Abstract
Background
The prognosis of brain injury caused by subarachnoid hemorrhage (SAH) is poor. Previous studies showed that abnormal function of RBPs might be involved in brain injury, neuroinflammation and further affect microglia homeostasis. However, no studies have systematically analyzed the genome-wide abnormal expression of RBPs genes in microglia during SAH.
Methods
RNA-seq data of microglia from the SAH mouse group (SAH) and control sham-operated mouse group (sham) were downloaded from the GEO database in GSE167957, including four samples from the sham group and four samples from the SAH group for subsequent analysis.Utilizing GO and KEGG functional enrichment analyses, we conducted a comprehensive study of differentially expressed genes (DEGs), alternative splicing patterns, and co-expression networks to gain deeper insights into the differential expression of RNA-binding proteins (RBPs) and differential alternative splicing events (ASEs) between the SAH (subarachnoid hemorrhage) and sham groups. This analysis aimed to elucidate the potential mechanisms underlying the aberrant expression of RBPs in microglia during brain injury caused by SAH.
Results
ASEs and co-expression analyses of differentially expressed RBPs and differential ASEs were carried out in microglia in terms of gene expression. GO and KEGG functional enrichment analysis showed that aberrantly expressed RBPs such as Mcm7, Mtdh, SRSF3, and Hnrnpa2b1 may affect and regulate downstream Csnk1d, Uckl1 and other protein phosphorylation-related genes by alterative splicing.
Conclusion
RBPs were aberrantly expressed in microglia during the development of brain injury secondary to SAH, regulating alterative splicing of downstream genes and influencing the progression of SAH brain injury in this study. This implies that RBPs are important for the identification of new therapeutic targets for brain injury after SAH.
Similar content being viewed by others
Introduction
Subarachnoid hemorrhage (SAH) is main cause of ruptured intracranial aneurysms (IAs) which is the subtype of stroke, with high rate of morbidity and mortality [1].Early brain injury (EBI) usually occurred after the bleeding at the first 72 h which was considered as the pathophysiological mechanism of delay brain ischemia and poor outcome [2]. It is still challenge to stable unruptured IAs to prevent SAH by exploring new methods. Therefore, it is necessary to clarify the mechanisms of EBI to interfere this model and to further improve the prognosis.
Neuroinflammation is one of most important mechanism involved in EBI after SAH, and microglia, as immunocyte in the central nervous system (CNS), is a key point to attend neuroinflammation [3]. Because of the discrepancy and functional complex of microglia, its detail mechanisms and potential functions are unclear [4, 5].
RBPs are commonly regarded as proteins that bind RNA via one or more globular RNA-binding domains (RBDs), and regulate gene expression and protein function [6]. Zhu et al [7] found that RBM3 had promoted neurogenesis after hypoxic-ischemic brain injury. Shen et al [8] reported that RBPs fox-1 homolog 1 (Rbfox-1) had contributed to protect brain injury secondary to intracerebral hemorrhage (ICH) through blocking micro-RNA-124. Therefore, RBPs with abnormal expression or function deficiency always leads to kinds of disease occurrence and development [9, 10]. Alterative splicing events (ASE) of precursor mRNA (pre-mRNA) are the source of protein diversity, which are associated with the brain development and disease progression [11, 12]. Previous study found that RBPs participate in regulation of ASEs to maintain microglia homeostasis in neuroinflammation [13,14,15].
In summary, RBPs abnormal expression is associated with EBI, neuroinflammation, or microglia. In addition, RBPs regulate ASEs through binding RNA, and further affect downstream ASEs abnormal expression, eventually maintain microglia homeostasis. However, there is no systematic analysis RBPs abnormal and ASEs expression pertaining to microglia in genome-wide of SAH-EBI. This study aims to review microglia RNA-seq in mice with respect to SAH and Sham group through downloading GSE167957 date packet, including four SAH samples and four Sham samples separately to clarify potential mechanism of RBPs abnormal expression of microglia in EBI secondary to SAH.
Materials and methods
Retrieval and process of public data
From the Sequence Read Archive (SRA), we downloaded Public sequence data files. The SRA Run files were transformed into the fastq format using the NCBI SRA Tool fastq-dump. After that, low-quality bases were filtered out of the raw reads using the FASTX-Toolkit (v.0.0.13; http://hannonlab.cshl.edu/fastx_toolkit/). Finally, we used FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc) to evaluate the clean reads’ quality.
Analysis of differentially expressed genes (DEG) and read alignment
HISAT2 was used to map clean readings to the human GRch38 genome [16]. For further analysis, only reads that were uniquely mapped were taken into account. For each gene, the read count and reads per kilobase of exon per million fragments mapped (RPKM) were calculated.Using RPKM, the expression levels of the genes were assessed.For performing the analysis of differential gene expression, the software DEseq2 is opted [17]. The scaling factor will be used by DEseq2 to represent the original reads and explain the variation in library depth.In DESeq2, the scaling factor is often calculated using a method called “median of ratios.” This method first computes the median read count for each gene across all samples, then calculates the geometric mean of these medians (also referred to as the “size factor”). The estimation of gene dispersion is then started by DEseq2 after that, which is further refined to generate improved dispersion estimates for more precise modeling of read counts. Finally, DEseq2 does hypothesis testing using the Wald test or likelihood ratio test and fits the model of negative binomial distribution.The results of the analysis can be used to determine if a gene is differentially expressed using fold change (FC) and false discovery rate (FDR), and DEseq2 can be used to investigate the differential expression between two or more samples. To perform FDR (False Discovery Rate) correction, using method is the Benjamini-Hochberg (B-H) procedure. FC: fold change, the absolute ratio of expression change, is a crucial metric.FDR stands for false discovery rate.The following criteria were used to determine whether a difference was significant: FC 2 or 1/2, FDR 0.05;
Identification of RBPs with differential expression
The analysis of differential gene expression was conducted using DEseq2 [17], a software program designed specifically for this purpose. Using this software, differentially expressed genes (DEGs) were searched for in the raw count data.To ascertain whether a gene was expressed differently, the findings were evaluated using the fold change (FC 2 or 1/2) and false discovery rate (FDR 0.05).Using a database of 1,914 RBPs compiled from two prior reports [6],all differentially expressed genes (DEGs) were stripped of the RNA binding proteins (RBPs) expression profile.
Analysis of alternative splicing
As previously mentioned, the ABLas pipeline was used to identify and measure the alternative splicing events (ASEs) and regulated alternative splicing events (RASEs) that took place between the samples [18, 19]. We summarize ten common ASEs types based on general knowledge of biology and transcriptomics, AS well as reference to the use of AS mentioned in other articles.Briefly, ABLas detected ten types of alternative splicing events (ASEs) by analyzing splice junction reads. These included alternative site (A5SS), exon skipping (ES), mutually exclusive exons (MXE), alternative site (A3SS), mutually exclusivepMXE), mutually exclusivepMXE), cassette exon, A5SS&ES, and A3SS&ES.
Co-expression analysis
All differentially expressed RBP and RASE underwent co-expression analysis.We computed the Pearson correlation ratio between RNA alternative splicing events (RASEs) and differentially expressed RNA-binding proteins (DERBPs). We then filtered out DERBP-RASE pairs that have a correlation factor in the absolute sense.
of 0.8 or higher and a P value of 0.01 or lower.
Analysis of functional enrichment
KOBAS 2.0 was used to find KEGG pathways and Gene Ontology (GO) keywords [20]. The enrichment of each phrase was determined using the hypergeometric test and the Benjamini-Hochberg FDR controlling technique.
Other statistical analysis
Through Principal Component Analysis (PCA) in R, samples were clustered using the first two components. This analysis was performed using the factoextra package (https://cloud.r-project.org/package=factoextra). Additionally, clustering based on Euclidean distance was carried out using the heatmap package (https://cran.r-project.org/web/packages/pheatmap/index.html) in R.
Results
Analysis of gene expression profile of microglia from mouse with SAH
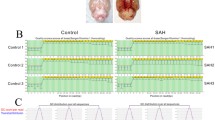
There was difference in gene expression between SAH and Sham based on four SAH and four Sham clustering analysis (Fig. 1A). And gene differential expression analysis showed that there were 1198 differential expression genes, including 879 genes upregulation, 319 genes downregulation, between SAH and Sham group, which indicated microglial gene abnormal expression (Fig. 1B). According to these differentially expression gene performed by cluster analysis, SAH and Sham were classified into two groups, and which further revealed gene abnormal expression in microglia of SAH (Fig. 1C). GO pathway analysis of abnormal expression gene in SAH, significant upregulation genes concentrated on top ten immune inflammation signaling pathways according to p value (Fig. 1D), while downregulation genes concentrated on top ten signaling pathways in terms of negative Transcriptional Regulation by RNA Polymerase II and Enhanced Control of Cellular Migration translation according to p value (Fig. 1E). Further KEGG pathway analysis of abnormal expression gene in SAH, significant upregulation genes concentrated on top ten immune inflammation signaling pathways according to p value (Fig.S1A), while downregulation genes concentrated on top ten signaling pathways according to p value (Fig.S1B).
RNA-seqanalysisofdifferentiallyexpressedgenes(DEGs)betweenSAHandSham samples (A) Heat map showing the clustering of the 8 samples by calculating the Pearson’s correlation. Coefficient susing gene expression (B) Bar plot showing the number of differentially expressed genes in SAN versus Sham group (C) Hierarchi calclustering heat map showing the expression pattern of DEGs among the samples (D) Bar plot showing most enriched GO biological processes of up-regulated genes (E) Bar plot showing most enriched GO biological processes of down-regulated genes
The ASEs in microglia from mouse with SAH
ASEs were done through clustering analysis on the basis of four SAH RNA-seq and four Sham RNA-seq. It could be seen that 4 SAH samples were clustered together, while 4 sham samples were clustered another, indicating that there were differences in the ASEs of the overall genes in SAH and sham (Fig. 2A). The T-test method was used to compare the ratio value of the same splicing event in the ASEs level changes between two samples, and the significant difference of ASEs event screening criterion was p-value ≤ 0.05, The results showed that a large number of A3SS and other ASEs were confirmed (Fig. 2B). Based on the PSI value of these splicing events in differential each sample further, the cluster analysis of the samples was carried out. It could be found that the samples in the SAH and sham groups were clustered into two groups also, respectively, indicating that many ASEs in SAH and sham were tremendous difference (Fig. 2C).
To identify the potential functions of these differentially AS events, further GO and KEGG analyses were performed when differentially AS events were extracted. The results of GO analysis showed that among the signal pathways enriched in genes where differential AS events were located in SAH and sham groups, the top 10 pathways according to p value included translation, RNA splicing, mRNA processing, and RNA polymerase II on transcription. Positive regulation, spliceosome-dependent mRNA splicing, mRNA export from the nucleus, chromatin organization, protein trafficking, DNA templated positive regulation of transcription and mRNA trafficking (Fig. 2D). While KEGG analysis showed that top 10 pathways according to p value included ribosome, mTOR signaling pathway, spliceosome, viral carcinogenesis, endocytosis Effects, rheumatoid arthritis, lysine degradation, metabolism of organic selenium compounds, cysteine and methionine metabolism, ferroptosis (Fig. 2E).
Further study of differently alternatively spliced genes (RASGs) and differentially expressed genes (DEGs) between sham and SAH groups revealed that gene expression and ASEs of 72 genes differed between SAH and sham groups. (Fig. S2A). The changed gene expression and AS were extracted, and then GO and KEGG analysis were performed. GO analysis showed that these genes were mainly enriched in the process of the immune system, innate immune response, Cell differentiation, transcriptional control via RNA polymerase II, and signal transduction (Fig. S2B). GO analysis showed that these genes were mainly enriched in pyruvate metabolism, carbon metabolism, metabolism of cysteine and methionine, glycolysis and glycogenesis, central carbon metabolism in cancer, PPAR signaling pathway, glucagon Signaling pathways, purine metabolism, asthma, hepatitis C (Fig. S2C). As a consequence, ASEs may affect neuroinflammation and microglial function [21].
Subarachnoid hemorrhages’ aberrant alternative splicing events being identified (A) PCA based on ratio values of all regulated alternative splicing events (RASEs). The ellipse for each group is the confidence ellipse. (B) The bar plot showing the number of all RASEs. X-axis: RASE number. Y-axis: the different types of AS events. (C) Heat map of all major RASEs based on a hierarchical grouping of ratio values. All samples that were AS filtered should have identifiable splice junctions, and at least 80% of those samples should have at least ten reads that support the splice junction. (D) The results of the regulated alternative splicing genes (RASGs) displayed in the scatter plot were the most enriched GO biological process results. (E) The most enriched KEGG pathways results of the RASGs were shown on the Scatter plot
Co-expression analysis of differentially RBPs and differentially ASEs in microglia
Owing to the functions of RBPs in ASEs regulation, 99 RBPs were aberrantly expressed in SAH group microglia by intersecting genes that are differently expressed in the SAH and sham groups with the reported genes for potential human RBPs (Fig. 3A), and might lead to abnormal function of these RBPs in microglia which in turn affected the ASEs of downstream target genes. According to the above 99 RBPs expression (FPKM) changes and Ratio value between FPKM and differential ASEs in SAH and sham groups, co-expression analysis was performed in all samples. and it was found that RBPs, such as Mcm7, might regulate multiple differential ASEs (Fig. 3B).
By extracting the top 10 RBPs with co-expression differential ASEs, it was found that these RBPs were abnormally up-regulated or down-regulated in SAH group compared with sham group, which probably affect and regulate the ASEs of downstream genes (Fig. 3C). Further extracting the top 10 RBPs with co-expressed differential ASEs, the genes where the co-expressing differential ASEs were further obtained using GO and KEGG functional analysis. GO functional analysis showed that the genes were mainly enriched in protein transport, chromatin organization, phosphorylation, peptidyl-serine phosphorylation, cell migration, endocytosis, protein ubiquitination, ubiquitination-dependent protein catabolism, and dephosphorylation (Fig. 3D).While KEGG analysis showed that the genes were mainly enriched in aldosterone-regulated sodium reabsorption, proteasome, endocrine resistance, insulin resistance, platinum resistance, organic selenium-containing compound metabolism, metabolic pathways, basal Transcription factors, viral carcinogenesis, carbon pool composed of folic acid (Fig. S3). Interaction network was constructed between differentially expressed RBPs and differentially ASEs, suggesting that abnormal up- or down-regulation of expression in these SAH groups would affect the ASEs of downstream related genes (Fig. 3E).
Co-disturbed network formation between SAH-RAS and RNA-binding proteins (RBP). (A) Venn diagram shows the intersection of differentially expressed genes with RNA binding proteins (DE RBPs). (B) Heatmap shows the top10 DE RBPs genes in samples. RBPs were filtered by which FPKM > = 1 in 80% samples. The z-score color range is indicated by the blue to red color key. (C) The scatter figure displays the number of co-expressed DERBP in B and RASEs by SAH in comparison to Sham samples. (D) The most enriched GO biological processes of RASGs co-disturbed with top10 DE RBPs. (E) The co-deregulation of alternative splicing network between top hub RBPs (the left most part, And the number of connections was indicated by the size of the circle) and RASs (the left-center portion). Co-disturbed RASGs’ top enriched GO biological process was highlighted in orange in the middle right portion of the image (rightmost portion). (F) The network shows phosphorylation term deregulated by RBPs. (G) Box plots shows the expression status of RBP (Mtdh and Mcm7 in the top panel) and splicing ratio profile of RASG in (F), showed in bottom panel
Previous studies [22, 23] showed that protein phosphorylation had an important function in microglia-mediated neuroinflammation. We further extracted the splicing events of two protein phosphorylation-related genes and the co-expressed RBPs, and constructed a co-expression network (Fig. 3F). It was found that abnormally expressed RBPs, such as Mcm7 and Mtdh, might regulate the downstream Csnk1d, Uckl1 and other protein phosphorylation-related ASEs (Fig. 3G).
Correlation analysis of RNA binding proteins (RBPs) with alternative splicing and differential alternative splicing of non-RBPs. (A) RBP and RASG’s overlap is depicted in a venn diagram. (B) The RBPs were extracted from Fig. 4A, and 80% of the samples should contain at least 10 readings that support splice junctions. and the average difference between Normal and Patient groups > = 0.15 Heatmap shows the RBP of the screened differential alternative splicing based on splicing ratio. (C) The RBP of the screened differential alternative splicing and the of non-RBP were analyzed for co expression, and the top 10 RBPs with the largest number of co expression were screened. Bar plot shows the most enriched GO biological process results of top10RBP regulated of non-RBP. (D) The network diagram shows the RBP of the screened differential alternative splicing regulated of non-RBP. (E) The transcripts for the gene are displayed below, and IGV-sashimi graphs demonstrate AS variations in SAH and Sham. The architecture of ASEs are shown schematically (right, top). Alternative exons are shown by blue boxes, constitutive exon sequences by white boxes, and intron sequences by horizontal lines. At the bottom of the right panel, the quantification of ASEs by RNA-seq is displayed. The formula alternative splice junction reads / (alternative splice junction reads + model splice junction reads) was used to determine the changing ratio of AS events in RNA-seq. Error bars represent mean ± SEM. ***P-value < 0.001, ** P-value < 0.01, * P-value < 0.05
Co-expression analysis of splicing-regulated RBPs and differential ASEs in microglia
Given that a large number of abnormally spliced genes enriched in pathways, such as RNA splicing and mRNA processing, 173 RBPs with significance changes were identified in SAH by intersecting the differentially spliced genes between the SAH and sham groups with the reported potential human RBPs genes (Fig. 4A), which might lead to abnormal function of these RBPs in microglia, and in turn affected ASEs of downstream target genes. We further screened these RBPs according to the different splicing body supporting reads (at least 6 samples greater than 10) and ratio variation (greater than 0.1), and obtained 30 differentially spliced RBPs, which were significant difference in SAH group compared to sham group. And the production of abnormal spliceosome proteins might lead to the loss of function of RBPs, which could affect the function of ASEs abnormalities of downstream genes (Fig. 4B).
Co-expression analysis was performed in PSI value of these differentially alternatively spliced RBPs and non-RBPs between SAH and sham group, and the co-expression differentially ASEs of these RBPs were identified, respectively. Ranked according to the number of co-expressed differentially spliced genes of each RBPs, the top 10 RBPs with co-expressed differentially spliced genes was extracted, which was performed by GO and KEGG functional analysis. The results of GO analysis showed that the co-expressed differentially ASEs were mainly enriched in the negative regulation of apoptosis, protein transport, transmembrane transport, phosphorylation, and the positive regulation of transcription by RNA polymerase II (Fig. 4C). and KEGG analysis showed that co-expressed differentially ASEs were mainly enriched in tuberculosis, Ferroptosis, nucleotide excision repair, Hedgehog signaling pathway, Notch signaling pathway, metabolism of amino sugars and nucleotide sugars, and N-Glycan biosynthesis, pyrimidine metabolism, metabolic pathways, and synaptic vesicle cycling (Fig. S4A).
We found that the potential downstream alternatively spliced genes regulated by abnormally spliced RBPs were also enriched in the phosphorylation pathway, and the interaction network was constructed between differentially alternatively spliced RBPs and spliced genes in phosphorylation pathway (Fig. 4D). which indicated RBPs with splicing regulation function, such as SRSF3, Hnrnpa2b1. (Fig. 4E)
Disscusion
In this study, we identified abnormally expressed or spliced RBPs in the microglia of SAH mice by analyzing differentially expressed genes and alternatively spliced genes in the microglia of SAH mice and Sham mice, and established aberrant expression, respectively. It was found that the abnormal expression or abnormal splicing of RBPs such as Mcm7, Mtdh, Srsf3, Hnrnpa2b1 and other RBPs may be variable by regulating downstream protein phosphorylation-related genes, such as Csnk1d and Uckl1, affecting the occurrence and development of microglia- mediated neuroinflammation after SAH.
RBPs are proteins that possess the ability to bind RNA molecules and thereby modulate gene expression and function. In the process of RNA transcription, modification, transport, translation, and degradation metabolism, various specific RBPs bind to them for transcriptional and post-transcriptional regulation. Abnormal expression or functional of RBPs are involved in the development of various diseases, which may be potential therapeutic targets.
The RBPs have an important function in brain injury. After hypoxic-ischemic, RBM 3 promotes neurogenesis through IMP 2-IGF2 signaling pathway in a niche-dependent manner, and inhibited oxygen-glucose deprivation / reoxygenation-induced apoptosis by promoting the formation of stress granules in PC12 cells and rat primary cortical neurons [24]. Rbfox1 was involvement in the expression of CaMKIIα secondary to brain injury induced by cerebral hemorrhage by blocking micro-RNA-124 [25].
The RBPs also have important function in neuroinflammation. Prdx-1 has reduced ICH-induced brain damage by targeting the mRNA stability associated with inflammation and apoptosis [26]. The decrease of TIA-1 has aggravated neuroinflammation in Tauopathy [27]. HuR also have promoted Th 17 cell differentiation, which could be targeted to reduce autoimmune neuroinflammation [28]. IMP 2 has driven the stromal-Th 17 cell circuitry in autoimmune neuroinflammation [29].
RBPs affect brain injury and neuroinflammation development by regulating ASEs. and Variable splicing events of RNA are an important source of protein diversity in living organisms. RBM-5 attenuates brain damage by regulating RNA variable splicing [11, 30]. Therefore, RBPs probably participate in regulating RNA splicing and affects neuroinflammation. The absence or abnormal function of RBPs may affect abnormal variable splicing of genes involved in neuroinflammation in the human brain group of Huntington disease, and RBPs are involved in the regulation of RNA variable splicing factor affecting the function of microglia. It is now recognized that ASEs abnormalities of RNA may affect brain development and the development of brain diseases.
This study discovered that the abnormal expression or abnormal splicing of RBPs such as Mcm7, Mtdh, Srsf3, Hnrnpa2b1 may be variable by regulating downstream protein phosphorylation-related genes, such as Csnk1d and Uckl1, affecting the occurrence and development of microglia-mediated neuroinflammation after SAH. However, the present study is limited to the absence of experimental evidence.
The utilization of Mcm7 proliferation index presents a more effective approach for identifying patients at risk of recurrence when compared to the conventional methods currently employed in clinical practice. This suggests that incorporating Mcm7 into diagnostic procedures has the potential to enhance the accuracy of prognosis prediction and treatment decision-making for meningioma patients [31]. Moreover, by directly targeting Mtdh, miR-30b-5p has a direct impact on glioma cell proliferation, highlighting its potential as a promising therapeutic target for treating glioma [32].
The smallest member of the SR protein family is called Ser/Arg-rich splicing factor 3 (SRSF3), is expressed abnormally in cancer cells and is regulated by a variety of variables and intricate mechanisms in eukaryotic cells. We place great importance on highlighting the detrimental effects of the increased expression of the SRSF3 oncogene in cancer development. Additionally, we delve into the mechanisms by which SRSF3 promotes the transformation of cells, as well as the potential implications of lowering SRSF3 expression as a strategy for developing anticancer drugs. The accumulation of research on SRSF3 has provided invaluable knowledge regarding its crucial role in sustaining biological functions, providing fresh targets for possible cancer treatments [33].
It should be emphasized that the presence and quantities of Hnrnpa2b1 have been found to be linked to cancer-associated fibroblasts in various types of cancers, including LUAD, adrenocortical carcinoma, and stomach adenocarcinoma. Furthermore, Hnrnpa2b1 actively participates in pathways related to spliceosome regulation and cell cycle control. And last, for diseases including lung squamous cell carcinoma, LUAD, esophageal carcinoma, breast invasive carcinoma, and liver hepatocellular carcinoma, Hnrnpa2b1 has a very high diagnostic importance.This thorough research highlighted the importance of Hnrnpa2b1 in the development of several cancer types [34].
In all, the findings of RBPs, including Mcm7, Mtdh, Srsf3, Hnrnpa2b1, may provide clues or further verification and revealing of the microglial cell-mediated neuroinflammation function and mechanism of RBPs in SAH, and are of great significance for the identification of new therapeutic targets for brain injury after SAH. Future research could reveal the pathophysiology of SAH-EBI to improve patient outcomes.
In the future, we will do specific experiments or studies that could address the current study’s limitations.We will increase the sample size to confirm this result.Through in vivo drug interventions, we will evaluate the therapeutic effect of targeting specific RBPs for the treatment of SAH. Utilizing cell models, we will delve into the functions of RBPs in SAH-related cellular signaling pathways.By employing techniques such as RNA interference (RNAi) or CRISPR-Cas9, we will specifically regulate the expression of RBPs and observe their impact on cellular functions.Combining clinical data, we will evaluate the potential application value of therapeutic strategies targeting RBPs in SAH patients.
Conclusions
RBPs were aberrantly expressed in microglia during the development of brain injury secondary to SAH, regulating alterative splicing of downstream genes and influencing the progression of SAH brain injury. This implies that RBPs are important for the identification of new therapeutic targets for brain injury after SAH.After conducting this investigation, we think we have identified a more reliable marker for brain damage to SAH formation than previous reports.If we verify the glial cell-mediated neuroinflammatory function and mechanism of RBPs in SHA, it is expected to be treated.
Limitations
The current study lacks experimental validation in mechanistic studies, which are crucial for understanding the underlying biological processes and molecular interactions. Without such validation, the conclusions drawn from the study may be limited in their generalizability and reliability. The study focuses on the role of RBP (RNA-binding protein) in aSAH (aneurysmal subarachnoid hemorrhage), but the specificity of RBP’s function in this context has not been thoroughly investigated. There may be other factors or pathways that contribute to the observed effects, and thus, the attribution of causality to RBP alone may be premature.The use of only 4 mice in the study is a significant limitation. The small sample size may not provide sufficient statistical power to detect subtle yet important differences or to generalize the findings to a larger population. A larger sample size would likely increase the robustness and reproducibility of the results.
Data availability
Public data were analyzed in this project. All data can be colocted from KEGG (http://www.genome.jp/kegg) and GEO databases (http://www.ncbi.nlm.nih.gov/geo).
References
Macdonald RL, Schweizer TA. Spontaneous subarachnoid haemorrhage. Lancet. 2017;389(10069):655–66.
Rass V, Helbok R. Early Brain Injury after Poor-Grade Subarachnoid Hemorrhage. Curr Neurol Neurosci Rep. 2019;19(10):78.
Lai N, Wu D, Liang T, Pan P, Yuan G, Li X, Li H, Shen H, Wang Z, Chen G. Systemic exosomal miR-193b-3p delivery attenuates neuroinflammation in early brain injury after subarachnoid hemorrhage in mice. J Neuroinflamm. 2020;17(1):74.
Zheng ZV, Lyu H, Lam SYE, Lam PK, Poon WS, Wong GKC. The dynamics of Microglial polarization reveal the Resident neuroinflammatory responses after subarachnoid hemorrhage. Translational Stroke Res. 2020;11(3):433–49.
Chen J, Zheng ZV, Lu G, Chan WY, Zhang Y, Wong GKC. Microglia activation, classification and microglia-mediated neuroinflammatory modulators in subarachnoid hemorrhage. Neural Regeneration Res. 2022;17(7):1404–11.
Hentze MW, Castello A, Schwarzl T, Preiss T. A brave new world of RNA-binding proteins. Nat Rev Mol Cell Biol. 2018;19(5):327–41.
Zhu X, Yan J, Bregere C, Zelmer A, Goerne T, Kapfhammer JP, Guzman R, Wellmann S. RBM3 promotes neurogenesis in a niche-dependent manner via IMP2-IGF2 signaling pathway after hypoxic-ischemic brain injury. Nat Commun. 2019;10(1):3983.
Shen F, Xu X, Yu Z, Li H, Shen H, Li X, Shen M, Chen G. Rbfox-1 contributes to CaMKIIalpha expression and intracerebral hemorrhage-induced secondary brain injury via blocking micro-RNA-124. J Cereb Blood flow Metabolism: Official J Int Soc Cereb Blood Flow Metabolism. 2021;41(3):530–45.
Holmqvist E, Vogel J. RNA-binding proteins in bacteria. Nat Rev Microbiol. 2018;16(10):601–15.
Diaz-Munoz MD, Turner M. Uncovering the role of RNA-Binding proteins in Gene expression in the Immune System. Front Immunol. 2018;9:1094.
Raj B, Blencowe BJ. Alternative splicing in the mammalian nervous system: recent insights into mechanisms and functional roles. Neuron. 2015;87(1):14–27.
Su CH, D D and, Tarn WY. Alternative splicing in neurogenesis and Brain Development. Front Mol Biosci. 2018;5:12.
Marchese E, Valentini M, Di Sante G, Cesari E, Adinolfi A, Corvino V, Ria F, Sette C, Geloso MC. Alternative splicing of neurexins 1–3 is modulated by neuroinflammation in the prefrontal cortex of a murine model of multiple sclerosis. Exp Neurol. 2021;335:113497.
Elorza A, Marquez Y, Cabrera JR, Sanchez-Trincado JL, Santos-Galindo M, Hernandez IH, Pico S, Diaz-Hernandez JI, Garcia-Escudero R, Irimia M, Lucas JJ. Huntington’s disease-specific mis-splicing unveils key effector genes and altered splicing factors. Brain. 2021;144(7):2009–23.
Lee J, Villarreal OD, Chen X, Zandee S, Young YK, Torok C, Lamarche-Vane N, Prat A, Rivest S, Gosselin D, Richard S. QUAKING regulates Microexon Alternative Splicing of the rho GTPase pathway and controls Microglia Homeostasis. Cell Rep. 2020;33(13):108560.
Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–60.
Love MI, Huber W, Anders S. Moderated estimation of Fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550.
Jin L, Li G, Yu D, Huang W, Cheng C, Liao S, Wu Q, Zhang Y. Transcriptome analysis reveals the complexity of alternative splicing regulation in the fungus verticillium dahliae. BMC Genomics. 2017;18(1):130.
Xia H, Chen D, Wu Q, Wu G, Zhou Y, Zhang Y, Zhang L. CELF1 preferentially binds to exon-intron boundary and regulates alternative splicing in HeLa cells. Biochim et Biophys acta Gene Regul Mech. 2017;1860(9):911–21.
Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011; 39(Web Server issue):W316–322.
Torres-Odio S, Key J, Hoepken HH, Canet-Pons J, Valek L, Roller B, Walter M, Morales-Gordo B, Meierhofer D, Harter PN, Mittelbronn M, Tegeder I, Gispert S, et al. Progression of pathology in PINK1-deficient mouse brain from splicing via ubiquitination, ER stress, and mitophagy changes to neuroinflammation. J Neuroinflamm. 2017;14(1):154.
Wittrahm R, Takalo M, Marttinen M, Kuulasmaa T, Makinen P, Kemppainen S, Martiskainen H, Rauramaa T, Pike I, Leinonen V, Natunen T, Haapasalo A, Hiltunen M. MECP2 increases the pro-inflammatory response of Microglial cells and phosphorylation at serine 423 regulates neuronal gene expression upon Neuroinflammation. Cells. 2021; 10(4).
Wang H, Huang M, Wang W, Zhang Y, Ma X, Luo L, Xu X, Xu L, Shi H, Xu Y, Wang A, Xu T. Microglial TLR4-induced TAK1 phosphorylation and NLRP3 activation mediates neuroinflammation and contributes to chronic morphine-induced antinociceptive tolerance. Pharmacol Res. 2021;165:105482.
Si W, Li Z, Huang Z. et al.RNA binding protein motif 3 inhibits oxygen-glucose Deprivation/Reoxygenation-Induced apoptosis through promoting stress granules formation in PC12 cells and rat primary cortical Neurons[J].Frontiers in cellular neuroscience, 2020, 14:559384.
Shen F, Xu X, Yu Z, et al. Rbfox-1 contributes to CaMKIIα expression and intracerebral hemorrhage-induced secondary brain injury via blocking micro-RNA-124[J]. Journal of Cerebral Blood Flow and Metabolism; 2021.
Yang GQ, Huang JC, Yuan JJ. ,Prdx1 reduces Intracerebral Hemorrhage-Induced Brain Injury via Targeting inflammation- and apoptosis-related mRNA Stability[J].Frontiers in neuroscience,2020.00181.
LeBlang CJ, Medalla M, Nicoletti NW, Hays EC, Zhao J, Shattuck J, Cruz AL, Wolozin B, Luebke JI. Reduction of the RNA binding protein TIA1 exacerbates Neuroinflammation in Tauopathy. Front Neurosci. 2020;14:285.
Chen J, Martindale JL, Abdelmohsen K. ,RNA-Binding protein HuR promotes Th17 cell differentiation and can be targeted to reduce autoimmune Neuroinflammation[J]. J Immunol, 2020:ji1900769.
Bechara R, Amatya N, Bailey RD, Li Y, Aggor FEY, Li DD, Jawale CV, Coleman BM, Dai N, Gokhale NS, Taylor TC, Horner SM, Poholek AC, Bansal A, Biswas PS, Gaffen SL. The m6A reader IMP2 directs autoimmune inflammation through an IL-17- and TNFα-dependent C/EBP transcription factor axis. Sci Immunol. 2021;6(61):eabd1287.
Bielli P, Pagliarini V, Pieraccioli M, Caggiano C, Sette C. Splicing dysregulation as oncogenic driver and passenger factor in brain tumors. Cells. 2019;9(1):10.
Hallén T, Olsson DS, Hammarstrand C, Örndal C, Engvall A, Ragnarsson O, Skoglund T, Johannsson G. MCM7 as a marker of postsurgical progression in non-functioning pituitary adenomas. Eur J Endocrinol. 2021;184(4):521–31.
Zhang D, Liu Z, Zheng N, Wu H, Zhang Z, Xu J. MiR-30b-5p modulates glioma cell proliferation by direct targeting MTDH. Saudi J Biol Sci. 2018;25(5):947–52.
Kim HR, Shin CH, Lee H, Choi KH, Nam DH, Ohn T, Kim HH. MicroRNA-1908-5p contributes to the oncogenic function of the splicing factor SRSF3. Oncotarget. 2017;8(5):8342–55.
Chen C, Huang L, Sun Q, Yu Z, Wang X, Bu L. HNRNPA2B1 demonstrates diagnostic and prognostic values based on Pan-cancer analyses. Comput Math Methods Med. 2022;2022:9867660.
Funding
This work was supported by the following projects: (1) Natural Science Fund of Jiangxi Province of China (Number:20192BAB215024), and (2) Natural Science Fund of Jiangxi Province of China (Number:20202BABL206055).
Author information
Authors and Affiliations
Contributions
P.X.Y.: Writing-Original Draft, Validation, Formal analysis, Conceptualization, Software, Methodology O.Y.H.Y.: Resources, Data Curation X.X.: Visualization, Investigation Z.X.B.: Formal analysis, Funding acquisition L.L.F.: Methodology, Writing - Review & Editing, Supervision, Project administrationAll authors reviewed the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it.The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Pan, X., Ouyang, H., Xiao, X. et al. Analysis of different expression RNA binding protein genes in mouse microglia cell from the brains of mice 72 h after subarachnoid hemorrhage or sham operation. BMC Med Genomics 17, 194 (2024). https://doi.org/10.1186/s12920-024-01972-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12920-024-01972-x