Abstract
Background
Algorithms that use administrative health and electronic medical record (EMR) data to determine cancer recurrence have the potential to replace chart reviews. This study evaluated algorithms to determine breast and colorectal cancer recurrence in a Canadian province with a universal health care system.
Methods
Individuals diagnosed with stage I-III breast or colorectal cancer diagnosed from 2004 to 2012 in Manitoba, Canada were included. Pre-specified and conditional inference tree algorithms using administrative health and structured EMR data were developed. Sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV) correct classification, and scaled Brier scores were measured.
Results
The weighted pre-specified variable algorithm for the breast cancer validation cohort (N = 1181, 167 recurrences) demonstrated 81.1% sensitivity, 93.2% specificity, 61.4% PPV, 97.4% NPV, 91.8% correct classification, and scaled Brier score of 0.21. The weighted conditional inference tree algorithm demonstrated 68.5% sensitivity, 97.0% specificity, 75.4% PPV, 95.8% NPV, 93.6% correct classification, and scaled Brier score of 0.39. The weighted pre-specified variable algorithm for the colorectal validation cohort (N = 693, 136 recurrences) demonstrated 77.7% sensitivity, 92.8% specificity, 70.7% PPV, 94.9% NPV, 90.1% correct classification, and scaled Brier score of 0.33. The conditional inference tree algorithm demonstrated 62.6% sensitivity, 97.8% specificity, 86.4% PPV, 92.2% NPV, 91.4% correct classification, and scaled Brier score of 0.42.
Conclusions
Algorithms developed in this study using administrative health and structured EMR data to determine breast and colorectal cancer recurrence had moderate sensitivity and PPV, high specificity, NPV, and correct classification, but low accuracy. The accuracy is similar to other algorithms developed to classify recurrence only (i.e., distinguished from second primary) and inferior to algorithms that do not make this distinction. The accuracy of algorithms for determining cancer recurrence only must improve before replacing chart reviews.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Cancer recurrence is the diagnosis of a second clinical episode of cancer after the first was considered cured. It occurs from residual microscopic disease which was not clinically detectable and is different from cancer progression which is due to the growth of known clinical disease. As novel cancer treatments and screening have been introduced and survival has improved, cancer recurrence has become an important outcome; it often results in additional treatment, is a predictor for subsequent mortality, and can be used to compare treatment effectiveness, measure recurrence-free survival, and plan and prioritize cancer control resources [1].
Since 1956, the Manitoba Cancer Registry (MCR) in the province of Manitoba, Canada has been legislated to collect, classify, and maintain population-based information about cancer cases including diagnosis date, histology, topography, stage, and treatment for the entire provincial population. The MCR has been consistently shown to be of very high quality, completeness, and histological verification [2]. Unfortunately, cancer registries, including the MCR, do not systematically identify recurrent cancers. Therefore, recurrence is determined using manual chart review. Manual chart reviews can provide detailed and reflective information. However, they have several important disadvantages. In order to complete a chart review in a timely manner, multiple abstractors are usually required which may introduce error and bias, especially if data abstraction processes are not clear and inter-rater reliability is low. Manually reviewing charts is also labour intensive, time consuming, and, hence, can be expensive.
An alternative method for identifying recurrence is to use existing structured health care data. Several prior studies have developed and validated algorithms for determining cancer recurrence using structured health data in the United States (US), Europe, and Canada [3,4,5,6,7,8,9,10]. Each of these studies has limitations including limited sources of data, narrowly defined populations, different definitions of recurrence, missing validation cohorts, and the use of suboptimal performance measures which do not account for prevalence. Based on the importance of determining recurrence, the inefficiencies of chart reviews, and the limitations of previous recurrence algorithm studies, our goal was to evaluate algorithms to determine recurrence in breast and colorectal cancer cohorts using administrative health and structured electronic medical record (EMR) data in a Canadian province with a universal health care system.
Methods
Setting
The province of Manitoba, located in central Canada, has a population of 1.37 million (as of 2019) [11]. Approximately 55% of the population live in the capital city of Winnipeg. Manitoba Health, Seniors and Active Living (MHSAL), the publicly funded provincial health insurance agency, provides comprehensive universal health coverage for hospitalizations, procedures, and physician visits for provincial residents. MHSAL maintains several electronic databases to monitor health care use and reimburse health care providers for services delivered. Since 1984, provincial residents have been assigned a personal health identification number (PHIN) which can be used to link provincial health information databases allowing health care utilization and outcomes to be tracked longitudinally.
Data sources
The MCR was used to identify individuals diagnosed with breast or colorectal cancer, cancer diagnosis date, age at diagnosis, cancer stage, estrogen receptor/progesterone receptor (ER/PR) and human epidermal growth factor receptor 2 (HER-2) status, date and type of cancer surgery, and date of the first radiation treatment for each course of radiation treatment. The CancerCare Manitoba (CCMB) electronic medical record is the record of clinical cancer interactions, investigations, and treatment, and was used to determine dates and types of systemic anti-cancer medical therapy as well as carcinoembryonic antigen (CEA) and cancer antigen 15–3 (Ca 15–3) blood test results.
We used three MHSAL administrative databases: the Manitoba Population Registry, the Medical Claims database, and the Drug Program Information Network (DPIN) database. The Manitoba Population Registry contains demographic, vital status, and migration information and was used to determine the start and end dates of provincial health coverage. The Medical Claims database is generated by claims filed by health care providers for reimbursement of service and includes services provided, diagnosis, provider, and service date. Medical Claims data were used to determine palliative care consultations. The DPIN database includes all prescriptions dispensed from outpatient pharmacies in Manitoba. DPIN data was used to determine capecitabine, a chemotherapy drug used to treat different cancers including breast and colorectal cancer. Laboratory data were obtained from Shared Health, Manitoba’s public sector laboratory, to identify CEA and Ca15–3 blood test results which were not already in the CCMB medical record. The accuracy and completeness of Manitoba Health’s administrative data has been previously established [12,13,14].
Study population
The study included individuals diagnosed with stage I-III colorectal cancer (International Classification of Diseases, Oncology 3rd edition (ICD-O-3) codes C18.0. C18.2–9, C19, C20, C26.0) or breast cancer (ICD-O-3 codes C50.0–6, C50.8–9). Stage IV cases, which have metastasis at diagnosis, were excluded as these individuals develop progression (i.e., worsening disease) rather than recurrence.
The study population was divided into a training cohort of individuals diagnosed from 2004 to 2007 and a validation cohort of individuals diagnosed from 2008 to 2012. Breast and colorectal cancers were analyzed separately. The breast cancer training cohort included cancers that were either ER negative, PR negative, or HER-2 positive because these cancers have a higher recurrence rate and therefore decreased the number of cases needed to review [15]. The colorectal cancer training cohort focused on stage II and III because they are expected to have higher rates of recurrence compared to stage I cancers [16]. The breast and colorectal cancer validation cohorts included individuals diagnosed with stage I-III cancers. However, the breast cancer cohort was oversampled with ER negative, PR negative, and HER-2 cases and the colorectal cancer cohort was oversampled with higher stages to ensure that enough recurrences were identified. The validation cohort included individuals diagnosed in later years to provide external validation, which is a more rigorous validation method than internal or apparent validation [17,18,19].
Study variables
Study variables are summarized in Table 1. Cancer recurrence included loco-regional (reappearance of cancer in the same region of the body or the lymph nodes) and distant (reappearance of cancer in another part of the body) recurrence. Surgery and radiation treatment data were linked by a tumour ID which identifies the treatment associated with a specific tumour. Therefore, if an individual had more than one cancer diagnosis, the treatment data could be linked to the appropriate cancer. The remaining variables could not be linked to a specific tumour. To increase accuracy in classifying the remaining variables, conditions were added. Surgery (mastectomy, lumpectomy, axillary lymph node dissection for breast cancer and bypass or resection surgery for CRC) beyond 12 months after diagnosis were included to capture local recurrence after the use of neoadjuvant treatments. New disease within 1 year is usually considered part of the primary diagnosis. Surgery for a non-breast site or liver or lung resections beyond 6 months were included to capture treatment for metastases after the initial treatment. The receipt of chemotherapy beyond 12 months of diagnosis and RT beyond 12 months for breast cancer were considered due to recurrence unless the treatments occurred after a second primary treated with surgery. Although chemotherapy treatment could not be linked to a specific tumour, the treatment site was identified which increased accuracy of correct association. A palliative care consult was considered due to recurrence if it was provided by an oncologist and was linked to a breast cancer (breast cancer cases only), colorectal cancer (colorectal cancer cases only), lung cancer, liver cancer, or undetermined metastases beyond 6 months after diagnosis to exclude any treatment discussions that may have occurred after diagnosis. Elevated blood markers (CEA > 10; Ca 15–3 > 50) often related to recurrence more than 12 months after diagnosis were considered due to recurrence unless they occurred within 3 months of another primary cancer diagnosis. Elevated blood markers prior to 12 months were not included to avoid initial elevations due to the original diagnosis or treatment.
Algorithm development and validation
A chart review was first conducted by trained research assistants to identify cancer recurrence. A duplicate chart review by a research assistant who did not conduct the initial chart review was conducted for a fraction of the cohort (10%) to evaluate inter-rater reliability. The algorithms were then developed by analyzing the same cohorts using two approaches: pre-specified variables and conditional inference trees. The pre-specified variable approach used variables and clinically meaningful cut offs determined prior to the start of the study. Variables and cut-offs were selected with information from previous studies and local cancer experts. For this algorithm, if an individual was positive for any of the variables included, they were predicted to have a recurrence. The conditional inference tree approach is an automated machine learning technique that explicitly states the algorithm that was developed, which is not achieved with other machine learning techniques. The conditional inference trees used the same variables as the pre-defined algorithm. However, trees were created based on the association between each covariate and the outcome of interest (i.e., recurrence). The ctree function within the party R package with a default setting (a quadratic test statistic, Bonferroni-adjusted p-values, and criterion p-value of 0.05) [20]. Validation cohorts were used to determine if the algorithms developed were generalizable to cancer cohorts independent of those analyzed as part of the algorithm development.
Performance metrics
Sensitivity (the percentage of individuals who had a recurrence that were correctly identified), specificity (the percentage of individuals who did not have a recurrence that were correctly identified), positive predictive value (PPV) (percentage of individuals predicted to have recurrence that truly have recurrence), negative predictive value (NPV) (percentage of individuals predicted to not have recurrence that truly do not have recurrence), correct classification (the percentage of individuals who were correctly classified as having a recurrence or not having a recurrence), and scaled Brier scores were calculated to determine algorithm accuracy. These classification measures are commonly used in the literature to describe the performance of algorithms and models. Brier scores measure the predictive accuracy by subtracting the predictive values from the outcome values (the average of squared differences between predicted values and outcome values). The Brier score was then scaled to the proportion of events (p) in the cohort 1-(Brier score/(mean(p)*(1-mean(p)))) where a value of 1 is perfect prediction, a value of 0 is chance, and a negative value is worse than chance) [21, 22]. Therefore, unlike measures like sensitivity, specificity, and correct classification, the scaled Brier score is adjusted for the prevalence of events in the cohort. This is advantageous in measuring accuracy over commonly used classification measures which ignore prevalence. Measures were unweighted for both training and validation cohorts. Weighted measures were also calculated for the validation cohort to account for oversampling. Confidence intervals were determined using the permutation method, including 1000 replications and reporting values at the 2.5th and 97.5th percentiles.
Results
Breast cancer
The breast cancer training cohort included 933 cases with 186 recurrences and the validation cohort included 1181 cases with 167 recurrences (Table 2). The mean age at diagnosis was 60.3 years (standard deviation (SD) 14.1) in the training cohort and 62.5 years (SD 13.9) in the validation cohort. In the training cohort, 36.2% were diagnosed at stage I, 45.7% at stage II, and 18.1% at stage III. The stage distribution was similar in the validation cohort. Two-hundred and seven charts were reviewed by a second research assistant and the Kappa statistic for recurrence status was 0.81.
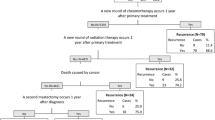
Table 3 shows the performance metrics for determining breast cancer recurrence for the training and validation cohorts, unweighted and weighted, using pre-specified variable and conditional tree algorithms. In the validation cohort, the pre-specified variable algorithm demonstrated the following weighted results: 81.1% sensitivity, 93.2% specificity, 61.4% PPV, 97.4% NPV, and 91.8% correct classification. The weighted scaled Brier score was 0.21 which demonstrates low accuracy. The conditional inference tree algorithm for the breast cancer training cohort is shown in Fig. 1. The percentage of individuals who were classified as having had a recurrence are shown for each node in the tree. For example, 70% of individuals who did not have chemotherapy but had radiation therapy greater than 12 months after diagnosis were classified as having had a recurrence (node 3, N = 53). Overall, the conditional inference tree algorithm demonstrated the following weighted results: 68.5% sensitivity, 97.0% specificity, 75.4% PPV, 95.8% NPV, and 93.6% correct classification. The weighted scaled Brier score was 0.39.
Colorectal cancer
The colorectal cancer training cohort included 620 cases with 126 recurrences and the colorectal cancer validation cohort included 693 cases with 136 recurrences (Table 2). The mean age at diagnosis was 69.4 years (SD 12.6) in the training cohort and 67.9 years (SD 12.3) in the validation cohort. In the training cohort, no individuals were included who were diagnosed at stage I; 46.0% were diagnosed at stage II and 54.0% were diagnosed at stage II. In the validation cohort, 19.9, 34.1, and 46.0% were diagnosed at stages I, II, and III, respectively. One-hundred and twenty-eight charts were reviewed by a second research assistant and the Kappa statistic for recurrence status was 0.73.
Table 4 shows the performance metrics for determining colorectal cancer recurrence for the training and validation cohorts, both unweighted and weighted, using the pre-specified variable and conditional tree algorithms. In the validation cohort, the pre-specified variable algorithm demonstrated the following weighted results: 77.7% sensitivity, 92.8% specificity, 70.7% PPV, 94.9% NPV, and 90.1% correct classification. The weighted scaled Brier score was 0.33. The conditional inference tree algorithm developed demonstrated the following weighted results: 62.6% sensitivity, 97.8% specificity, 86.4% PPV, 92.2% NPV, and 91.4% correct classification (Fig. 2). The weighted scaled Brier score was 0.42.
Discussion
Main findings
We found that algorithms using administrative health and structured EMR data to determine breast and colorectal cancer recurrence had high to moderate sensitivity and PPV, high specificity, NPV, and correct classification but low accuracy after adjusting for the prevalence of the outcome in the cohort. As expected, training cohort results were higher than validation cohort results because the algorithms were optimized on the training cohorts. We chose to include breast and colorectal cancers as these sites have relatively high survival rates, are the second and third most commonly diagnosed cancers in Manitoba (which makes chart reviews even more costly and time-consuming), and are historically more likely than aggressive cancers with poorer survival to have recurrences that can be effectively treated. Whether or not these algorithms can replace chart reviews for determining cancer recurrence necessitates weighing the costs required to conduct a chart review with the benefit of quickly applying an algorithm with less than optimal accuracy.
Comparison with other studies
Prior studies that evaluated cancer recurrence algorithms using structured data found moderate to high sensitivities and specificities but have several important limitations. Lamont et al. [3] used Medicare claims data to measure disease-free survival in individuals ≥65 years of age diagnosed with breast cancer (N = 45, 12 recurrences and 2 deaths) . Algorithm sensitivity and specificity were 83 and 97% respectively. Rasmussen et al. [6] used national data in Denmark to identify breast cancer recurrence (n = 471, 149 recurrences). Sensitivity was 97%, specificity was 97%, and PPV was 94%. Xu et al. [9] developed algorithms to identify breast cancer recurrence among women ≤40 years of age or those who received neoadjuvant chemotherapy in Alberta (N = 598, 121 recurrences) . Sensitivity values ranged from 75 to 94% and specificity values ranged from 94 to 98%. Chubak et al. [5] developed several algorithms to determine second breast cancer events and recurrence only among women diagnosed with stage I or II breast cancer (n = 3152, 407 breast cancer events). Sensitivity values ranged from 69 to 99% and specificity values ranged from 81 to 99%. These four studies generally demonstrated better accuracy compared to our study. However, one study included a single small training cohort [3] and two studies did not distinguish between recurrence and second breast cancer primary (i.e., a new primary cancer unrelated to the prior cancer) [6, 9]. This distinction is important in order to use the algorithms to evaluate outcomes such as the effectiveness of treatments in preventing a cancer recurrence. We attempted to distinguish recurrence from a second primary in our chart reviews, although this was difficult in some cases. In addition, one study’s results were based on a training cohort which would have produced overly optimistic results [23] and another excluded patients with second primary non-breast tumours [9] which may have introduced bias.
Chubak et al. [5] noted that their algorithms to classify only recurrence generally demonstrated accuracy that were not as high as their algorithms to classify second breast cancer events . Several recurrence-only studies have generally demonstrated lower accuracy in comparison to the previously mentioned studies, as well as similar accuracy to our study results. A recent study (2016) developed a medical claims-based algorithm to identify ovarian cancer recurrence (N = 94, 32 recurrences) [4]. Sensitivity was 100% and specificity was 89% but only a training cohort was assessed and the cohort size was small. A large US study (2014) evaluated multiple recurrence algorithms for each cancer site of lung, colorectal, breast, and prostate (N = 6227, 736 recurrences) [7]. Sensitivity ranged from 6 to 85% and specificity ranged from 70 to 97%. In 2017, the study was extended to include additional data to be used in the lung and colorectal cancer algorithms: sensitivity ranged from 72 to 91% and specificity ranged from 86 to 98% [8]. Cairncross et al. [10] randomly selected 200 women (26 recurrences) diagnosed with cancer and who had ever had a pregnancy between 2003 and 2012. Sensitivity was 81%, specificity was 81%, PPV was 39%, and NPV was 97%.
Importantly, none of the prior studies used metrics that are optimal to measure algorithm performance such as the scaled Brier score. Sensitivity and specificity are useful because they provide context about how an algorithm can be improved by identifying areas of weakness. For example, some recurrences in our study were missed because the individual did not receive treatment, which decreased sensitivity. This could have been improved if cause of death data were available for the study period. In addition, chemotherapy for a second primary was often found among false positives, which decreased specificity. This could have been improved if chemotherapy data could have been linked to individual tumours rather than to only individuals. However, sensitivity and specificity ignore the rate of events in a cohort which make assessing the overall performance of an algorithm challenging. For example, a specificity or correct classification of 95% will have a high rate of false positives if the rate of event is low (e.g., 1%) but would be substantially better with a higher rate of events (e.g., 50%). The scaled Brier score, which is a summary measure that accounts for the rate of events in a cohort, does not have this limitation. Moreover, if a proposed algorithm is expected to replace a chart review, metrics of accuracy should also indicate the amount of measurement error involved. The scaled Brier score, which has a similar interpretation to the R2, indicates random association with a value of 0 and perfect prediction with a value of 1. This provides more informative output to describe accuracy than measures that only use subsections of the cohort (e.g., sensitivity and specificity).
Other methods, such as those that use natural language processing (NLP) to capture recurrence from unstructured EMR data, have been used to determine breast cancer recurrence with sensitivities from that range from 83 to 92% [24,25,26,27]. These results are not very different that those that used structured administrative data and therefore, may also not be accurate enough at this time to replace a chart review. Another option is to use recurrence algorithms as a screening tool to reduce the number of charts that need to be manually reviewed. However, more research is needed to create algorithms with higher sensitivities to evaluate this possibility.
Strengths and limitations
We used data from previously validated, high-quality, complete, population-based administrative health databases [12, 13, 28, 29]. However, our gold standard was a chart review which is subject to human error. The inter-rater reliability was strong for breast cancer and moderate for colorectal cancer [30]. Therefore, there was some disagreement among the chart reviewers about what constitutes recurrence. When investigating samples of false positives and negatives in the training cohort, some misclassifications of recurrence status had occurred. This was often due to the difficulty in distinguishing recurrence from a second primary, which is expected because it is sometimes challenging for physicians to definitively make this determination. We also found that additional chemotherapy may have been due to a second cancer primary and not cancer recurrence leading to false positive cases. Like some prior studies, our definition of recurrence was not time dependent. We chose to not include this because this would only lead to poorer results.
Conclusions
Our algorithms that used structured administrative health and EMR data to determine recurrence in breast and colorectal cancer cohorts had moderate sensitivity and PPV, high specificity, NPV, and correct classification but low overall accuracy. These results and a review of other studies suggest that more accurate algorithms to capture recurrence-only events are required to replace chart reviews.
Availability of data and materials
The data that support the findings of this study are not publicly available to ensure and maintain the privacy and confidentiality of individuals’ health information. Requests for may be made to the appropriate data stewards (Manitoba Health, Seniors and Active Living’s Health Information Privacy Committee and CancerCare Manitoba’s Research and Resource Impact Committee).
Abbreviations
- CCMB:
-
CancerCare Manitoba
- CEA:
-
Carcinoembryonic antigen
- DPIN:
-
Drug Program Information Network
- EMR:
-
Electronic medical record
- ER/PR:
-
Estrogen receptor/progesterone receptor
- HER-2:
-
Human epidermal growth factor receptor 2
- ICD-0-3:
-
International Classification of Diseases, Oncology 3rd edition
- MCR:
-
Manitoba Cancer Registry
- MHSAL:
-
Manitoba Health Seniors and Active Living
- NPV:
-
Negative predictive value
- PHIN:
-
Personal Health Information Number
- PPV:
-
Positive predictive value
- SD:
-
Standard deviation
- US:
-
United States
References
Yu X. In: Feuerstein M, Ganz P, editors. Epidemiology of Cancer recurrence, second primary Cancer, and comorbidity among Cancer survivors. New York: Springer; 2011.
North American Association of Central Cancer Registries. APPENDIX C - Data Quality Indicators by Year and Registry. In: Hotes Ellison J, Wu XC, McLaughlin C, Lake A, Firth R, et al., editors. Cancer In North America: 1999–2003 Volume One: Incidence. Springfield: North American Association of Cancer Registries Inc.; 2006. p. II-325.
Lamont EB, Hernon JE, Weeks JC, Henderson C, Earle CR, Schilsky RL, et al. Measuring disease-free survival and cancer relapse using medicare claims from CALGB breast cancer trial participants (Companion to 9344). J Natl Cancer Inst. 2006;98(18)1335-8.
Livaudais-Toman J, Franco R, Prasad-Hayes M, Howell EA, Wisnivesky J, Bickell NA. A validation of administrative claims data to measure ovarian cancer recurrence and secondary debluking surgery. EGEMS. 2016;4(1):1208.
Chubak J, Yu O, Pocobelli G, Lamerato L, Webster J, Prout MN, et al. Administrative data algorithms to identify second breast cancer events following early-stage invasive cancer. J Natl Cancer Inst. 2012;104(12):931–40. https://doi.org/10.1093/jnci/djs233.
Rasmussen LA, Jensen H, Flytkjaer Virgilsen L, Beck Jellesmark Thorsen L, Vrou Offersen B, Vedsted P. A validated algorithm for register-based identification of patients with recurrence of breast cancer - based on Danish Breast Cancer Group (DBCG) data. Cancer Epidemiol. 2019;59:129–34.
Hassett MJ, Ritzwoller DP, Taback N, Carroll N, Cronin AM, Ting GV, et al. Validating billing/encounter codes as indicators of lung, colorectal, breast, and prostate cancer recurrence using 2 large contemporary cohorts. Med Care. 2014;52(10):e65–73. https://doi.org/10.1097/MLR.0b013e318277eb6f.
Hassett MJ, Uno H, Cronin AM, Hornbrook MC, Ritzwoller DP. Detecting lung and colorectal cancer recurrence using structured clinical/administrative data to enable outcomes research and population health management. Med Care. 2017;55(12):e88–98. https://doi.org/10.1097/MLR.0000000000000404.
Xu Y, Kong S, Cheung WY, Bouchard-Fortier A, Dort JC, Quan H, et al. Development and validation of case-finding algorithms for recurrence of breast cancer using routinely collected administrative data. BMC Cancer. 2019;19(210):210.
Cairncross ZF, Nelson G, Shack L, Metcalfe A. Validation in Alberta of an administrative data algorithm to identify cancer recurrence. Curr Oncol. 2020;27(3):e343–e6. https://doi.org/10.3747/co.27.5861.
Manitoba Health, Seniors and Active Living. Population Report, June 1, 2019. Winnipeg: Manitoba Health, Seniors and Active Living; 2019.
Robinson JR, Young TK, Roos LL, Gelskey DE. Estimating the burden of disease. Comparing administrative data and self-reports. Med Care. 1997;35(9):932–47. https://doi.org/10.1097/00005650-199709000-00006.
Roos LL, Mustard CA, Nicol JP, McLarran DF, Malenka DJ, et al. Registries and administrative data: organization and accuracy. Med Care. 1993;31(3):201–12. https://doi.org/10.1097/00005650-199303000-00002.
Roos LL, Walld R, Uhanova J, Bond R. Physician visits, hospitalizations, and socioeconomic status: ambulatory case sensitive condintions in a Canadian setting. Health Serv Res. 2005;40(4):1167–85. https://doi.org/10.1111/j.1475-6773.2005.00407.x.
Arvold ND, Taghaian AG, Niemierko A, Abi Raad RF, Srreedhara M, Nguyen PL, et al. Age, breast cancer subtype approximation, and local recurrence after breast-conserving therapy. J Clin Oncol. 2011;29(29):3885–91. https://doi.org/10.1200/JCO.2011.36.1105.
Siegel R, Miller K, Fedewa S, Ahnen D, Meester R, Barzi A, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67(3):177–93. https://doi.org/10.3322/caac.21395.
Chatfield C. Model uncertainty, data mining and statistical inference. J R Stat Soc A. 1995;158(3):419–66. https://doi.org/10.2307/2983440.
Efron B. Estimating the error rate of a prediction rule: improvement on cross-validation. J Am Stat Assoc. 1983;78(382):316–31. https://doi.org/10.1080/01621459.1983.10477973.
Harrell FE, Lee K, Mark D. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing erros. Stat Med. 1996;15(4):361–87. https://doi.org/10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4.
Hothorn T, Hornik K, Zeileis A. Unbiased recursive partitioning: a conditional inference framework. J Comput Graph Stat. 2006;15(3):651–74. https://doi.org/10.1198/106186006X133933.
Steyerberg EW, Vickers AJ, Cook NR, Gerds T, Gonen M, Obuchowski N, et al. Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology. 2010;21(1):128-38.
Steyerberg EW. Clinical prediction models: a practical approach to development, validation, and udating. New York: Springer-Verlag; 2009. https://doi.org/10.1007/978-0-387-77244-8.
Albrechtsen S, Rasmussen M, Thoresen S, Irgens LM, Iversen OE. Pregnancy outcomes in women before and after cervical conisation: population based cohort study. BMJ. 2008;337(sep18 1):a1343. https://doi.org/10.1136/bmj.a1343.
Banerjee I, Bozkurt S, Caswell-Jin JL, Kurian AW, Rubin DL. Natural language processing approaches to detect the timeline of metastatic recurrence of breast cancer. JCO Cllin Cancer Inform. 2019:1–12.
Carrell DS, Halgrim S, Tran D-T, Buist DS, Chubak J, Chapman WW, et al. Using natural language processing to improve efficiency of manual chart abstraction in research: the case of breast cancer recurrence. Am J Epidemiol. 2013;179(6):749–58.
Ling AY, Kurian AW, Caswell-Jin JL, Sledge GW Jr, Shah NH, Tamang SR. Using natural language processing to construct a metastatic breast cancer cohort from linked cancer registry and electronic medical records data. JAMIA Open. 2019;2(4):528–37. https://doi.org/10.1093/jamiaopen/ooz040.
Zeng Z, Espino S, Roy A, Li X, Khan SA, Clare SE, et al. Using natural language processing and machine learning to identify breast cancer local recurrence. BMC Bioinformatics. 2018;19(S17):498–508. https://doi.org/10.1186/s12859-018-2466-x.
Roos LL, Traverse D, Turner D. Delivering prevention: the role of public programs in delivering care to high-risk populations. Med Care. 1999;37(6):JS264–JS78.
McPhee SJ, Nguyen TT, Shema SJ, Nguyen B, Somkin C, Vo P, et al. Validation of recall of breast and cervical cancer screening by women in an ethcinally diverse population. Prev Med. 2002;35(5):463–73. https://doi.org/10.1006/pmed.2002.1096.
McHugh ML. Interrater reliability: the kappa statistic. Biochem Med. 2012;22(3):276–82.
Acknowledgements
We gratefully thank from the CancerCare Manitoba Foundation for their support and CancerCare Manitoba, Manitoba Health, Seniors and Active Living, and Shared Health for the provision of data. We also thank Zack Hall, LoriAnn Love, Coreen Hildebrand, and Megan Noonan for conducting the chart reviews and Elizabeth Harland for study coordination.
Funding
This work was financially supported by the CancerCare Manitoba Foundation. The funding body played no role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
PL contributed conceptualization, methodology, formal analysis, investigation, data curation, writing – review & editing, visualization, supervision, funding acquisition. MP contributed conceptualization, methodology, resources, writing – review & editing, funding acquisition. HS contributed conceptualization, methodology, resources, writing – review & editing, funding acquisition. KD contributed conceptualization, methodology, writing – original draft, writing – review & editing, visualization, supervision, funding acquisition. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the University of Manitoba’s Health Research Ethics Board, Manitoba Health’s Health Information and Privacy Committee, and CancerCare Manitoba’s Research and Resource Impact Committee. Because data were de-identified, informed consent was not required.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Lambert, P., Pitz, M., Singh, H. et al. Evaluation of algorithms using administrative health and structured electronic medical record data to determine breast and colorectal cancer recurrence in a Canadian province. BMC Cancer 21, 763 (2021). https://doi.org/10.1186/s12885-021-08526-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-021-08526-9