, de Mas N, Ramsey P, Henderson G. Efficient biological process characterization by definitive-screening designs: the formaldehyde treatment of a therapeutic protein as a case study. Biotechnol Lett. 2013;35(3):323–9. https://doi.org/10.1007/s10529-012-1089-y.
Herzer S, Bhangale A, Barker G, Chowdhary I, Conover M, O’Mara BW, et al. Development and scale-up of the recovery and purification of a domain antibody Fc fusion protein-comparison of a two and three-step approach. Biotechnol Bioeng. 2015;112(7):1417–28. https://doi.org/10.1002/bit.25561.
Pan X, Streefland M, Dalm C, Wijffels RH, Martens DE. Selection of chemically defined media for CHO cell fed-batch culture processes. Cytotechnology. 2017;69(1):39–56. https://doi.org/10.1007/s10616-016-0036-5.
Bailey JE, Ollis DF. Biochemical engineering fundamentals. 2nd ed. New York: McGraw-Hill College; 1986.
Ju LK, Chase GG. Improved scale-up strategies of bioreactors. Bioprocess Eng. 1992;8(1):49–53. https://doi.org/10.1007/bf00369263.
Burdick RK, LeBlond DJ, Pfahler LB, Quiroz J, Sidor L, Vukovinsky K, et al. Statistical applications for chemistry, manufacturing and controls (CMC) in the pharmaceutical industry. New York: Springer international publishing; 2017.
Fidaleo M, Lavecchia R, Petrucci E, Zuorro A. Application of a novel definitive screening design to decolorization of an azo dye on boron-doped diamond electrodes. Int J Environ Sci Technol (Tehran). 2016;13(3):1735–472. https://doi.org/10.1007/s13762-016-0933-3.
CMC BiotechWorking Group. A-Mab Case Study. 2009. http://www.casss.org/associations/9165/ files/A-Mab Case Study Version 2-1.pdf. Accessed 23 Feb 2019.
Ratanji KD, Derrick JP, Dearman RJ, Kimber I. Immunogenicity of therapeutic proteins: influence of aggregation. J Immunotoxicol. 2014;11(2):99–109. https://doi.org/10.3109/1547691X.2013.821564.
Shields RL, Lai J, Keck R, O’Connell LY, Hong K, Meng YG, et al. Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human Fcgamma RIII and antibody-dependent cellular toxicity. J Biol Chem. 2002;277(30):26733–40. https://doi.org/10.1074/jbc.M202069200.
Yu M, Brown D, Reed C, Chung S, Lutman J, Stefanich E, et al. Production, characterization, and pharmacokinetic properties of antibodies with N-linked mannose-5 glycans. mAbs. 2012;4(4):475–87. https://doi.org/10.4161/mabs.20737.
Levin AD, Wildenberg ME, van den Brink GR. Mechanism of action of anti-TNF therapy in inflammatory bowel disease. J Crohns Colitis. 2016;10(8):989–97. https://doi.org/10.1093/ecco-jcc/jjw053.
Acknowledgments
The authors would like to thank Dr. Khursheed Anwer from Celsion Corporation for manuscript review.
Funding
This study was supported by National Science and Technology Major Projects for “Major New Drugs Innovation and Development” (Grant No. 2018ZX09736008) and Science Program of Zhejiang province, China (Grant No. 2017C03003).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Nie, L., Gao, D., Jiang, H. et al. Development and Qualification of a Scale-Down Mammalian Cell Culture Model and Application in Design Space Development by Definitive Screening Design. AAPS PharmSciTech 20, 246 (2019). https://doi.org/10.1208/s12249-019-1451-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1208/s12249-019-1451-7