Abstract
Background
Studies have shown that m6A modification is related to the occurrence and development of papillary thyroid carcinoma (PTC). The disorder of succinic acid metabolism is associated with the occurrence and development of various tumors. However, there are few studies based on m6A and succinate metabolism-related genes (SMRGs) in PTC.
Methods
The TCGA-Thyroid carcinoma (THCA), GSE33630, 1159 SMRGs, and 23 m6A regulatory factors were collected from the online databases. Subsequently, the differentially expressed genes (DEGs) were selected between PTC (Tumor) and Normal samples. The overlapping genes among the DEGs, m6A, and SMRGs were applied to screen the biomarkers. Using the 3 machine-learning algorithms, the biomarkers were determined based on the overlapping genes. Next, the biomarkers were evaluated by the ROC curve and expression analysis in TCGA-THCA and GSE33630. Then, the overall survival (OS) differences were compared between the high-and low-expression biomarkers. Finally, immune infiltration analysis, molecular regulatory network, and drug prediction were performed based on the biomarkers.
Results
In TCGA-THCA, there were 2800 DEGs between and Normal samples, and then 7 overlapping genes were obtained. Importantly, ADK, TNFRSF10B, CYP7B1, FGFR2, and CPQ were determined as biomarkers with excellent diagnostic efficiency (AUC > 0.7). In PTC samples, ADK and TNFRSF10B were high-expressed while CYP7B1, FGFR2, and CPQ were low-expressed. Especially, the high-expression groups of ADK had a better prognosis, while the high-expression groups of CYP7B1, FGFR2, and CPQ had a worse prognosis. Afterward, immune infiltration analysis found that 16 immune cells had infiltration differences between the Tumor and Normal samples. Finally, transcription factor SP1 could regulate CYP7B1 and TNFRSF10B. Moreover, Navitoclax was a potential drug for PTC patients.
Conclusion
Overall, we described 5 biomarkers associated with adverse prognosis of PTC, including ADK, TNFRSF10B, CYP7B1, FGFR2, and CPQ. All these biomarkers were involved in succinate metabolism and m6A modification of RNA. This set of biomarkers should be explored further for their diagnostic value in PTC. Investigations into the mechanistic role of alteration of succinate metabolism and m6A modification of RNA pathways in the pathophysiology of PTC are warranted.
Similar content being viewed by others
Background
During the past decade, thyroid cancer incidence has increased, making it one of the most common malignant tumors in the endocrine system [1]. Papillary thyroid carcinoma (PTC) is the most common subtype of all thyroid cancers. Generally, it is characterized by a high degree of differentiation, slow clinical course, and low specific mortality. However, a considerable number of patients still experience persistent or recurrent diseases, with an incidence rate of 14–30% [2,3,4]. Most patients with thyroid cancer have a good prognosis, but some thyroid cancers are aggressive and prone to recurrence and metastasis. In addition, the accompanying over-diagnosis and over-treatment are important problems [5]. At present, the most reliable method to evaluate PTC is fine needle aspiration cytology (FNA), but FNA still has its technical limitations. Less than 20% of surgically removed nodules are malignant. Therefore, in order to improve the diagnostic accuracy of PTC and avoid unnecessary surgery, more and more researchers focus on the exploration of molecular markers of PTC [6, 7].
In eukaryotes, m6A is the most common form of mRNA modification. Studies have shown that m6A exists widely in transcriptome, and has modified more than 7600 genes and 300 non-coding RNAs [8]. Studies have shown that the changes and defects of the m6A regulatory factor gene are closely related to the malignant progress of various cancers [9, 10]. However, although m6A modification is involved in tumorigenesis [11], proliferation [12], differentiation, invasion, and metastasis of different types of tumors, the role of m6A modification and the potential regulatory mechanism in PTC have not been fully explored.
Succinic acid is produced by succinyl coenzyme A through the TCA cycle enzyme succinyl coenzyme A synthetase, which plays an important role in mitochondrial energy metabolism [13]. Abnormal mitochondrial metabolism may lead to cytoplasmic and/or extracellular succinic acid accumulation. It is reported that LPS-induced macrophage activation will trigger the release of succinic acid, thus increasing the production of IL-1b and promoting inflammation [14]. It is reported that the accumulation of cytoplasmic succinic acid in cancer cells caused by TCA circulatory dysfunction promotes tumorigenesis [15]. Another study shows that comparative metabonomics determines that succinic acid is a new factor secreted by cancer cells, which triggers TAM polarization and promotes cancer metastasis in vitro and in vivo [16]. Succinic acid enhances the migration of macrophages and the migration and invasion of cancer cells, which is helpful for cancer metastasis. These new findings indicate that succinic acid is a carcinogenic metabolite and may be a valuable target for cancer chemoprevention and treatment.
In this study, bioinformatics technology was used to explore the biological function and potential mechanism of m6A and succinate metabolism-related biomarkers in the occurrence and development of PTC, so as to provide some reference for clinical diagnosis and treatment of PTC.
Materials and methods
Data sources
In this study, PTC patients of dataset were achieved by the TCGA online tool, namely TCGA-Thyroid carcinoma (THCA) dataset, containing 510 PTC (Tumor) and 58 Normal samples, and it was utilized as the training set. The external validation set GSE33630 had 60 Tumor samples and 45 Normal samples. Moreover, a total of 1159 succinate metabolism-related genes (SMRGs) were acquired (Relevance score > 7) by the GeneCards online tool (https://pathcards.genecards.org/). There were 23 m6A regulatory factors to the related reference article [17].
Differential expression analysis and the WGCNA
In our study, DEGs between PTC (Tumor) and Normal samples were acquired by the DESeq2 (v 1.36.0) [18] package (|Log2FC| > 1 and p.adj < 0.05). Moreover, a heat map and a volcano map of DEGs were plotted by pheatmap (v 0.7.7) and ggplot2 (v 3.3.0) [19] packages, respectively. The expression levels of 23 m6A regulatory factors were compared between the tumor and normal controls in TCGA-THCA (Wilcoxon test, p < 0.05). Meanwhile, the m6A regulatory factors were used as the gene set, and all samples were scored by GSVA, and the difference in the GSVA score between Tumor and Normal groups was analyzed. Furthermore, the WGCNA was performed on the samples to screen out the critical module. Firstly, outlier samples were eliminated to secure the precision of the analysis by sample clustering. An appropriate soft threshold (β) was selected to make the engagement among genes conformed to the scale-free distribution to the maximum extent. Then, these genes were classified into several modules using dynamic tree cutting. The GSVA score of the m6A regulatory factor was used as a trait, and the relationships between the modules and trait were computed. Next, the module that most relevant to the trait was defined as the critical module.
Screening of overlapping genes
The SMRGs, DEGs, and the genes in the critical module were crossed to achieve the overlapping genes. In addition, for further studying the related biological functions and signaling pathways of the overlapping genes, the Gene ontology (GO) enrichment analysis (p.adjust < 0.05) was conducted by the clusterProfiler (v 3.8.1) package [20].
Identification and verification of biomarkers
The Least absolute shrinkage and selection operator (LASSO), Random Forest (RF), and Extreme Gradient Boosting (XGBoost) algorithms were implemented on the overlapping genes to acquire the feature genes, respectively. Furthermore, the feature genes identified in those three algorithms were crossed to acquire the biomarkers. To analyze the diagnostic performance of each biomarker, the ROC curves for biomarkers were drawn in the TCGA-THCA dataset and validation set GSE33630, respectively. The expression levels of biomarkers were compared between the tumors and normal samples in both TCGA-THCA and GSE33630 (Wilcoxon test, p < 0.05). Furthermore, according to the above biomarkers, a nomogram for forecasting disease probability rates of PTC patients was created. Moreover, the calibration curve was drawn to evaluate the precision of the prediction model in the TCGA-THCA dataset and validation set GSE33630. Subsequently, the decision curve analysis (DCA) and ROC curve of the nomogram were performed in the training set. The chromosomal localization of biomarkers was performed by biomaRt (v 2.52.0) [21].
The gene-gene interaction (GGI) network
Based on the 23 m6A regulatory factors and biomarkers, the top 20 genes that interacted with these genes were predicted from the GeneMANIA website (http://genemania.org). Next, a GGI network was constructed based on the above genes using the Cytoscape (v 3.10.0) software.
The survival analysis
Firstly, the expression levels of each biomarker were calculated in 510 PTC samples from the TCGA-THCA dataset. The optimal thresholds of the expression were calculated by the ‘surv_cutpoint()’ function of the survminer (v. 0.4.9) package. Based on the optimal threshold of the biomarkers, the 510 PTC samples were divided into high- and low-expression groups. Then, the overall survival (OS) differences were compared between the high- and low-expression groups by the K-M curves (Wilcoxon test, p < 0.05).
Enrichment analysis of biomarkers
In order to find the regulatory pathways and related molecular functions enriched by various biomarkers, the GSEA was conducted using the clusterProfiler (v 3.8.1) package [20] (p.adjust < 0.05).
Immune infiltration analysis
In order to further explore the immune infiltration condition of PTC, the CIBERSORT algorithm was implemented on the samples in GSE32918 dataset to estimate the abundance of 22 different cell types of the immune cells between the Tumor and Normal groups. In addition, the differential immune cells between the Tumor and Normal groups were computed using the Wilcoxon test method. The Pearson correlation method was performed to analyze the association between immune cells. Moreover, the relationships between biomarkers and differential immune cells were computed by the Pearson method.
The construction of a regulatory network and the sensitivity analysis of drugs
The miRNAs corresponding to the above biomarkers were forecasted using the miRTarBase (http://miRTarBase.mbc.nctu.edu.tw/) and TarBase (http://www.microrna.gr/tarbase) online databases. The miRNAs in those two databases were crossed to acquire the common miRNAs (co-miRNAs) of each biomarker. The lncRNAs corresponding to the above miRNAs were acquired based on the miRTarBase and TarBase online databases. Subsequently, the competitive endogenous RNA (ceRNA) network was created. Moreover, the transcription factors (TFs) of the biomarkers were forecasted by the TRRUST (www.grnpedia.org/trrust) online database. The TF-mRNA regulatory network was created. Besides, the drug sensitivity of biomarkers was analyzed based on the GSCALite online database (http://bioinfo.life.hust.edu.cn/web/GSCALite/).
Results
Acquisition of DEGs and critical module
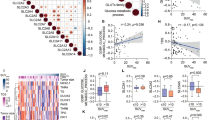
There were 2800 DEGs between PTC (Tumor) and Normal samples (Fig. 1A, Supplementary Table 1). The expression heat map of PTC-associated DEGs is shown in Fig. 1B. In TCGA-THCA, there were 20 differentially expressed m6A regulatory factors between the tumor and normal samples. Among them, the expression levels of 17 m6A regulatory factors were much lower in tumors than those of normal samples (Supplementary Fig. 1). Likewise, there was a significant difference in GSVA scores for m6A regulatory factors between the Tumor and Normal groups (Fig. 1C). In addition, the sample clustering result demonstrated that there were three outlier samples, and the remaining samples were used for subsequent analysis (Fig. 1D). When the soft threshold was 18, the genes conformed to a scale-free distribution to the greatest extent possible (Fig. 1E). A total of 13 modules were identified after merging (Fig. 1F). The MEblack module had the highest and most significant correlation with the trait, thus we identified it as the critical module (Cor = 0.67 and p.value < 0.05). There were 5254 genes in the module which were utilized for subsequent analysis (Fig. 1G).
Determination of DEGs and m6A score-related genes in PTC. (A) Volcano plot of DEGs between PTC (Tumor) and Normal samples. (B) Heat map of PTC-associated DEGs. (C) GSVA scores for m6A regulatory factors between the Tumor and Normal groups. (D) The sample clustering plot to remove outlier samples. (E) Screening of the optimal soft-threshold values. (F) The dynamic cut tree after merging modules. (G) Module-trait relationship heatmap
A total of 7 overlapping genes were identified
According to the intersection, 7 overlapping genes (FGFR2, CPQ, etc.) were identified (Fig. 2A). The enrichment analysis demonstrated that overlapping genes mainly participated in ‘prostate gland epithelium morphogenesis’, ‘death receptor activity’, etc. GO items (Fig. 2B, Supplementary Table 2).
Five biomarkers were screened out
According to the RF algorithm, 5 feature genes such as CYP7B1, FGFR2, ADK, CPQ, and TNFRSF10B were identified by the RF algorithm (Fig. 3A). There were six feature genes including FGFR2, CPQ, TNFRSF10B, GLS2, ADK, and CYP7B1 were detected by the LASSO algorithm (Fig. 3B). The top five feature genes (ADK, TNFRSF10B, CYP7B1, FGFR2, and CPQ) were acquired using the XGBoost algorithm (Fig. 3C). Furthermore, 5 biomarkers including ADK, TNFRSF10B, CYP7B1, FGFR2, and CPQ were achieved by the intersection (Fig. 3D). In the TCGA-THCA dataset, the AUC values for all biomarkers were above 0.8. Moreover, in the validation set GSE33630, the AUC values of three biomarkers (ADK, TNFRSF10B, and FGFR2) were greater than 0.9, and the AUC value of CPQ was above 0.7, demonstrating that the screened biomarkers had high diagnostic value (Fig. 3E-F). In TCGA-THCA and GSE33630, the expression levels of the 5 biomarkers had significant differences between the tumors and normal (p < 0.05). Compared with the normal samples, the expression levels of TNFRSF10B and ADK were significantly higher in tumors, and the other 3 biomarkers were the opposite (Fig. 3G-H). In addition, a nomogram for forecasting survival rates of PTC patients was created based on ADK, TNFRSF10B, CYP7B1, FGFR2, and CPQ (Fig. 4A). The chromosome mapping results showed that the positions of ADK and FGFR2 were on 10-chromosome, and TNFRSF10B, CYP7B1, and CPQ were located on 8-chromosome (Fig. 4B). The calibration curves and the DCA curve indicated that the predictive ability of the model was favorable (Fig. 4C-E). The AUC value of the nomogram was 0.9774, which demonstrated that the effectiveness of the nomogram (Fig. 4F). Moreover, a GGI network was generated based on the 23 m6A regulatory factors, 5 biomarkers, and the top 20 genes. Obviously, we found that ADK-RBKS, RBM15-DIDO1, YTHDF2-YTHDF3, etc., interacted with each other (Supplementary Fig. 2).
Determination of m6A-succinic acid-related biomarkers in PTC. (A) Importance score of variables from random forest model. (B, C) LASSO regression was performed. (D) The XGBoost model based on the SHAP algorithm. (E) Five biomarkers detected by Venn diagram. (F, G) Diagnostic value of hub genes. ROC curves of hub genes in the training dataset (F) and validation dataset (G). (H, I) The expression levels of biomarkers in the training dataset (H) and validation dataset (I)
The nomogram prediction model for PTC based on the nomogram prediction model. (A) Construction of the nomogram prediction model. (B) Chromosome mapping of five genes that encode biomarkers. (C-E) Calibration curves (C-D) and decision curve analysis (DCA) (E) of the nomogram. (F) The AUC value of the Nomogram in the ROC curve is 0.9774
The survival analysis of biomarkers
The low-expression group of CYP7B1, FGFR2, and CPQ had a higher survival probability (p < 0.05), and the low-expression group of ADK had a lower survival probability (p > 0.05) (Fig. 5A-D). There was no significant difference between the high- and low-expression groups of TNFRSF10B (Fig. 5E).
The GSEA of the biomarkers
We performed GSEA on the above biomarkers, and according to the GO enrichment analysis, we found that ADK and TNFRSF10B were mainly associated with ‘cadherin binding’, ‘macroautophagy’, and so on GO items. CYP7B1, FGFR2, and CPQ were mainly involved in ‘cilium organization’, ‘MHC protein complex’, etc. GO items (Fig. 6, Supplementary Tables 3–7). Moreover, the KEGG enrichment analysis demonstrated that ADK and TNFRSF10B were mainly enriched in ‘Bacterial invasion of epithelial cells’, and ‘Endocytosis’ etc. KEGG pathways. CYP7B1, FGFR2, and CPQ mainly participated in the ‘mRNA surveillance pathway’, ‘Lysine degradation’, and so on KEGG pathways (Fig. 7, Supplementary Tables 8–12).
Immune infiltration analysis between Tumor and normal samples
The abundance of immune cells in Tumor and Normal samples was shown by the heat map (Fig. 8A). Furthermore, 16 differential immune cells (Neutrophils, Macrophages M0, Eosinophils, etc.) between Tumor and Normal groups were acquired (Fig. 8B). There was the highest positive correlation (Cor = 0.48, p < 0.05) between Monocytes and Eosinophils, and Plasma cells had the highest negative correlation (Cor = -0.38, p < 0.05) with memory resting CD4 T cells (Fig. 8C). We found that CYP7B1 was significantly negatively correlated with regulatory T cells (Tregs) (Cor = -0.33, p < 0.05), and there was a highest positive relationship between FGFR2 and Eosinophils (Cor = 0.3, p < 0.05) (Fig. 8D).
Immune cell infiltration analysis. (A) Heatmap of immune cell abundances. (B) Boxplot of immune cell abundances between Tumor and Normal groups (* p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001). (C) Heatmap of immune cell correlation analysis. (D) Heatmap of Pearson correlation between the biomarkers
The construction of ceRNA and TF-mRNA networks and the drug sensitivity analysis
The ceRNA network was created including 5 biomarkers, 36 miRNAs, and 146 lncRNAs. We found that CYP7B1 was regulated by hsa-miR-17-5p, and hsa-miR-335-5p could be regulated by C15orf32 (Fig. 9A, Supplementary Table 13). In addition, a total of 25 TFs were forecasted based on two biomarkers (CYP7B1 and TNFRSF10B), such as TP53, STAT3, MYC, and so on (Fig. 9B). Among them, the SP1 regulated both CYP7B1 and TNFRSF10B biomarkers. Moreover, TNFRSF10B had the highest positive association with Navitoclax, and there was the highest negative correlation between ADK and AICAR (Fig. 9C).
Discussion
About 80% of thyroid cancers are PTC, a relatively inert tumor with a long-term survival rate exceeding 95%. However, there are some types of PTC that are extremely invasive, and their overall survival rate as well as disease-free survival rate are poor [22, 23].
More than 100 chemical modifications of RNA have been identified in organisms, including N1-methyladenosine (m1A), N6-methyladenosine (m6A), and 5-methylcytosine (m5C). M6A is the most abundant mRNA and internal modification of long-chain noncoding RNA (lncRNA) in most eukaryotes. In addition, m6A was significantly clusters around the stop codon and 3’ untranslated region (3’UTR), mainly on the RRACH motif. In different tumors, the effect of m6A modification is different. The change of m6A also affects the progress of tumors, including proliferation, growth, invasion, and metastasis [24,25,26]. Studies have shown that m6A can inhibit the development of PTC by modifying and regulating APOE expression [27], and METTL3 can inhibit the development of PTC through m6A-mediated neutrophil infiltration [28].
Succinic acid is a typical respiratory metabolite. As an intermediate of the TCA cycle, it is usually confined to the mitochondrial matrix. In normal cells, succinate dehydrogenase catalyzes the transformation of succinic acid into fumaric acid in the TCA cycle and electron transfer in ETC (electron transfer chain). When cells are under stress, such as hypoxia, hyperglycemia, and endotoxemia, the TCA cycle is damaged, leading to the accumulation of succinic acid in the mitochondrial matrix. Excessive succinic acid leaks into the cytoplasm and is secreted into the extracellular space cytoplasm, and the accumulation of extracellular succinic acid promotes cancer growth through different mechanisms [29]. Studies have shown that the succinate pathway can be used as a diagnostic marker in head and neck squamous cell carcinoma [30]. Metastatic cancer nodules in the lungs of mice receiving succinate are higher than those of mice receiving normal saline [31]. In addition, the imbalance of succinate metabolism is related to the occurrence and development of gastrointestinal stromal tumors, gastric cancer, ganglioma, hepatocellular carcinoma, ovarian cancer, and prostate cancer [32,33,34,35,36,37], but there are few reports exploring the potential mechanism of succinate metabolism-related genes in PTC based on bioinformatics technology.
Based on the transcriptome information and clinical information of thyroid papillary carcinoma data set in TCGA database and GEO database, this study conducted bioinformatics analysis, and obtained the following results: First, the differential expression analysis of TCGA-THCA data set was carried out, and 2,800 DEGs were screened out, including 1,730 up-regulated DEGs and 1,070 down-regulated DEGs. Through WGCNA analysis, the module (MEblack) with the highest correlation with the GSVA score of the m6A regulatory factor was found, which contained 5254 module genes. Then, after crossing the DEGs with the module gene (m6a correlation) and succinic acid-related genes, seven intersection genes were obtained, the enrichment analysis of the intersection genes was carried out, and the enrichment items of the intersection genes were found. Then, five biomarkers (ADK, TNFRSF10B, CYP7B1, FGFR2, and CPQ) were further screened out by three machine learning models, and the ROC curve of the biomarkers was verified (diagnostic value verification). The results showed that the biomarkers could effectively predict papillary thyroid cancer. The nomogram model was constructed by using biomarkers, and the calibration curve of the nomogram model was constructed. The results showed that the prediction effect of nomogram model was decent.
Adenosine is a key regulator of metabolism and immune checkpoint, which is related to the escape of tumors from host immune system. The receptor-independent epigenetic effect of adenosine has been confirmed, and adenosine has additional adenosine receptor-independent activities, including the special emerging role of adenosine kinase (ADK), which exists in the cytoplasm (ADK-S) and nucleus (ADK-L) isoforms. Cytoplasmic ADK-S provides AMP with the main metabolic pathway of adenosine clearance through adenosine phosphorylation under physiological conditions [38]. Nuclear ADK-L is biochemically directly linked to S-adenosylmethionine (SAM)-dependent methylation pathway, which drives DNA and histone methylation [39].
Tumor necrosis factor (TNF) is a proinflammatory cytokine involved in cancer progression and development. The TNF family inhibits tumor formation through apoptosis, but TNF imbalance promotes metastasis, migration and invasion of tumor cells [40]. TNFRSF10B, a tumor suppressor gene located on chromosome 8. Mutations in candidate genes lead to the deletion of chromosome P-arm, which is very common in head and neck tumors. TNFRSF10B inhibits tumor formation through apoptosis, but deregulates and promotes metastasis, migration, and invasion of tumor cells [38]. The decreased expression of CYP7B1 triggers the accumulation of 27-hydroxycholesterol (27-HC), and CYP7B1 is down-regulated in breast cancer compared with normal breast tissue [41, 42]. According to these findings, compared with benign tumors, CYP7B1 is strongly down-regulated in invasive tumor tissues (PTC high risk and PDTC/ATC). These data indicate that the CYP7B1 may promote the development and progress of thyroid cancer [43]. Moreover, a common TF between the TNFRSF10B and CYP7B1, specificity protein 1 (SP1), is ubiquitously expressed in mammalian cells [44]. Meng et al. found that the binding of the SP1 to the TNFRSF10B promoter was enhanced in the presence of poly (ADP-ribose) polymerase (PARP) inhibitors [45]. And the regulation of CYP7B1 transcription by SP1 plays an important role in regulating the level of oxysterol in cholesterol metabolism [46].
FGFR2 (fibroblast growth factor receptor 2), also known as CD332, is located on chromosome 10. FGFR2 plays an important role in embryonic development, tissue repair, and angiogenesis. Like other members of the fibroblast growth factor receptor family, these receptors activate tyrosine kinase activity by binding to their ligands. These signal molecules regulate cell division, growth, and differentiation. FGFR2 is highly expressed in various human malignant tumors, such as gastric cancer, lung cancer, breast cancer, ovarian cancer, and endometrial cancer. Previous studies have shown that the up-regulation of FGFR2 partially reversed the inhibitory effect of miR-1266 overexpression on cell growth and progress, and miR-1266 can inhibit cell proliferation and the progress of thyroid papillary carcinoma by targeting FGFR2 [47].
CPQ is a regulatory factor related to phagocytosis, which is positively correlated with macrophages in TC tissue and is considered as a potential prognostic indicator [48].
These 5 biomarkers including ADK, TNFRSF10B, CYP7B1, FGFR2, and CPQ were identified by K-M analysis, and we found the low-expression group of CYP7B1, FGFR2, and CPQ had a higher survival probability, and the low-expression group of ADK had a lower survival probability. Interestingly, lower expression of CYP7B1, FGFR2, and CPQ was observed in thyroid cancers, while a low-expression group of CYP7B1, FGFR2, and CPQ had a high overall survival. We infer that CYP7B1, FGFR2, and CPQ have inhibitory roles in the transition of normal cells to early thyroid cancer in thyroid cells. In thyroid cancer, CYP7B1, FGFR2, and CPQ may promote or inhibit certain pathways leading to tumor progression. Also, higher expression of ADK was observed in thyroid cancers, while a high-expression group of ADK had a high overall survival. This is the opposite of the previous article, the ADK gene plays a catalytic role in the occurrence of tumors, while the ADK gene plays an inhibitory role in the development of tumor cells. What’s more, the expression level of TNFRSF10 was higher in tumors than that of normal samples. However, differences in TNFRSF10 expression do not affect the survival of thyroid cancer. It is speculated that the TNFRSF10 gene only affects the occurrence of thyroid cancer and does not worsen cancer. The mechanism of action of these biomarkers in the progression of PTC requires further study.
We performed GSEA on the above biomarkers, and according to the GO enrichment analysis, we found that ADK and TNFRSF10B were mainly associated with ‘cadherin binding’, ‘macroautophagy’, and so on GO items. CYP7B1, FGFR2, and CPQ were mainly involved in ‘cilium organization’, ‘MHC protein complex’, etc. GO items. Moreover, the KEGG enrichment analysis demonstrated that ADK and TNFRSF10B were mainly enriched in ‘Bacterial invasion of epithelial cells’, and ‘Endocytosis’ etc. KEGG pathways. CYP7B1, FGFR2, and CPQ mainly participated in ‘mRNA surveillance pathway’, ‘Lysine degradation’, and so on KEGG pathways.
CD4 + T and B cells are reported to be positively correlated with reduced tumor sizes in PTC [49]. Increased tissue infiltration of Treg cells was positively correlated with advanced thyroid cancer stage, whereas NK-cell infiltration was negatively correlated, indicating that NK and Treg cells might be important regulators of PTC progression [50,51,52]. High tumor-infiltrating CD8 + Tc e l l density was associated with a favorable prognosis in thyroid cancer patients [49, 53].
In our study, 16 differential immune cells (Neutrophils, Macrophages M0, Eosinophils, etc.) between Tumor and Normal groups were acquired. We found that CYP7B1 was significantly negatively correlated with regulatory T cells (Tregs) (Cor = -0.33), and there was a highest positive relationship between FGFR2 and Eosinophils (Cor = 0.3).
We found that CYP7B1 was regulated by hsa-miR-17-5p, and hsa-miR-335-5p could be regulated by C15orf32. Research has shown that hsa-miR-17-5p may be a positive regulator of NFE2L2 Nuclear Factor (Erythroid-derived 2)-Like 2, which is involved in angiogenesis and oxidative stress, can lead to thyroid cancer progression [54].
We found that TNFRSF10B had the highest positive association with Navitoclax, and there was the highest negative correlation between ADK and AICAR. Navitoclax (ABT-263) is a Small-molecule BH3 mimetics, disrupt BCL-2/BCL-XL interactions with proapoptotic proteins such as BIM, thereby inducing apoptosis [55]. Study shows that the combination of vemurafenib and Navitoclax can induce substantial cell death and exert cytotoxic effects in BRAFV600E-positive PTC. Moreover, the combination of vemurafenib and navitoclax requires lower doses to obtain a therapeutic effect compared to the doses with single drug treatments; therefore, it may be applied as a safer and more efficient therapeutic strategy in BRAFV600E-positive PTC [56].
AICAR exerts its action by activating the energy sensor (AMP-activated protein kinase) AMPK, which plays a central role in the control of cell growth, proliferation, and autophagy through the regulation of mTOR (mammalian target of rapamycin) [57, 58]. AMPK agonists, such as AICAR, are regarded as potential therapeutic molecules in cancer therapy [59, 60]. Study demonstration of the inhibition of CXCL8 secretion exerted by AICAR in TPC-1 and BCPAP indicating that the anticancer properties of AICAR are, at least in part, mediated by its ability to reduce the protumorigenic effects of CXCL8 [61].
Conclusion
In the current study, there were 5 succinate metabolism and m6A-related biomarkers including ADK, TNFRSF10B, CYP7B1, FGFR2, and CPQ associated with PTC, providing a reference for the clinical diagnosis and treatment of PTC. This study provides ideas for further study on the diagnostic value and potential mechanism of genes related to m6A and succinic acid metabolism in PTC. Further experimental studies should be carried out to elucidate the mechanisms involved in this observation.
Data availability
Publicly available datasets were used for analysis in this study. These data can be obtained from the Gene Expression Omnibus database (GEO, https://www.ncbi.nlm.nih.gov/geo/ and TCGA database (https://portal.gdc.cancer.gov).
Abbreviations
- PTC:
-
Papillary thyroid carcinoma
- DEGs:
-
Differentially expressed genes
- WGCNA:
-
Weighted gene co-expression network analysis
- CeRNA:
-
Competing endogenous RNAs
- TF:
-
Transcription factors
- FNA:
-
Fine needle aspiration cytology
- TCA:
-
Tricarboxylic acid cycle
- LPS:
-
Lipopolysaccharide
- TAM:
-
Tumor-associated macrophage
- TCGA:
-
The Cancer Genome Atlas
- GSVA:
-
Gene Set Variation Analysis
- GO:
-
Gene ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- LASSO:
-
Least absolute shrinkage and selection operator
- XGBoost:
-
Extreme Gradient Boosting
- ROC:
-
Receiver operator characteristic curve
- THCA:
-
Thyroid carcinoma
- DCA:
-
Decision curve analysis
- GSEA:
-
Gene Set Enrichment Analysis
- GSE:
-
GEO Series
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Mazzaferri EL, Jhiang SM. Long-term impact of initial surgical and medical therapy on papillary and follicular thyroid cancer.Am. J Med. 1994;97:418–28.
Grogan RH, Kaplan SP, Cao H, Weiss RE, Degroot LJ, Simon CA, Embia OM, Angelos P, Kaplan EL, Schechter. R.B.A study of recurrence and death from papillary thyroid cancer with 27 years of median follow-up. Surgery. 2013;154:1436–47.
Dong W, Horiuchi K, Tokumitsu H, Sakamoto A, Noguchi E, Ueda Y, Okamoto T. Time-varying pattern of mortality and recurrence from papillary thyroid Cancer: lessons from a long-term Follow-Up. Thyroid. 2019;29:802–8.
Lim ZR, Dalmasol, Zhang, Wei W, Vaccarellas. Mapping over-diagnosis of diseases and diseases. Lancet Diabetesendocrinol. 2021;9(6):330–2.
Liu Y, Geng H, Liu X, Cao M, Zhang X. A meta-analysis of circulating microRNAs in the diagnosis of papillary thyroid carcinoma. PLoS ONE. 2021;16(5):e0251676.
Chen Y, Dong B, Huang L, Huang H. Serum microRNAs as biomarkers for the diagnosis of papillary thyroid carcinoma: a meta-analysis. Bosn J Basic Med Sci. 2022;22(6):862–71.
Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell. 2012;149(7):1635–46.
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27(5):626–41.
Pan Y, Ma P, Liu Y, Li W, Shu Y. Multiple functions of m(6)a RNA methylation in cancer. J Hematol Oncol. 2018;11(1):48.
Gilbert WV, Bell TA, Schaening C, Messenger. RNA modifications: form, distribution, and function. Science. 2016;352(6292):1408–12.
Alarcón CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. HNRNPA2B1 is a mediator of m(6)A-Dependent Nuclear RNA Processing events. Cell. 2015;162(6):1299–308.
Tretter L, Patocs A, Chinopoulos C. Succinate, an intermediate in metabolism, signal transduction, ROS, hypoxia, and tumorigenesis. Biochim Biophys Acta. 2016;1857(8):1086–101.
Littlewood-Evans A, Sarret S, Apfel V, Loesle P, Dawson J, Zhang J, Muller A, Tigani B, Kneuer R, Patel S, et al. GPR91 senses extracellular succinate released from inflammatory macrophages and exacerbates rheumatoid arthritis. J Exp Med. 2016;213:1655–62.
King A, Selak MA, Gottlieb E. Succinate dehydrogenase and fumarate hydratase: linking mitochondrial dysfunction and cancer. Oncogene. 2006;25(34):4675–82.
Wu Jing-Yiing,Huang Tsai-Wang,Hsieh Yi-Ting et al. Cancer-Derived Succinate Promotes Macrophage Polarization Cancer Metastasis via Succinate Receptor Mol Cell, 2020, 77: 213–e2275.
Zhang XZ, Shizhen,Yan, Xinyu et al. m6A regulator-mediated RNA methylation modification patterns are involved in immune microenvironment regulation of periodontitis.[J].J Cell Mol Med, 2021, 25: 3634–3645.
Robinson Mark D,McCarthy, Davis J. Smyth Gordon K,edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. [J] Bioinf. 2010;26:139–40.
Ito K. Murphy D,application of ggplot2 to Pharmacometric Graphics.[J]. CPT Pharmacometrics Syst Pharmacol. 2013;2:e79.
Wu Tianzhi,Hu Erqiang,Xu Shuangbin et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data.[J].Innovation (Camb) et al. 2021, 2: 100141.
Kinsella Rhoda J, Syed, et al. J] Database (Oxford). 2011;2011:bar030. Ensembl BioMarts: a hub for data retrieval across taxonomic space.[.
Donaldson LB, Yan F, Morgan PF, Kaczmar JM, Fernandes JK, Nguyen SA, Jester RL, Day TA. Hobnail variant of papillary thyroid carcinoma: a systematic review and meta-analysis. Endocrine. 2021;72(1):27–39.
Yan T, Qiu W, Weng H, Fan Y, Zhou G, Yang Z. Single-Cell Transcriptomic Analysis of Ecosystems in Papillary thyroid carcinoma progression. Front Endocrinol (Lausanne). 2021;12:729565.
Liu ZX, Li LM, Sun HL, Liu SM. Link between m6A modification and cancers. Front Bioeng Biotechnol. 2018;6:89.
Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, Jaffrey SR. M(6)a RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537(7620):369–73.
Yang Y, Hsu PJ, Chen YS, Yang YG. Dynamic transcriptomic m(6)a decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018;28(6):616–24.
Huang J, Sun W, Wang Z, Lv C, Zhang T, Zhang D, Dong W, Shao L, He L, Ji X, Zhang P, Zhang H. FTO suppresses glycolysis and growth of papillary thyroid cancer via decreasing stability of APOE mRNA in an N6-methyladenosine-dependent manner. J Exp Clin Cancer Res. 2022;41(1):42.
He J, Zhou M, Yin J, Wan J, Chu J, Jia J, Sheng J, Wang C, Yin H, He F. METTL3 restrains papillary thyroid cancer progression via m6A/c-Rel/IL-8-mediated neutrophil infiltration. Mol Ther. 2021;29(5):1821–37.
Kuo CC, Wu JY, Wu KK. Cancer-derived extracellular succinate: a driver of cancer metastasis. J Biomed Sci. 2022;29(1):93.
Terra X, Ceperuelo-Mallafré V, Merma C, Benaiges E, Bosch R, Castillo P, Flores JC, León X, Valduvieco I, Basté N, Cámara M, Lejeune M, Gumà J, Vendrell J, Vilaseca I, Fernández-Veledo S, Avilés-Jurado FX. Succinate pathway in Head and Neck squamous cell carcinoma: potential as a diagnostic and prognostic marker. Cancers (Basel). 2021;13(7):1653.
Wu JY, Huang TW, Hsieh YT, Wang YF, Yen CC, Lee GL, Yeh CC, Peng YJ, Kuo YY, Wen HT, Lin HC, Hsiao CW, Wu KK, Kung HJ, Hsu YJ, Kuo CC. Cancer-Derived Succinate promotes macrophage polarization and Cancer metastasis via succinate receptor. Mol Cell. 2020;77(2):213–e2275.
Flavahan WA, Drier Y, Johnstone SE, Hemming ML, Tarjan DR, Hegazi E, Shareef SJ, Javed NM, Raut CP, Eschle BK, Gokhale PC, Hornick JL, Sicinska ET, Demetri GD, Bernstein BE. Altered chromosomal topology drives oncogenic programs in SDH-deficient GISTs. Nature. 2019;575(7781):229–33.
Mu X, Zhao T, Xu C, Shi W, Geng B, Shen J, Zhang C, Pan J, Yang J, Hu S, Lv Y, Wen H, You Q. Oncometabolite succinate promotes angiogenesis by upregulating VEGF expression through GPR91-mediated STAT3 and ERK activation. Oncotarget. 2017; 21;8(8):13174–13185.
Bardella C, Pollard PJ, Tomlinson I. SDH mutations in cancer. Biochim Biophys Acta. 2011;1807(11):1432–43.
Tseng PL, Wu WH, Hu TH, Chen CW, Cheng HC, Li CF, Tsai WH, Tsai HJ, Hsieh MC, Chuang JH, Chang WT. Decreased succinate dehydrogenase B in human hepatocellular carcinoma accelerates tumor malignancy by inducing the Warburg effect. Sci Rep. 2018;15(1):3081.
Zhang J, Zhang Q, Yang Y, Wang Q. Association between Succinate receptor SUCNR1 expression and Immune infiltrates in Ovarian Cancer. Front Mol Biosci. 2020;7:150.
Weber A, Klocker H, Oberacher H, Gnaiger E, Neuwirt H, Sampson N et al. Succinate Accumulation Is Associated with a Shift of Mitochondrial Respiratory Control and HIF-1α Upregulation in PTEN Negative Prostate Cancer Cells. Int J Mol Sci. 2018;19(7).
Boison D. Adenosine kinase: exploitation for therapeutic gain. Pharmacol Rev. 2013;65(3):906–43.
Tahir Rana Adnan,Sehgal Sheikh Arslan,Khattak Naureen Aslam. Tumor necrosis factor receptor superfamily 10B (TNFRSF10B): an insight from structure modeling to virtual screening for designing drug against head and neck cancer. Theor Biol Med Model. 2013;10:38.
Schabath MB, Giuliano AR, Thompson Z, Fenstermacher D, Jonathan K, Sellers TA, Haura E. TNFRSF10B polymorphisms and haplotypes predicts survival in non-small cell lung cancer patients. Cancer Res. 2012;72:4506–20.
Wu Q, Ishikawa T, Sirianni R, Tang H, McDonald JG, Yuhanna IS, et al. 27-Hydroxycholesterol promotes cell-autonomous, ER-positive breast cancer growth. Cell Rep. 2013;5(3):637–45.
Gibson DA, Collins F, Cousins FL, Esnal Zufiaurre A, Saunders P. The impact of 27-hydroxycholesterol on endometrial cancer proliferation. Endocr Relat Cancer. 2018;25(4):381–91.
Revilla G, Pons MP, Baila-Rueda L, García-León A, Santos D, Cenarro A, et al. Cholesterol and 27-hydroxycholesterol promote thyroid carcinoma aggressiveness. Sci Rep. 2019;9(1):10260.
Li L, He S, Sun JM, Davie JR. Gene regulation by Sp1 and Sp3. Biochem Cell Biol. 2004;82(4):460–71. https://doi.org/10.1139/o04-045.
Meng XW, Koh BD, Zhang JS, et al. Poly(ADP-ribose) polymerase inhibitors sensitize cancer cells to death receptor-mediated apoptosis by enhancing death receptor expression. J Biol Chem. 2014;289(30):20543–58. https://doi.org/10.1074/jbc.M114.549220.
Wu Z, Chiang JY. Transcriptional regulation of human oxysterol 7 alpha-hydroxylase gene (CYP7B1) by Sp1. Gene. 2001;272(1–2):191–7. https://doi.org/10.1016/s0378-1119(01)00541-8.
Fu Y-T, Zheng H-B, Zhang D-Q, et al. MicroRNA-1266 suppresses papillary thyroid carcinoma cell metastasis and growth via targeting FGFR2. Eur Rev Med Pharmacol Sci. 2018;22:3430–8.
Hou Changran,Wu Mengmeng,Zhang Haojie. The specific phagocytosis regulators could predict recurrence and therapeutic effect in thyroid cancer: a study based on bioinformatics analysis. Med (Baltim). 2023;102:e33290.
Cunha LL, Morari EC, Guihen AC, Razolli D, Gerhard R, Nonogaki S, et al. Infiltration of a mixture of immune cells may be related to good prognosis inpatients with differentiated thyroid carcinoma. Clin Endocrinol (Oxf). 2012;77(6):918–25.
Gogali F, Paterakis G, Rassidakis GZ, Liakou CI, Liapi C. CD3(-)CD16(-)CD56(bright) immunoregulatory NK cells are increased in the tumor microenvironment and inversely correlate with advanced stages in patients with papillary thyroid cancer. Thyroid. 2013;23(12):1561–8.
Gogali F, Paterakis G, Rassidakis GZ, Kaltsas G, Liakou CI, Gousis P, et al. Phenotypical analysis of lymphocytes with suppressive and regulatory properties(Tregs) and NK cells in the papillary carcinoma of thyroid. J Clin Endocrinol Metab. 2012;97(5):1474–82.
French JD, Weber ZJ, Fretwell DL, Said S, Klopper JP, Haugen BR. Tumor associated lymphocytes and increased FoxP3 + regulatory T cell frequency correlate with more aggressive papillary thyroid cancer. J Clin Endocrinol Metab. 2010;95(5):2325–33.
Yang Z, Wei X, Pan Y, Xu J, Si Y, Min Z, et al. A new risk factor indicator for papillary thyroid cancer based on immune infiltration. Cell Death Dis. 2021;12(1):51.
Stuchi LP, Castanhole-Nunes M, Maniezzo-Stuchi N, Biselli-Chicote PM, Henrique T, Padovani Neto JA et al. VEGFA and NFE2L2 Gene expression and regulation by MicroRNAs in thyroid papillary Cancer and colloid goiter. Genes (Basel). 2020;11(9).
Tse C, Shoemaker AR, Adickes J, Anderson MG, Chen J, Jin S, Johnson EF, Marsh KC, Mitten MJ, Nimmer P, Roberts L, Tahir SK, Xiao Y, Yang X, Zhang H, Fesik S, Rosenberg SH, Elmore SW. ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res. 2008;68:3421–8.
Jeong JH, Oh JM, Jeong SY, Lee SW, Lee J, Ahn BC. Combination treatment with the BRAF(V600E) inhibitor vemurafenib and the BH3 mimetic navitoclax for BRAF-Mutant thyroid carcinoma. Thyroid. 2019;29(4):540–8.
Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9:563–75.
Green AS, Chapuis N, Lacombe C, Mayeux P, Bouscary D. Tam-Burini J.LKB1/AMPK/mTOR signaling pathway in hematological malignancies: from metabolism to cancer cell biology. Cell Cycle. 2011;10:2115–20.
Guan TJ, Qin FJ, Du JH, Geng L, Zhang YY, Li M. AICAR inhibits proliferation and induced S-phase arrest, and promotes apoptosis in CaSki cells. Acta Pharmacol Sin. 2007;28:1984–90.
Rosilio C, Lounnas N, Nebout M, et al. The metabolic perturbators metformin, phenformin and AICAR interfere with the growth and survival of murine PTEN-deficient T cell lymphomas and human T-ALL/T-LL cancer cells. Cancer Lett. 2013;336:114–26.
Awwad O, Coperchini F, Pignatti P, et al. The AMPK-activator AICAR in thyroid cancer: effects on CXCL8 secretion and on CXCL8-induced neoplastic cell migration. J Endocrinol Invest. 2018;41:1275–82.
Acknowledgements
Not applicable.
Funding
This work was supported by The Construction Fund of Medical Key Disciplines of Hangzhou Project (No.20200486), Biomedical and Health Industry Development Support Science and Technology Project in Hangzhou (No.2021WJCY043, No.2022WJC008), Medical and Health Technology Project of Hangzhou (No.A20210059).
Author information
Authors and Affiliations
Contributions
Minyu Li and Xiaodan Fu performed the primary data analysis and wrote the main manuscript. Tianhan Zhou completed the summary and graphical visualization of the results. Hui Han designed the study and reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Ethics approval was not required because our study was based on a public database and did not involve animal or human samples.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, M., Fu, X., Zhou, T. et al. Biomarkers related to m6A and succinic acid metabolism in papillary thyroid carcinoma. BMC Med Genomics 17, 199 (2024). https://doi.org/10.1186/s12920-024-01975-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12920-024-01975-8