Abstract—
Identification of new protein-protein interactions (PPI) and characterization of quantitative parameters of complex formation represent one of central tasks of modern protein interactomics. This study is a logical continuation of the cycle of our previous works aimed at the PPIs between components of the cytochrome P450-dependent monooxygenase system. Using an optical biosensor employing surface plasmon resonance (SPR biosensor), a comparative analysis characterizing kinetic and equilibrium parameters of complex formation between the membrane-bound hemoprotein cytochrome b5 and cytochrome P450s was performed using two different protocols for protein immobilization: (1) covalent non-oriented immobilization on the carboxymethyl dextran chip and (2) non-covalent oriented immobilization in the lipid environment. In the case of the second protocol, PPIs were characterized by 2.5-fold higher affinity due to a decrease in rate dissociation constants values of the studied complexes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Currently, optical biosensors operating on the surface plasmon resonance effect (SPR biosensors) are widely used for analysis of various intermolecular interactions, including protein-protein interactions (PPIs). In the latter case, the most common mode of protein ligand immobilization consists in covalent non-oriented immobilization on an optical chip with a carboxylated dextran surface for the free amino groups of the protein with the formation of an amide bond [1, 2]. Oriented ligand protein immobilization is less common; it includes special tags (biotin, histidine tag) or certain sites of the secondary structure (e.g., a transmembrane domain). The study of PPIs involving membranes or membrane-bound proteins containing one or more transmembrane domains represents a non-trivial task. Its solution often requires modeling the membrane environment (lipid bilayer) with a protein built into it. Membrane proteins were immobilized into model lipid membranes placed onto a solid carrier, for example, studies on an optical chip surface with or without chemical modification [3, 4], studies used Langmuir−Blodgett and Langmuir−Schaefer techniques [5, 6], as well as adsorption and fusion of monolayer liposomes [5–7]. Moreover, many factors can influence the lipid bilayer formation: the surface quality of the chip; composition, pH and ionic strength of the buffer; temperature; lipid composition; size, homogeneity and concentration of liposomes; the presence of divalent ions [7, 8]. SPR studies of interactions of membrane proteins in the lipid environment (including as part of model proteoliposomes) [9–14] have shown the importance of weakly controlled factors, which lead to a significant variation in the results obtained. These factors include the features of liposome suspension preparation, detergents used and purification methods (this influences liposome size heterogeneity); non-oriented insertion of proteins into liposomes; the effect of lipid composition on the stability of proteoliposomes; the need of additional approaches to control protein incorporation into liposomes.
The aim of this SPR-based study was to investigate PPIs involving proteins of the P450-dependent monooxygenase system (cytochrome P450 (CYP3A4 and CYP3A5) and cytochrome b5 (CYB5A)) under conditions of a bilayer lipid membrane. For this purpose we have used different methods of protein immobilization on standard optical chips from GE Healthcare (USA): covalent immobilization on CM chips with a surface coated with carboxymethylated dextran and non-covalent oriented immobilization in a bilayer lipid membrane on an L1 chip. This study employed the main advantage of the SPR biosensor: real-time operation monitoring of all stages of the experiments: liposome binding, liposome fragmentation and formation of a planar lipid bilayer membrane from them, assessment of the uniformity of the lipid bilayer (internal SPR control) and non-covalent oriented immobilization of CYB5A in bilayer possessing a single C-terminal membrane domain.
MATERIALS AND METHODS
Recombinant Proteins
The preparations of CYP3A4, CYP3A5, CYB5A (the SDS-PAGE purity >95%) were obtained by their heterologous expression as hexa-His tagged proteins in E. coli followed by affinity chromatography purification on a Ni-NTA agarose column as described earlier [15, 16]. CYB5A was obtained as a full-sized molecule, and CYP3A4 and CYP3A5 were shortened by their membrane domain [17]. BSA was obtained from AppliChem (Germany), and polyclonal antibodies to BSA (pAb to BSA) from Sigma (USA).
Reagents
The following buffer solutions and reagents for optical biosensor analysis were obtained from GE Healthcare: HBS-N buffer (150 mM NaCl, 10 mM Hepes, pH 7.4), 10 mM acetate buffer (pH 5.0), aqueous solution 0.1 M NHS (N-hydroxy-succinimide) and 0.4 M EDC (1-ethyl-3-(3-dimethyl-aminopropyl) carbodiimide hydrochloride). CHAPS (3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate) and the total soybean lipid fraction were obtained from (Merck, USA). Other reagents of analytical purity were obtained from Russian suppliers.
Liposome Sample Preparation
Phospholipids were isolated from the total soybean lipid fraction by precipitation with cold acetone. The total soybean lipid fraction (5 g) was dissolved in a minimum amount of hexane at room temperature and then, an excess of acetone, cooled in the freezer (–18°C), was added to the solution. After precipitation of the phospholipid fraction, the supernatant was discarded and traces of acetone were removed on a rotary evaporator. The dissolution and precipitation procedure was repeated 3 times. The obtained fraction of soybean phospholipids was dissolved in chloroform and stored at –40°C. For liposomes preparation, an aliquot of the phospholipid solution in chloroform was dried on a rotary evaporator in a round bottom flask. The resultant mass of phospholipids was weighed. The phospholipids were then dispersed in HBS-N buffer at a concentration of 100 mg/mL by shaking with glass beads. Then, the liposome suspension was dispersed on ice on a Sonoplus hd 3110 ultrasonic homogenizer (Bandelin, Germany) for 2 min at 20% of the maximum ultrasonic emission power. Size-homogeneous liposomes were obtained by repeated filtration of the liposome suspension through a polycarbonate filter (pore diameter 100 nm) using the LipoFast-Basic kit (Avestin, Canada).
Dynamic Light Scattering
The size of the obtained liposomes was estimated using the dynamic light scattering method on a ZetaSizer MicroV zeta sizer (Malvern, USA) at 25°C for 1 min with measurement frequency of 333,600/s. Standard 1× phosphate buffer (PBS buffer) was used as the medium. The liposome suspension was diluted 1000 times with PBS buffer. The measurements were carried out in a 2 μL quartz cuvette zmv 1002 (Malvern). Figure 1 shows an example of the evaluation of liposome size populations by means of the MicroV zeta sizer.
Distribution of liposome diameters according to dynamic light scattering: (a) intensity distribution of diameters; (b) volume distribution of diameters; (c) correlation function. Additional measurement data: the maximum peak for calculation of volume distribution: 127 nm, peak width 82 nm; the maximum peak for calculation of intensity distribution: 215 nm, peak width 98 nm; the average diameter for Z-coefficient calculation: 174 nm, PdI 0.173, the shift along the ordinate axis according to the correlogram was 0.88.
Surface Plasmon Resonance (SPR)
PPIs registration was performed using a Biacore X100 optical biosensor (GE Healthcare). For the analysis, standard optical chips CM5 and L1 types (GE Healthcare) were used. All procedures for immobilization of cytochrome b5 (CYB5A) on a CM5 chip and registration of its complex formation with cytochromes P450 were performed according to the procedures described previously [17, 18], using a biosensor channel without an immobilized protein as a control channel.
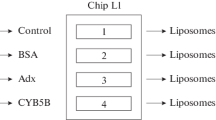
Figure 2 shows the general scheme of the experimental design. One of the biosensor channels was used as a control (without incorporation of CYB5A into the lipid bilayer). The L1 chip was washed with 50 mM NaOH for 30 s at a flow rate of 30 μL/min, and then with 2 M NaCl containing 0.5% CHAPS (w/w). After that a liposome suspension (2 mg phospholipids/mL) was injected into the biosensor channel for 18 min at a flow rate of 5 μL/min, followed by injection of 50 mM NaOH for 30 s at a flow rate of 30 μL/min. A test for non-specific binding of BSA (0.5 mg/mL in HBS-N buffer for 10 min at a flow rate of 10 μL/min) and injection of anti-BSA pAb (Sigma) at a concentration of 10 μg/mL under the same conditions in HBS-N buffer were used as the internal SPR control of the uniformity of liposome distribution of the L1 chip surface, as well as the assessment of the presence of areas uncovered by liposomes. CYB5A immobilization was performed by injecting 12.56 μM CYB5A in HBS-N buffer for 5 min at a flow rate of 10 μL/min. For the PPI analysis, samples of the CYP3A4 analytes (at concentrations of 0.5 μM; 0.75 μM; 1 μM) and CYP3A5 (at concentrations of 0.1 μM; 0.25 μM; 0.5 μM) were sequentially injected through the biosensor control channel and a working channel with a CYB5A incorporated in the lipid bilayer for 10 min at a flow rate of 5 μL/min. Dissociation of protein complexes was recorded for at least 15 min after the end of analyte injection. The removal of liposomes and proteins from the surface of the L1 chip after each biosensor cycle was performed by 4-fold injection of 2 M NaCl solution containing 0.5% CHAPS (w/w) for 30 s at a flow rate of 30 μL/min and then a single injection of 50 mM NaOH solution under the same conditions.
RESULTS AND DISCUSSION
A comparative SPR analysis of the interaction of the CYB5A with cytochromes P450 was performed using two different methods of CYB5A immobilization: (1) covalent immobilization (in an aqueous environment); (2) incorporation into a bilayer lipid membrane formed on the optical chip surface. Cytochromes P450 truncated by their membrane domains were used as analyte proteins. The first method employed the previously used covalent non-oriented immobilization of membrane proteins CYB5A, CYB5B, and CPR on the surface of a CM type chip coated with carboxylated dextran [17, 19, 20]. Such simple immobilization method is preferable for screening studies and for determining the temperature dependence of the PPIs for calculating their thermodynamic parameters (∆G, ∆H, –T∆S) [17, 20]. In the second method, non-covalent oriented protein immobilization was achieved by incorporation of the CYB5A membrane domain into a planar lipid bilayer; this approach basically mimics real conditions for PPIs involving membrane proteins. However, this approach is much more complex, labor-, resource- and time-consuming.
Figure 2 shows scheme of experimental design and the relationship of the approaches used in this work. A new essential element, which differs this scheme from traditional experimental schemes with proteoliposomes [9–14] and lipid-specific optical chips [21–23], is real-time monitoring of all processes of formation of a bilayer lipid membrane on a special L1 chip, including control liposomes binding on a chip surface and homogeneity of the formed lipid bilayer (BSA—anti-BSA pAb system). In the case of a presence of defective areas in the structure of the lipid bilayer, nonspecific adsorption of BSA occurs on the open hydrophobic surface of the L1 chip in these places and this is recorded as a significant increase in the biosensor signal. Figure 3 shows a typical sensogram of CYB5A immobilization. One can see reasonable homogeneous distribution of liposomes and the lipid bilayer formation on the surface of the L1 chip, as evidenced by the absence of a noticeable increase in the biosensor signal during BSA injection. It should be noted that subsequent injection of anti-BSA pAb is necessary to eliminate a false negative result (the lack of SPR evidence for BSA binding on a chip with a lipid bilayer due to possible BSA incorporation into the membrane during a partial substitution of lipids). Thus, the low values of BSA binding signals, as well as the lack of biosensor signal amplification after injection of anti-BSA pAb confirm the absence of hydrophobic regions of the L1 chip surface uncovered by the lipid bilayer. According to Table 1, the variation of the biosensor signals during repeated injections of liposome and CYB5A preparations was quite low (1−4%). This suggests good reproducibility of the planar bilayer formation procedure and non-covalent oriented immobilization of the CYB5A membrane protein.
Figures 4 and 5 show typical sensograms of complex formation CYB5A/CYP3A4 and CYB5A/CYP3A5 in the bilayer lipid membrane formed on an L1 chip. The experimentally obtained Kd values for the complexes CYB5A/CYP3A4 and CYB5A/CYP3A5 (Table 2) using the standard amino coupling protocol for covalent immobilization of CYB5A on the CM5 chip are within the range of previously published values for these protein pairs [18, 24]. Table 2 shows that Kd values for the complexes CYB5A/CYP3A4 and CYB5A/CYP3A5 in the case of non-covalent oriented immobilization of CYB5A in the lipid bilayer were 2.5 times lower than the Kd values obtained using the CM5 chip. The increase in the PPI affinity is mainly determined by the higher stability of the complexes (decrease in koff values, Table 2). This may be a consequence of the following factors: (1) chemical modification of a protein during covalent immobilization on carboxylated dextran; (2) non-oriented immobilization for amino groups on the surface of a protein can limit the mobility of interacting proteins. Therefore, СYB5A molecules in the lipid bilayer membrane can have more degrees of freedom for conformational changes during their interactions with cytochrome P450, which, in general, can affect stability of СYB5A/CYPs complexes. This is consistent with results of the evaluation of the PPI stoichiometry. On the CM5 chip, in fact, only one of seven immobilized CYB5A molecules is available for binding to cytochromes P450, while in the case of the L1 chip with a lipid bilayer this number increases to 3−4 of 10. Therefore, in the case of non-covalent oriented immobilization of CYB5A in the bilayer lipid membrane formed on the L1 chip, the total contribution of different factors favored the formation of tighter CYB5A complexes with CYP3A4 and 3A5.
CONCLUSIONS
The comparative SPR experiments have shown that using non-covalent oriented immobilization of cytochrome b5 by its incorporating into the bilayer lipid membrane formed on the surface of the L1 chip it is possible to obtain better data on its PPIs. The affinity of the interaction of cytochrome b5, immobilized in the membrane environment, with cytochromes P450 3A4 and 3A5 was 2.5 times higher as compared with similar interactions in the aqueous environment. However, such a system cannot yet be used for thermodynamic studies of PPIs involving membrane proteins. Therefore, it is too early to abandon the traditionally used scheme of covalent immobilization of a protein ligand for free amino groups on chips coated with carboxylated dextran, which lacks temperature-dependent side effects [17, 19, 20].
REFERENCES
Nguyen, H.H., Park, J., Kang, S., and Kim, M., Sensors (Basel), 2015, vol. 15, pp. 10 481–10 510. https://doi.org/10.3390/s150510481
Singh, P., Sensors Actuators B: Chemical, 2016, vol. 229, pp. 110–130. https://doi.org/10.1016/j.snb.2016.01.118
Galdiero, S., Falanga, A., Cantisani, M., Vitiello, M., Morelli, G., and Galdiero, M., Int. J. Mol. Sci., 2013, vol. 14, pp. 18 758–18 789. https://doi.org/10.3390/ijms140918758
Maynard, J.A., Lindquist, N.C., Sutherland, J.N., Lesuffleur, A., Warrington, A.E., Rodriguez, M., and Oh, S.-H., Biotechnol. J., 2009, vol. 4, pp. 1542–1558. https://doi.org/10.1002/biot.200900195
Hardy, G.J., Nayak, R., and Zauscher, S., Curr. Opin. Colloid Interface Sci., 2013, vol. 18, pp. 448–458. https://doi.org/10.1016/j.cocis.2013.06.004
Merz, C., Knoll, W., Textor, M., and Reimhult, E., Biointerphases, 2008, vol. 3, FA41. https://doi.org/10.1116/1.2896119
Seantier, B. and Kasemo, B., Langmuir, 2009, vol. 25, pp. 5767–5772. https://doi.org/10.1021/la804172f
Mashaghi, A., Swann, M., Popplewell, J., Textor, M., and Reimhult, E., Anal. Chem., 2008, vol. 80, pp. 3666–3676. https://doi.org/10.1021/ac800027s
Bécsi, B., Kiss, A., and Erdődi, F., Chem. Phys. Lipids., 2014, vol. 183, pp. 68–76. https://doi.org/10.1016/j.chemphyslip.2014.05.009
Lee, T.-H., Hirst, D.J., and Aguilar, M.-I., Biochim. Biophys. Acta, 2015, vol. 1848, pp. 1868–1885. https://doi.org/10.1016/j.bbamem.2015.05.012
Olguín, Y., Villalobos, P., Carrascosa, L.G., Young, M., Valdez, E., Lechuga, L., and Galindo, R., Anal. Bioanal. Chem., 2013, vol. 405, pp. 1267–1281. https://doi.org/10.1007/s00216-012-6523-4
Patching, S.G., Biochim. Biophys. Acta, 2014, vol. 1838, pp. 43–55. https://doi.org/10.1016/j.bbamem.2013.04.028
Scalise, M., Pochini, L., Giangregorio, N., Tonaz-zi, A., and Indiveri, C., Pharmaceutics, 2013, vol. 5, pp. 472–497. https://doi.org/10.3390/pharmaceutics5030472
Seelheim, P. and Galla, H.-J., Biochem. Biophys. Res. Commun., 2013, vol. 431, pp. 519–523. https://doi.org/10.1016/j.bbrc.2013.01.026
Gilep, A.A., Guryev, O.L., Usanov, S.A., and Estabrook, R.W., J. Inorg. Biochem., 2001, vol. 87, pp. 237–244. https://doi.org/10.1016/S0162-0134(01)00333-6
Sergeev, G.V., Gilep, A.A., and Usanov, S.A., Biochemistry (Moscow), 2014, vol. 79, pp. 406–416. https://doi.org/10.1134/S0006297914050046
Yablokov, E., Florinskaya, A., Medvedev, A., Sergeev, G., Strushkevich, N., Luschik, A., Shkel, T., Haidukevich, I., Gilep, A., Usanov, S., and Ivanov, A., Arch. Biochem. Biophys., 2017, vol. 619, pp. 10–15. https://doi.org/10.1016/j.abb.2017.02.006
Gnedenko, O.V., Yablokov, E.O., Usanov, S.A., Mukha, D.V., Sergeev, G.V., Bulko, T.V., Kuzikov, A.V., Moskaleva, N.E., Shumyantseva, V.V., Ivanov, A.S., and Archakov, A.I., Chemical Physics Letters, 2014, vol. 593, pp. 40–44. https://doi.org/10.1016/j.cplett.2013.12.041
Ershov, P.V., Yablokov, E.O., Florinskaya, A.V., Mezentsev, Y.V., Kaluzhskiy, L.A., Tumilovich, A.M., Gilep, A.A., Usanov, S.A., and Ivanov, A.S., J. Steroid Biochem. Mol. Biol., 2019, vol. 187, pp. 124–129. https://doi.org/10.1016/j.jsbmb.2018.11.009
Yablokov, E.O., Sushko, T.A., Ershov, P.V., Florinskaya, A.V., Gnedenko, O.V., Shkel, T.V., Grabovec, I.P., Strushkevich, N.V., Kaluzhskiy, L.A., Usanov, S.A., Gilep, A.A., and Ivanov, A.S., Biochimie, 2019, vol. 162, pp. 156–166. https://doi.org/10.1016/j.biochi.2019.04.020
Kamisaka, Y., Goto, R., Shibakami, M., Yoshioka, K., and Sato, Y., Biosci. Biotechnol. Biochem., 2011, vol. 75, pp. 1135–1139. https://doi.org/10.1271/bbb.110034
Smith, E., Vekaria, R., Brown, K.A., and Longstaff, C., Mol. Cell. Biochem., 2013, vol. 382, pp. 193–201. https://doi.org/10.1007/s11010-013-1735-2
Wikström, A. and Deinum, J., Anal. Biochem., 2007, vol. 362, pp. 98-107. https://doi.org/10.1016/j.ab.2006.12.009
Ivanov, A.S., Medvedev, A., Ershov, P., Molnar, A., Mezentsev, Y., Yablokov, E., Kaluzhsky, L., Gnedenko, O., Buneeva, O., Haidukevich, I., Sergeev, G., Lushchyk, A., Yantsevich, A., Medvedeva, M., Kozin, S., Popov, I., Novikova, S., Zgoda, V., Gilep, A., Usanov, S., Lisitsa, A., and Archakov, A., Proteomics, 2014, vol. 14, pp. 2261–2274. https://doi.org/10.1002/pmic.201400117
Funding
The work on the SPR analysis of protein-protein interactions was supported by the Russian Foundation for Basic Research (project no. 19-04-00485 “The SPR analysis of intermolecular interactions involving membrane proteins incorporated in a lipid bilayer membrane”).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
COMPLIANCE WITH ETHICAL STANDARDS
This article does not contain any research involving humans or using animals as experimental objects.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
Additional information
Translated by A. Medvedev
Abbreviations used: BSA—bovine serum albumin; CYB5A—microsomal cytochrome b5; СYP—cytochrome P450; kon—the association rate constant; koff—the dissociation rate constant; Kd—the dissociation constant; pAb—polyclonal antibodies; PPI—protein-protein interactions; RU—resonance units, 1 RU corresponds to the binding of 1 pg of material per 1 mm2 of the chip surface; SDS-PAGE—polyacrylamide gel electrophoresis in the presence of sodium dodecyl sulfate, SPR—surface plasmon resonance.
Rights and permissions
About this article
Cite this article
Kaluzhskiy, L.A., Ershov, P.V., Kurpedinov, K.S. et al. SPR Analysis of Protein-Protein Interactions Involving Cytochromes P450 and Cytochrome b5 Integrated into Lipid Membrane. Biochem. Moscow Suppl. Ser. B 14, 168–173 (2020). https://doi.org/10.1134/S1990750820020067
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1990750820020067