Abstract
I suggest a common population genetic mechanism that explains persistence of biological species as consistently reproducing groups of similar organisms: genetic renewal due to genetic drift or selection, which restricts genetic diversity of populations. In contrast to concepts explaining species integrity via interbreeding, the concept of drift- and selection-induced genetic renewal explains species existence not only for sexually reproducing organisms, but also for asexual, or agamous, organisms. I redefine concepts of population, isolation and species in terms of genetic renewal. The proposed concept of renewing species develops Alan Templeton’s cohesion species concept.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Biological species are distinguishable groups of genetically similar organisms persisting for many generations and distinguishable from other such groups. Species are found in all sorts of organisms, regardless of their reproduction mode and the way they pass genes to next generations: in sexuals, asexual multicellulars (Fontaneto and Barraclough, 2015), and in prokaryotes (Rosselló-Móra and Amann, 2015), many of the later capable of conjugation and other kinds of lateral gene transfer (LGT). But so far, there is no convincing universal concept explaining why species are ubiquitously found in this variety of organisms. This article offers such an explanation.
Existing species concepts focus on many aspects of the species phenomena. As pointed out by Wilkins (Wilkins, 2011) upon studying 27 different species concepts, most of them do not actually aim to explain “universal mechanics” of species existence, reasons for species cohesion, that is, a force that sustains species integrity for many generations. Most concepts focus on other important biological questions. Some help “identify” species, line out their borders, attribute given individuals to the same or different species (e.g. the morphological and taxonomical species). Other concepts explore properties of an evolving species as a “black box”, regardless of the forces that bring such a species into existence (e.g. the evolutionary species).
Few concepts aim their scope at “species mechanics.” All of them explain species cohesion via elementary population-genetic processes. Let’s look at these concepts.
Mayr’s biological species (Mayr, 1942) and Paterson’s recognition species (Paterson, 1985) concepts explain species cohesion via interbreeding. These explanations are therefore applicable only to sexuals, and not to organisms with other kinds of gene transfer: asexuals with strictly vertical gene transfer or irregular LGT. Nonetheless, asexual and sexual species as categories do not consistently differ from each other: “…the asexual world is for the most part just as well (or even better) subdivided into easily defined biological taxa as is the sexual world” (Templeton, 1989). For instance, species of asexual bdelloid rotifers are better distinguished (Fontaneto and Barraclough, 2015) then monogonont rotifers, which do occasionally interbreed (Holman, 1987). This hints at common mechanisms underlying species in sexuals and asexuals.
Ecological species concepts, apart from providing tools for attributing organisms to the same or different species (i.e. identifying species boundaries), also propose a population-genetic species cohesion mechanism: selection in favor of adaptation to a specific ecological niche, which leads to genetic similarity of conspecifics in their habitat. This explanation of species has the advantage of being applicable to both sexuals and asexuals. However, I suggest a simpler explanation of organism similarity in one ecological niche, which does not even involve selection: reproducing in an environment with limited capacity is itself sufficient to provide similarity. My concept is the development of Alan Templeton’s cohesion species concept.
Cohesion species (Templeton, 1989) unites biological and ecological concepts to give a common explanation for species cohesion in sexuals and asexuals. The concept doesn’t propose a single common cohesion mechanism, but it shows similarity of mechanisms that take effect in sexuals and asexuals.
I will outline cohesion concept cornerstones. This is not essential for understanding further explanations, but the cohesion species concept is the basement for renewal species, and I will explain in short the part related to the new concept. You can learn more about cohesion species in Templeton’s original work (Templeton, 1989).
In Templeton’s words, “the cohesion concept species is the most inclusive population of individuals having the potential for phenotypic cohesion through intrinsic cohesion mechanisms,” which are outlined further in that work. The effect of these mechanisms creates a genetic community, a field of action for population-genetic forces: “the cohesion concept of species defines a species as an evolutionary lineage through the mechanisms that limit the populational boundaries for the action of such basic microevolutionary forces as gene flow (in Templeton’s article, this term has a non-conventional meaning of interbreeding, e.g. see table 2 of the article), natural selection, and genetic drift” (Templeton, 1989, pp. 12, 20).
Several intrinsic cohesion mechanisms sustain species (Templeton, 1989, Table 2, the original numbering in the list is preserved). In different conditions, various cohesion mechanisms come into play, but any of them is capable of creating a species:
(I) In sexuals, Templeton outlines the mechanisms of genetic exchangeability, “the factors that define the limits of spread of new genetic variants through gene flow” (sensu interbreeding):
(A) genetic identity via interbreeding;
(B) lack of genetic identity with other groups due to sexual isolation.
(II) For asexual organisms, the cohesion is achieved via mechanisms of demographic exchangeability, “the factors that define the fundamental niche and the limits of spread of new genetic variants through genetic drift and natural selection”:
(A) cohesion via genetic drift (identity via descent from a common ancestor);
(B1) cohesion via selective fixation of a favorable variant;
(B2) disruption of demographic exchangeability due to natural selection (adaptive transitions).
Templeton explains existence of species by the action of these cohesion mechanisms and views speciation as their evolution.
I will show that just one of these mechanisms is sufficient to explain species: cohesion via genetic drift, point IIA in the list above and in (Templeton, 1989, Table 2).
RENEWAL COHESION IN ASEXUALS
Time of the Most Recent Common Ancestor T(MRCA) Constantly Shifts into the Future
This chapter is an overview of known findings of Neutral Theory and Coalescent Theory (Futuyama, 2005; Hedrick, 2009). It does not contain any new scientific information, but consolidates statements essential for further reading.
Let’s look at genetic diversity accumulation in an isolated asexual population with the following properties:
—the effective population size Ne is constant in generations;
—generations do not overlap;
—organisms multiply asexually, transferring a copy of their genome to their descendants (with introducing new mutations);
—the population is isolated, i.e. over time, there is no influx of individuals from other populations;
—the population originates from a single common ancestor and then exists in isolation for a relatively large timespan, \( \gg \)Ne generations.
Suppose then that we have been observing the population throughout its whole history, and for every extant or deceased individual, we know precisely, who is their parent.
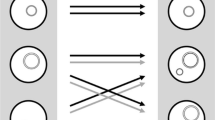
In Fig. 1a there is an example of such a population and size Ne = 4. Individuals of the population and their common ancestors are mapped on a phylogenetic tree. The tree shows the following:
The dynamics of T(MRCA), time of the most recent common ancestor, on the phylogenetic tree: (a) in generation t1, the MRCA of all organisms existed T(MRCA)1 generations ago; (b) before generation t2, lines B and D came extinct. As a result, the time of the new most recent common ancestor, T(MRCA)2, is later than T(MRCA)1 in generation t1.
—the founder of the population lived T(F) generations ago;
—the most recent common ancestor of B and C lived T(B,C) generations ago. In other words, B and C coalesce T(B,C) generations ago;
—similarly, A, B and C coalesce T(A,B,C) generations ago;
—all individuals coalesce at the most recent common ancestor (MRCA) of the entire population, which existed T(MRCA) generations ago.
Any ancestor of the MRCA is a common ancestor for the entire population as well, e.g. the population founder living T(F) generations ago. But they are not the most recent ones.
In each generation, some individuals leave more than one descendant propagating to reproduction, while others fail to produce a single one. This may occur both due to selection favoring certain genetic variants or purely by chance (genetic drift). As a result, from time to time, whole genetic lineages disappear from the population. Figure 1b shows the same population at a later generation t2. In the time span between generations t1 and t2, descendants of B and D came extinct. Their place was taken by descendants of A and C. This does not necessarily mean that A and C are better adapted: elimination could be accidental, i.e. caused solely by genetic drift.
This is how whole phylogenetic tree branches spanning from the MRCA, with all their individuals, occasionally get extinct from the population. The ex‑MRCA is no more the most recent one. Another individual becomes the population’s MRCA, the one marking the first split of phylogenetic tree of the surviving individuals. The new MRCA is always a descendant o the old one, and the new TMRCA is always later than the previous one.
Thus, from time to time, TMRCA shifts into a more recent generation.
Cohesion via Genetic Renewal
According to the Coalescent Theory, the average TMRCA of a population is statistically determined by temporal dynamics of effective population size (Ne). For example, if Ne is constant, TMRCA of a haploid population on average is 2Ne, e.g. see formulas for the average coalescence time in (Templeton, 2005; Hedrick, 2009).
Genetic diversity of the asexual population described above is determined by the number of mutations accumulated in branches of the population’s phylogenetic tree, between the MRCA and current generation’s individuals. But since TMRCA periodically shifts forward in time, genetic differences between individuals in a population do not increase infinitely: the influx of new mutations is balanced by loss of whole branches of the phylogenetic tree. Diversity is fixed around a value determined by Ne and the mutation rate µ (see the formulas for mutational-drift equilibrium, e.g. in (Gillespie, 1998; Futuyama, 2005)). Therefore, such a population will evolve as a group of genetically ever-similar organisms, without accumulating genetic diversity above a certain threshold.
In our model, we assumed that the entire population descends from a single common ancestor. Even if this is not the case and there is more than one ancestor, after a sufficiently long time (\( \gg \)Ne) all individuals of the population are likely to be descendants of a single founder due to accidental loss of descendants of other founders. Right from the moment of this loss, a population evolves as a group of genetically close descendants of one common ancestor.
Cohesion via genetic renewal, or renewal cohesion, is the restriction of genetic diversity of a group of related individuals as a result of their recent origin from a common ancestor (in Templeton, 1989, Table 2, this kind of cohesion is listed under the number IIA). Under certain conditions, the emergence of renewal cohesion is inevitable:
(1) Genotypes of individuals in the group are reproduced by copying without recombination.
(2) The group of individuals is isolated from other such groups.
You may view genetic renewal in two temporal aspects:
—In retrospect, all currently existing genotypes of a population converge to their MRCA. The convergence time has a certain statistical distribution determined by temporal dynamics of Ne and has an average value the order of Ne.
—In perspective, any genotype is a potential common ancestor of the whole group of genotypes in some future generation, provided that the group is not dissected by some impenetrable barrier. For different genotypes, the probability of this may be different due to unequal fitness.
Selection Does Not Prevent Loss of Genetic Diversity
For renewal cohesion to take the effect (i.e., for genetic diversity to be lost due to genetic renewal), genetic drift alone is sufficient, even in the absence of selection. But if selection is in place, can it counteract renewal cohesion? In case of selection favoring certain genotypes, the answer is negative. On the contrary, selection increases the likelihood of rapid loss of less adapted genotypes and may only reduce population’s TMRCA, since the increase of fitness variation in comparison to neutrality decreases Ne (Lee, 1978), on which TMRCA directly depends.
The mode of selection that prevents loss of genetic diversity is balancing selection, e.g. when the least frequent genotype gets a selective advantage. This kind of selection can sustain coexistence of several genotypes in a population for indefinitely long time, provided that Ne is large enough so that accidental loss of one balancing genotype due to genetic drift is unlikely.
However, in a population of asexuals, genotypes sustained by balancing selection start to evolve as separate isolated subpopulations. In each of them, TMRCA keeps shifting forward in time, while the common MRCA of subpopulations remains fixed at the same historical moment. Subpopulations evolve as if they were separated by an impenetrable barrier. As a result, the level of genetic differences accumulates between the divided subpopulations, and genetic renewal no longer maintains genetic similarity between them. In other words, in a population of asexuals, balancing selection does not prevent renewal cohesion, but rather leads to dissection of the population into several subpopulations.
No Difference between Renewal Cohesion Induced by Genetic Drift and Selection
In an asexual population, genetic renewal due to drift does not differ in its effect from genetic renewal due to selection: in both cases, only a part of individuals leaves offspring that will survive to reproduce. By pure chance (drift) or via selection, slower or faster, genotypes are still lost from the population. The consequence for genetic diversity is the same: one and only one of extant genotypes will become an ancestor of the entire population in some future generation.
Renewal Isolation
One of conditions for renewal cohesion is isolation (see condition 2 above). In our explanatory model population above, we introduced isolation as a pre-condition. In natural conditions, this kind of isolation providing the effect of cohesion renewal exists as a variety of degrees.
Renewal isolation between groups of related asexual organisms is their state when none of them can evolutionarily displace the other, or, in other words, these groups are not connected via renewal cohesion. The degree of renewal isolation may vary from complete lack of isolation (an analogue of panmixia in sexuals) to complete renewal isolation, when individuals of one group are not able to leave offspring in the habitat or ecological niche of the other. Complete renewal isolation between two closely related groups means no renewal cohesion between them. The groups evolve independently and accumulate genetic differences between each other. Factors leading to isolation may be:
—Physical barriers that prevent exchange of individuals between populations.
—Ecological barriers, when individuals of isolated populations are adapted to different ecological conditions and occupy different ecological niches. In this case, individuals of one population are unable to dwell in the other’s habitat and cannot leave progeny that would effectively compete with the aboriginal population.
The Renewing Population
As an evolving unit, a population is a group of organisms united via renewal cohesion. Renewal cohesion comes into effect because a population occupies a certain ecological niche. Boundaries of that niche provide population’s isolation from others, and the “space” within these boundaries creates space for action of renewal cohesion.
However, this would not be correct to attribute all organisms occupying the same ecological niche to the same population. Two unrelated groups may temporarily co-inhabit an ecological niche and compete, still this doesn’t make them one population. Therefore, to define a population as an evolving unit, it is important that its individuals are recent descendants of a common ancestor, with their TMRCA of the same order of magnitude as the population’s Ne.
So, a renewing population in asexuals is a group of individuals that: (1) occupy a certain ecological niche and (2) descend from a recent common ancestor.
RENEWAL COHESION IN SEXUALS
We have defined renewal cohesion for asexuals, with their genotypes reproduced by copying, and in which all existing individuals are descendants of one common ancestor. However, the concept of renewal cohesion is not directly applicable to sexuals due to the fact that their genomes recombine. You cannot pick a particular individual as a single common ancestor for all extant organisms of a sexual species, because each individual genome, instead of being consistently reproduced by copying, maintains its integrity only for the time span between two recombination events. So, does renewal cohesion apply to in sexuals at all?
Renewal Cohesion at a Non-Recombining Locus
The concept of renewal cohesion is indeed not applicable to life forms with recombining genomes, but it is suitable for describing evolutionary processes in these genomes’ non-recombining parts. Take mtDNA, which is reproduced by vertical copying, so that renewal cohesion takes its effect in a species’ mtDNA pool. At any moment, you can define the MRCA for all species’ mtDNAs: a specific lineage, which once in the past was a part of some individual genome.Footnote 1
Similarly, we can look at genetic diversity of any non-recombining region of a recombining genome. For example, non-recombining parts of nuclear chromosomes in humans. At any given time, all haplotypes (alleles) of such a non-recombining locus coalesce to their MRCA variant. Alleles in a non-recombining locus form a good renewing pool with properties of a renewing population. Mind that MRCA variants for the today’s allelic diversity of different non-recombining loci existed in different individuals, who lived at different times.
Presenting a Recombining Genome as a Set of Non-Recombining Fragments
You can attribute any region of a recombining genome to a specific renewing pool of haplotypes.
Consider an SNP (single nucleotide polymorphism). It is reproduced via simple copying of its parent version. Alleles of an SNP locus derive from their ancestral allele due to a single nucleotide substitution. And although it is often impossible to figure out which of the extant alleles was the ancestor and whether it is still present in the population, this is definite that the whole existing allele diversity of this SNP had a single ancestral variant. Thus, the pool of alleles of an SNP locus behaves as renewing, and their genetic diversity is limited due to recent origin from their common ancestor. The observed genetic diversity is even more limited in comparison to that of long asexual genomes, due to an additional factor of recurrent mutations: since an SNP locus has only four possible states (four nucleotides), and newly emerging alleles are likely to coincide with already existing ones, we cannot don’t distinguish recurrent mutations.
Now we can view the whole genome as a set of “atomic” non-recombining loci. Some may consist of one or several nucleotides: SNPs, micro-insertions, micro-deletions, tandem repeats, etc. Others span considerably: non-recombining parts of human Y‑chromosome are tens of millions of nucleotide pairs long. Each non-recombining locus forms a renewing pool, with genetic diversity limited by renewal cohesion and statistically determined by population’s demographic parameters. For example, human autosomal loci represented by 2Ne copies (where Ne is the effective size of the human population) generally coalesce after separation of human and chimpanzee lines. Coalescence time for X-linked loci, loci of the non-recombining part of Y-chromosome and mtDNA is on average even more recent due to the smaller effective size of pools of corresponding alleles (Fig. 2).
Distribution of T(MRCA) for 25 non-recombining human loci. The value of T(MRCA) is statistically determined by historical dynamics of the effective size of the allele pool in a locus. For autosomes, this number is 2Ne, for X-linked loci it is about (3/2)Ne, for mtDNA and Y-chromosomes it is about Ne (where Ne is human effective population size). Fig. 3 from (Templeton, 2005) is reproduced.
This lets us conclude that in sexuals, as well as in asexuals, the force that limits genetic diversity is the renewal cohesion, too. Interbreeding does not limit genetic diversity by itself, it only determines boundaries of non-recombining regions in the genome by crossover spots. Those non-recombining regions are separately affected by renewal cohesion, which is exactly the same kind of cohesion, that takes effect in asexual populations.
Epistasis and Recessive-Dominant Gene Interactions
Do gene interactions (recessive-dominant and epistatic) in a recombining genome affect cohesive renewal? Gene interactions can affect fitness variance and, consequently, the process of selection. But as we have seen earlier, selection, if anything, only contributes to accelerating genetic renewal. Thus, gene interactions in recombining genomes do not prevent renewal cohesion from affecting genetic diversity of recombinant loci.
Reproductive Isolation and Renewal Isolation
In a panmictic population of sexual organisms, pools of alleles in atomic loci behave as renewing. When the population deviates from panmixia with a certain level of reproductive isolation, the corresponding level of renewal isolation is introduced in allele pools of atomic loci. Upon reaching the marginal state of complete reproductive isolation, in all atomic loci of the genome, complete renewal isolation is introduced. Thus, reproductive isolation leads to the emergence of renewal isolation, which induces the accumulation of genetic differences between the divided populations.
An atomic locus avoids the effect of renewal cohesion, when there is balancing selection in effect between its alleles, in the same way as we have discussed earlier in asexual genomes. As a result, allele TMRCA in loci with balancing selection may be significantly older than TMRCA of other loci. If alleles of such a locus coalesce at an earlier point in time than the establishment of reproductive isolation of the species from its sister species, allelic variety of these loci in sister species may be similar.
RENEWING SPECIES AND SPECIATION
Speciation Induced by Genetic Renewal
A natural species usually constitutes a connected system of populations, or to say, a divided super-population. A species is often viewed as the most inclusive super-population with its parts unified by some mechanisms of cohesion. As we have seen, genetic renewal is a sufficient mechanism of such cohesion, capable of continuously uniting individuals into populations for many generations. This leads us to an explanation of the species phenomena based on the definition of renewal population given above.
Arenewal species is the most inclusive group of individuals, which are united by renewal cohesion, i.e. (1) are reproduced in a specific ecological niche and (2) are genetically similar due to recent origin of their genomes (as whole uninterrupted DNA stretches or in parts) from genomes of ancestral individuals. Definition of renewal species describes the conditions, in which a group of individuals forms a “species-like” evolving group: a reproducing and identifiable group of genetically similar individuals. Applied to sexuals, the concept of renewal species is reduced to Mayr’s biological concept, with the environment of atomic non-recombining alleles (their “ecological niche”) comprising not just of ecological factors in their common sense, but also of genes in other loci of the species gene pool.
By including an “ecological part”, the renewal species definition resembles ecological species concepts. For example, according to Van Valen ecospecies, “a species is a lineage (or a closely related set of lineages) which occupies an adaptive zone minimally different from that of any other lineage in its range and which evolves separately from all lineages outside its range” (Van Valen, 1976, p. 233). However, ecological species concepts usually assume that conspecifics experience a similar selection process, which forces them to adapt to the same niche. This provides genetic stability of a species, and its organisms remain similar for generations. For the renewal species concept, the only thing important is that the boundaries of the occupied ecological niche define the boundaries for the action of renewal cohesion (i.e., first of all, of genetic drift).
To what degree may ecological niches of different populations of a species be unlike so that they still make a whole species? If an individual from one population has a potential to become an ancestor of all individuals in the habitat of another population (i.e., there is a renewal cohesion between these populations), then the populations belong to the same species.
Speciation may occur due to the emergence of renewal isolation due to physical, environmental or, in case of sexuals, reproductive barriers between groups of individuals.
Renewing Species and Lateral Gene Transfer (LGT)
Populations of asexuals with purely vertical inheritance (such as the model asexual population discussed above) are a rare, perhaps an exceptional phenomenon, since LGT is widespread in natural populations. How does this affect renewal cohesion and speciation?
In terms of genetic lineages, the LGT effect is that certain loci in gene pool have non-typical histories: most loci of the acceptor lineage coalesce in one population, and the newly acquired genes coalesce with genes in another population. In reality, LGT occurs often enough that there can be many groups of alleles in one genome with histories different from the rest of the genepool and coalescing in different vertically unrelated groups. As a result, attribution of a group to super-species level taxa faces conceptual difficulties.
The good news is that at the species level, LGT does not prevent us from identifying a species, because in its effect on renewal cohesion, LGT is conceptually no different from any other kind of mutation. The degree of change in species identity depends on how the acquisition of gene changes its adaptive zone. The effect of LGT, just like the effect of mutations, varies from complete absence of phenotypic manifestation to significant changes in the adaptive zone (e.g. the emergence of resistance to antibiotics or the ability to produce a specific amino acid). If genes received via LGT change boundaries of the occupied ecological niche, the pool of modified individuals acquires a new species identity in the concept of renewal species, in the same way as this happens as a result of regular mutations.
Renewal Species among Other Species Concepts
The renewal species concept is a theoretical generalization offering new explanation of mechanics for species persistence. You cannot apply this concept practically for species identification or definition of species boundaries, just like in case of Mayr’s biological species concept, you cannot directly determine the potential ability of individuals to intercross, unless they factually do. The value of the biological concept is not that it offers empirical tools for identifying conspecifics, but that it offers a logical explanation of why sexual species exist. The value of the renewal species concept in the same, for a broader case of species with any mode of reproduction, sexual and asexual.
CONCLUSIONS
In renewal species concept, the existence of species as stable populations of genetically similar individuals, regardless of their mode of reproduction, is explained by renewal cohesion. In an asexual species, renewal cohesion maintains similarity of individuals due to the origin of their genomes from a common ancestral genome as such. In recombinant genomes of sexual organisms, renewal cohesion acts similarly, but at the level of individual non-recombining loci.
The renewal species concept develops Templeton’s cohesive species concept. The new concept differs by identifying just a single mechanism of cohesion as sufficient: renewal cohesion.
This is possible that a phenomenon similar to renewal species arises in any evolving system of replicators, not necessarily genetic. Examples of such systems are many cultural phenomena: languages and dialects, musical and artistic styles, types of corporate and industrial culture, traditions of raising children, versions of chronicles and military regulations, versions of the machine code, and many others. These phenomena show pronounced discontinuity in their diversity pattern, similar to biodiversity discontinuity between species. In renewal cohesion mechanics, there is nothing incompatible with nature of these phenomena, although grounds for identifying the level of “atomic non-intermixing elements” analogous to atomic non-recombining alleles are not that evident in them. Looking for such an analogy may be a subject of future research.
Notes
For simplicity, in this example with mtDNA, I don’t consider interspecific mtDNA introgression. We will take a look at what happens in case of such introgression and, in general, in case of lateral gene transfer (LGT), in the end of the article.
REFERENCES
Fontaneto, D. and Barraclough, T.G., Do species exist in asexuals? Theory and evidence from bdelloid rotifers, Integr. Comp. Biol., 2015, vol. 55, no. 2, pp. 253–263.
Futuyama, J., Evolution, Sunderland: Sinauer, 2005.
Gillespie, J.H., Population Genetics: A Concise Guide, Baltimore: Johns Hopkins Univ. Press, 1998.
Hedrick, P., Population Genetics and Ecology, Levin, S., Ed., Princeton: Princeton Univ. Press, 2009, pp. 109–116.
Holman, E.W., Recognizability of sexual and asexual species of rotifers, Syst. Zool., 1987, vol. 36, pp. 381–386.
Li, C.C., The First Course in Population Genetics, Pacific Grove, CA: Boxwood, 1976.
Mayr, E., Systematics and the Origin of Species, New York: Columbia Univ. Press, 1942.
Paterson, H.E.H., The recognition concept of species, in Species and Speciation, Vrba, E., Ed., Transvaal Museum Monograph no. 4, Pretoria: Transvaal Mus., 1985, pp. 21–29.
Rosselló-Móra, R. and Amann, R., Past and future species definitions for Bacteria and Archaea, Syst. Appl. Microbiol., 2015, vol. 38, no. 4, pp. 209–216.
Templeton, A., The meaning of species and speciation: a genetic perspective, in Speciation and Its Consequences, Daniel, O. and Endler, J.A., Eds., Sunderland: Sinauer, 1989, pp. 3–27.
Templeton, A., Haplotype trees and modern human origins, Am. J. Phys. Anthropol., 2005, vol. 41, pp. 33–59.
Valen van, L., Ecological species, multispecies, and oaks, Taxon, 1976, vol. 25, pp. 233–239.
Wilkins, J., Philosophically speaking, how many species concepts are there? Zootaxa, 2011, vol. 2765, pp. 58–60.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest. The author declares that he has no conflict of interest.
Statement of the welfare of animals. This article does not contain any studies involving animals or human participants.
Rights and permissions
About this article
Cite this article
Pshenichnov, A. The Renewing Species. A Common Population-Genetic Explanation of Species Phenomena for Sexual and Asexual Organisms. Biol Bull Rev 9, 385–392 (2019). https://doi.org/10.1134/S2079086419050074
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S2079086419050074