Abstract
Extracellular deposition of amyloid-β as neuritic plaques and intracellular accumulation of hyperphosphorylated, aggregated tau as neurofibrillary tangles are two of the characteristic hallmarks of Alzheimer’s disease1,2. The regional progression of brain atrophy in Alzheimer’s disease highly correlates with tau accumulation but not amyloid deposition3,4,5, and the mechanisms of tau-mediated neurodegeneration remain elusive. Innate immune responses represent a common pathway for the initiation and progression of some neurodegenerative diseases. So far, little is known about the extent or role of the adaptive immune response and its interaction with the innate immune response in the presence of amyloid-β or tau pathology6. Here we systematically compared the immunological milieux in the brain of mice with amyloid deposition or tau aggregation and neurodegeneration. We found that mice with tauopathy but not those with amyloid deposition developed a unique innate and adaptive immune response and that depletion of microglia or T cells blocked tau-mediated neurodegeneration. Numbers of T cells, especially those of cytotoxic T cells, were markedly increased in areas with tau pathology in mice with tauopathy and in the Alzheimer’s disease brain. T cell numbers correlated with the extent of neuronal loss, and the cells dynamically transformed their cellular characteristics from activated to exhausted states along with unique TCR clonal expansion. Inhibition of interferon-γ and PDCD1 signalling both significantly ameliorated brain atrophy. Our results thus reveal a tauopathy- and neurodegeneration-related immune hub involving activated microglia and T cell responses, which could serve as therapeutic targets for preventing neurodegeneration in Alzheimer’s disease and primary tauopathies.
Similar content being viewed by others
Main
To explore the disease microenvironment in the presence of amyloid-β or tau deposition, we systematically compared the immunological milieux in the brains of the amyloid-β-depositing mice APP/PS1-21 (A/PE4) and 5xFAD (5xE4)7,8,9,10, and tauopathy (TE4) mice11 that express human APOE4 (E4). The pathologies in these models mirror amyloid deposition and tau aggregation with neurodegeneration, respectively12. We observed significant brain regional atrophy by 9.5 months but not at 6 months of age in TE4 mice (Fig. 1a). In addition, brain atrophy was not present in A/PE4 or 5xE4 mice by 9.5 months of age despite high levels of amyloid-β deposition in the brain (Fig. 1a and Extended Data Fig. 1a). The atrophy in the TE4 mice at 9.5 months primarily occurred in regions that developed the most tauopathy (that is, the hippocampus, piriform–entorhinal cortex and amygdala) and was accompanied by significant lateral ventricular enlargement (Fig. 1a–d and Extended Data Fig. 1b–d). The thickness of the granule cell layer in the dentate gyrus as assessed by NeuN staining was noticeably decreased in TE4 mice, and the thickness correlated highly with hippocampal volume (Extended Data Fig. 1e–g). Consistent with the neuronal loss, positive staining for myelin basic protein, which is present around intact axons, was altered in TE4 mice at 9.5 months (Extended Data Fig. 1h,i). Both TE4 and TE3 (expressing human APOE3) mice developed prominent brain atrophy with greater atrophy in the TE4 mice (Extended Data Fig. 1j–l). Additionally, male mice tended to have higher levels of brain atrophy than that of females (Extended Data Fig. 1m–o). For further exploration of mechanisms of brain atrophy and neurodegeneration, we focused on male mice for the remainder of the experiments.
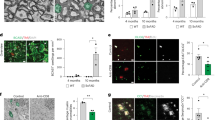
a, Representative images of 6-month-old E4 and TE4, and 9.5-month-old E4, TE4, A/PE4 and 5xE4 mouse brain sections stained with Sudan black. Scale bar, 1 mm. b–d, Volumes of hippocampus (b), piriform–entorhinal cortex (piri–ent ctx) (c) and posterior lateral ventricle (d) in 6-month-old E4 and TE4, and 9.5-month-old E4, TE4, A/PE4, 5xE4 and WT mice (6-month E4: n = 3; 6-month TE4: n = 7; 9.5-month E4: n = 15; 9.5-month TE4: n = 13; 9.5-month A/PE4: n = 7; 9.5-month 5xE4: n = 6; 9.5-month WT: n = 6). Data are mean ± s.e.m.; ***P < 0.0001 for 9.5-month TE4 versus A/PE4; TE4 versus 5xE4; TE4 versus E4; and TE4 versus WT (one-way analysis of variance (ANOVA) with Tukey’s post hoc test). e, Fluorescence-activated cell sorting of CD45total and/or CD45hi cells from brain parenchyma and meninges from E4, A/PE4 and TE4 mice for immune scRNA-seq. f, CD45total immune cells from brain parenchyma assigned into 12 cell types as visualized by uniform manifold approximation and projection (UMAP) plots. DCs, dendritic cells; ILCs, innate lymphocyte cells. g, Bar plot showing the proportions of the 12 cell types of immune cells in the brain parenchyma. Data are mean ± s.e.m.; two biologically independent samples were used, and samples were sequenced in n = 2 batches from the E4 and TE4 groups. Prolif., proliferating. h, CD45total immune cells from meninges assigned into 12 cell types as visualized by UMAP plots. i, Bar plot showing the proportions of the 12 cell types of immune cell in the meninges. Data are mean ± s.e.m.; two biologically independent samples were used, and samples were sequenced in n = 2 batches from the E4 and TE4 groups.
Dysregulated innate and adaptive immune responses contribute to some neurodegenerative diseases13,14. Neuroinflammation is present in the brain of individuals with Alzheimer’s disease, and many studies focus on the cellular and molecular changes and the role of microglia, a key component of the innate immune response in the brain during the development and progression of Alzheimer’s disease15. Microglia are brain-resident cells, which may lead to a pro- or anti-inflammatory milieu within the brain together with monocytes, monocyte-derived macrophages and dendritic cells16,17,18. T cells and natural killer (NK) cells, if present, are more directly linked with cytotoxicity, and could potentially contribute to neuronal loss in a pro-inflammatory environment19,20,21,22. Recent studies found an increase of T cells in the cerebrospinal fluid, leptomeninges and hippocampus in patients with AD23,24 and in mouse models25,26.
Brain and immune cells continuously surveil the environment and make on-demand adjustment to maintain their homoeostasis6,27. State and functional mapping of these cell types in single-cell resolution provide a foundation for understanding the brain in health and disease28. To fully map the innate and adaptive immune responses in the presence of amyloid-β or tau pathology, we generated a cellular and molecular atlas of the meningeal and parenchymal immune cell niche through immune single-cell RNA sequencing (scRNA-seq) on sorted total CD45+ cells (CD45total) from meninges and CD45total and CD45-high cells (CD45hi) cells from the brain parenchyma in APOE4-knock-in (E4), A/PE4 and TE4 male mice at 9.5 months with matched genetic backgrounds (Fig. 1e, Extended Data Fig. 2a and Supplementary Table 1). Unsupervised clustering identified 12 robust cell types of CD45total in the parenchyma of E4, A/PE4 and TE4 mice—microglia, T cells, neutrophils, proliferating cells, B cells, dendritic cells, NK cells, macrophages, γδ T cells, innate lymphocyte cells, mast cells and FN1+ macrophages (Fig. 1f). Unexpectedly, the percentage of the immune cells represented by T cells was strongly increased in TE4 mice as compared with A/PE4 and E4 mice (Fig. 1g and Supplementary Table 3). In fluorescence-activated cell sorting analysis, the proportion of CD45hi cells, which mainly represents the adaptive immune cell populations and innate immune cells such as dendritic cells and macrophages, was enriched in the brain parenchyma of 9.5-month-old TE4 mice (Fig. 1e and Extended Data Fig. 2a–d). Consistent with scRNA-seq data, a significant increase in CD4+ and CD8+ T cells was observed in TE4 versus E4 mice, and CD8+ cells were the more abundant population (Extended Data Fig. 2b). The meninges are a triple-layer structure enveloping the brain and are an immune blood–brain interface29. During ageing and neurodegenerative diseases, dysfunctional lymphatic vessels lead to impaired drainage, which seems to result in dysregulated immune cell trafficking30,31. Distinct cell types were observed in CD45total populations, and the diversity and relative abundance were consistent with observations in previous studies32 (Fig. 1h,i and Extended Data Fig. 2e). In addition, the peripheral T cell composition as assessed in the spleen was not significantly changed in TE4 mice as compared to E4 and A/PE4 mice (Extended Data Fig. 3a,b). Together, these results reveal comprehensive and distinct innate and adaptive immune niches present in the parenchyma and an increased proportion of T cells in the presence of tauopathy and neurodegeneration.
Increased T cells with tau pathology
To further investigate the apparent expansion of the T cell population observed in our scRNA-seq data, we carried out immunohistochemical analyses of the parenchyma from TE4, A/PE4 and 5xE4 mice using antibodies to cluster of differentiation 3 (CD3) and ionized calcium-binding adaptor molecule 1 (IBA1, also known as AIF1), pan markers for T cells and microglia, respectively. We found that T cell numbers were significantly elevated in 9.5-month-old TE4 mice, but not in 9.5-month-old E4 controls or in 6-month-old TE4 mice (Fig. 2a,b and Supplementary Video 1). Increased T cell numbers were also found in TE3 mice (Extended Data Fig. 3c) and tau mice expressing mouse APOE, suggesting a link between T cells and tau-mediated neurodegeneration rather than it requiring a specific APOE isoform. Notably, T cell numbers were not obviously increased in amyloid-depositing A/PE4 and 5xE4 mice at 9.5 months of age or even at 19 months of age compared with those for TE4 mice (Fig. 2a–c). Of note, CD3 staining was primarily present in the hippocampus and piriform–entorhinal cortex, which are regions with accumulation of hyperphosphorylated tau and neuronal loss, indicating a possible detrimental role for T cells in tau-dependent neurodegeneration (Extended Data Fig. 3d). In accordance with the increase in the number of infiltrated T cells, microglia numbers were also significantly elevated in 9.5-month-old TE4 mice in regions with brain atrophy (Fig. 2a,d). The number of T cells showed a positive correlation with the number of microglia (Fig. 2e) and negatively correlated with the granule cell layer thickness in the dentate gyrus (Fig. 2f). To assess whether the T cells were localized in the brain parenchyma as opposed to within the vasculature, we co-stained brain vessels by retro-orbital injection of lectin–dye and for CD3, and noted that CD3+ cells were not present in the lumen of blood vessels (Fig. 2c). Furthermore, transmission electron microscopy also revealed that T cells were in the parenchyma adjacent to other cells in the brain (Extended Data Fig. 3e). To determine whether a similar tau-pathology-associated increase of T cell numbers is present in the parenchyma in human Alzheimer’s disease, we carried out immunohistochemical analyses in brain samples of patients with Alzheimer’s disease (superior frontal gyrus) of low (I–II) and high (VI) Braak stages (Fig. 2g and Supplementary Table 2). In line with the amount of phosphorylated tau (p-tau) pathology, CD3+ T cell numbers were strongly elevated in the superior frontal gyrus from Braak stage VI versus Braak stage I or II cases (Fig. 2g–i). By contrast, in these samples, overall amyloid-β deposition was similar in brain tissues of both low and high Braak stages (Fig. 2g,j). Together, these data demonstrate that increased parenchymal T cell numbers are present in brain regions with tauopathy but not in those with amyloid deposition alone in both humans and mice.
a, IBA1 and CD3 staining in 6-month-old E4 and TE4, and 9.5-month-old E4, TE4, A/PE4 and 5xE4 mice in the dentate gyrus. Scale bar, 20 μm. b, Quantification of numbers of CD3+ T cells in the dentate gyrus (DG) per 0.3 mm2 (6-month E4: n = 3; 6-month TE4: n = 7; 9.5-month E4: n = 15; 9.5-month TE4: n = 13; 9.5-month A/PE4: n = 7; 9.5-month 5xE4: n = 6). ***P < 0.0001 for 9.5-month TE4 versus A/PE4; TE4 versus 5xE4; and TE4 versus E4 (one-way ANOVA with Tukey’s post hoc test). c, Vessel and CD3 staining in 9.5-month-old TE4, and 19-month-old A/PE4 and 5xE4 mice, Scale bar, 20 μm. d, Quantification of the area covered by IBA1 in the dentate gyrus (6-month E4: n = 3; 6-month TE4: n = 7; 9.5-month E4: n = 15; 9.5-month TE4: n = 13; 9.5-month E3: n = 5; 9.5-month TE3: n = 6). ***P < 0.0001 for 9.5-month TE4 versus 9.5-month E4; *P = 0.0276 for 9.5-month TE3 versus 9.5-month E3 (one-way ANOVA with Tukey’s post hoc test). e, Correlation between the area covered by IBA1 and number of CD3+ T cells. n = 49 biologically independent animals from d. Pearson correlation analysis (two-sided); R2 = 0.902, P < 0.0001. f, Correlation between the number of CD3+ T cells with the granule cell layer thickness in the dentate gyrus. n = 49 biologically independent animals from d. Pearson correlation analysis (two-sided); R2 = 0.7454, P < 0.0001. g, CD3, amyloid-β (Aβ) and AT8 staining in brain sections from patients with Alzheimer’s disease (superior frontal gyrus) of low Braak stage I or II and high Braak stage VI. Scale bar, 10 μm. h, Quantification of the area covered by AT8 in g (Braak I or II: n = 3; Braak VI: n = 7). n refers to staining quantification from brain of individual humans. *P = 0.0195 (unpaired two-tailed Student’s t-test). i, Quantification of numbers of CD3+ T cells per square millimetre in g. *P = 0.0277 (unpaired two-tailed Student’s t-test). j, Quantification of the area covered by amyloid-β in g (Braak I or II: n = 3; Braak VI: n = 7). n refers to the number of individual human brains for which staining was quantified. NS (not significant), P = 0.1522 (unpaired two-tailed Student’s t-test). Data are mean ± s.e.m.
T cells shift states with tau pathology
To depict the cellular and molecular signatures of the T cells in the presence of amyloid-β or tau pathology, we assessed T cell populations from immune scRNA-seq data from CD45hi cells in the parenchyma and CD45total cells in the meninges in E4, A/PE4 and TE4 mice. T cells were categorized into 15 subgroups across all samples on the basis of expression of featured genes (Fig. 3a, Extended Data Fig. 4a and Supplementary Table 3). Cell population analysis revealed population differences between parenchyma and meninges. Naive CD8+ T cells (subgroup 11), FOLR4+CD4+ T cells (subgroup 4) and regulatory T (Treg) cells (subgroup 13) were highly enriched in meninges, but effector CD8+ T cells (subgroups 3, 8 and 10) were preferentially enriched in brain parenchyma (Fig. 3b). These results suggest that brain-border and brain-resident T cells are functionally different in accordance with their immune niche. Interaction between the T cell receptor (TCR) and antigens presented by the major histocompatibility complex (MHC) is critical to adaptive immunity33. T cells clonally expand when they recognize cognate antigen34. We next carried out single-cell TCR sequencing (scTCR-seq) on T cells, which showed unique T cell clonal enrichment in the parenchyma with tauopathy and neurodegeneration (Extended Data Fig. 4b–d). We evaluated TCR repertoires among CD4+ T cell subsets and observed an increased clonality in CD4+ T cells in TE4 mice that was concentrated within the activated CD4+ T cells (NKG7+CCL5+ and CXCR6+CCR8+CD4+ T cells; Fig. 3c–e). Similar to what we found in CD4+ T cells, the results of paired TCRα–TCRβ repertoire analysis revealed TCR clonal expansion in CD8+ T cells in TE4 mice (Fig. 3f). Unsupervised clustering identified ten robust cell types in CD8+ T cells (Fig. 3g, Extended Data Fig. 4a and Supplementary Table 3). Activated CD8+ T cells (CD11c+KLRE1+ and ISG15+CD8+ T cells) were more abundant in TE4 mice, whereas the fraction of TOX+PDCD1+CD8+ exhausted T cells was slightly decreased, suggesting a potential role for activated CD8+ T cells in mediating neuronal loss in tauopathy (Fig. 3h). Pseudotime analysis of CD8+ T cells found a range of T cell states indicative of a dynamic shift from activated to exhausted states (Fig. 3i). We also observed an increased clonality in activated and exhausted CD8+ T cells in TE4 mice (Fig. 3j). Together, these data illustrate that T cells in the brain parenchyma dynamically shift from activated to exhausted states with unique TCR clonal expansion in both CD4+ and CD8+ populations in the brain in a mouse model of tauopathy.
a, Total T cells from brain parenchyma and meninges assigned into 15 categories as visualized by UMAP plots. b, Bar plot showing the percentages of T cells subgroups in E4, A/PE4 and TE4 mice. Data are mean ± s.e.m.; two biologically independent samples were used, and samples were sequenced in n = 2 batches from the E4 and TE4 groups. c, Scatter plot illustrating differential TRAV and TRBV pairing expression in CD4+ T cells in TE4 versus E4 (x axis) and A/PE4 versus E4 (y axis) mice. Avg, average. FC, fold change. d, CD4+ T cells from brain parenchyma assigned into six cell types as visualized by a UMAP plot. e, Representative TRAV–TRBV (TRAV6N-6–TRBV13-1) pairing projection in CD4+ T cells in E4, A/PE4 and TE4 mice. f, Scatter plot illustrating differential TRAV and TRBV pairing expression in CD8+ T cells in TE4 versus E4 (x axis) and A/PE4 versus E4 (y axis) mice. g, CD8+ T cells from brain parenchyma assigned into ten cell types as visualized by UMAP plot. h, Percentages of activated (cell types 3 and 6) and exhausted (cell type 1) CD8+ T cells in E4, A/PE4 and TE4 mice. Data are mean ± s.e.m.; two biologically independent samples were used and samples were sequenced in n = 2 batches from the E4 and TE4 groups. i, Trajectory analysis showing naive CD8+ T cells demarcated into three paths: cell type 6, ITGAX+KLRE1+; cell type 3, ISG15+; and cell type 1, TOX+PDCD1+. j, Representative TRAV–TRBV (TRAV16–TRBV13-1) pairing projection in CD8+ T cells in E4, A/PE4 and TE4 mice.
Interaction of microglia and T cells
We next explored the unique but complex immune hubs in the parenchyma of tauopathy brains, which lead to T cell homing and activation. Notably, CCL3, CCL4 and CXCL10, chemokines previously reported to be associated with T cell chemotaxis and brain infiltration, were increased in the brain lysates of TE4 mice compared to those of E4 and TEKO (tau(P301S) and Apoe-KO) mice25,35 (Extended Data Fig. 5). Microglia are responders to neuroinflammation or damage, and they rapidly adapt their phenotypes and functions in response to the dynamic brain milieu36. Typical functions of microglia such as phagocytosis and cytokine production have been well characterized in models of neurodegeneration including AD37,38; however, whether they exert their effects through their interactions with T cells is largely unknown. We subgrouped microglia (cell type 0, Fig. 1f) from the CD45total population of E4, A/PE4 and TE4 mice and obtained three subgroups with distinguishing markers associated with homoeostatic microglia (HOM), disease-associated microglia (DAM) and interferon (IFN)-activated microglia (Fig. 4a,b). Notably, the percentages of the DAM and IFN subgroups were strongly elevated in TE4 mice, whereas the percentage of the HOM subgroup decreased (Fig. 4c and Supplementary Table 3). We found that genes related to antigen presentation, complement response and cytokines, metabolism and oxidative stress, together with lysosomal enzymes, were upregulated in TE4 mice to a greater extent than in A/PE4 mice, for which the expression levels were higher than those in the control mice (Fig. 4d). Classically, MHC class I and II proteins enable antigen presentation to CD8+ T cells and CD4+ T cells, respectively. MHC class I proteins are expressed by all nucleated cells, whereas MHC class II proteins are expressed only by antigen-presenting cells (APCs), such as dendritic cells, macrophages, B cells and microglia39. By co-staining for the perivascular macrophage marker MRC1 mannose receptor C (CD206) in addition to MHC class II proteins, IBA1 and GFAP, we found that MHC class II proteins were primarily present in IBA1+ microglia in the brain parenchyma in regions with neurodegeneration (Extended Data Fig. 6a). Indeed, in line with the increase in the number of parenchymal T cells, we found that the number of microglia positive for MHC class II proteins was significantly elevated in brain regions with tau pathology in TE4 mice (Extended Data Fig. 6b,c). DAM and its subtypes have been well characterized in amyloid models40. Here, in TE4 mice, we found that microglia positive for αX integrin (also known as CD11c), a representative marker for triggering receptor expressed on myeloid cells 2 (TREM2)-dependent type 2 DAM, physically co-localized with CD8+ T cells (Supplementary Video 2). Notably, CD11c was also strongly increased in TE4 hippocampus as compared to the levels in A/PE4 and E4 control mice (Extended Data Fig. 6d,e). These results highlight a tight correlation with microglia positive for MHC class II proteins, CD11c+ microglia, T cells and neurodegeneration. Sparse microglia positive for MHC class II proteins and CD11c+ microglia were also found co-localized with parenchymal plaques in A/PE4 mice (Extended Data Fig. 6b,d). Apoe deletion rescued brain atrophy in P301S mice, and the numbers of microglia positive for MHC class II proteins and CD11c+ microglia as well as T cell numbers were significantly decreased (Extended Data Fig. 6b–g). The higher inflammatory reactivity associated with tau-mediated neurodegeneration and APOE were also confirmed by assessment of inflammatory cytokines in brain tissue from TE4 and TEKO mice (Extended Data Fig. 5). Together, these data demonstrate that parenchymal microglia, in the presence of tauopathy, shift their transcriptomic and phenotypical states from homoeostatic to disease-associated, IFN-activated states positive for CD11c and MHC class I proteins, with an accompanying increase in the number of inflammatory chemokines and cytokines.
a, Total microglia assigned into three subgroups (HOM, DAM and IFN-activated microglia) as visualized by UMAP plot. b, Heat map showing representative markers specifically expressed in the three subgroups. c, Bar plot showing the percentages of the three subgroups of microglia in E4, A/PE4 and TE4 mice. Data are mean ± s.e.m.; two biologically independent samples were used, and samples were sequenced in n = 2 batches from the E4 and TE4 groups. d, Differentially expressed genes in microglia subgroups. e, IBA1 and CD3 staining in 9.5-month-old TE4 mice. PLX, PLX3397. Ctrl, control. Scale bar, 20 μm. f, P2RY12, MHC class II protein and IBA1 staining in 9.5-month-old TE4 mice. Scale bar, 50 μm. g, CD11c, CD8 and IBA1 staining in 9.5-month-old TE4 mice. Scale bar, 50 μm. h–k, Quantification of the areas covered by IBA1, P2RY12, MHC class II proteins and CD11c in 9.5-month-old mice (TE4 Ctrl: n = 5; TE4 PLX: n = 11). ***P < 0.0001, ***P < 0.0001, **P = 0.0031 and ***P < 0.0001 for TE4 PLX versus TE4 Ctrl for IBA1, P2RY12, MHC class II proteins and CD11c, respectively (unpaired two-tailed Student’s t-test). l, Representative images of brain sections from 9.5-month-old mice stained with Sudan black. Scale bar, 1 mm. m–o, Volumes of hippocampus (m), piriform–entorhinal cortex (n) and posterior lateral ventricle (o) in 9.5-month-old mice (E4 PLX: n = 12; TE4 Ctl: n = 5; TE4 PLX: n = 11). **P = 0.0078 and **P = 0.0012 for TE4 Ctrl versus TE4 PLX for volumes of hippocampus and posterior lateral ventricle, respectively (one-way ANOVA with Tukey’s post hoc test). p, Quantification of the number of CD3+ T cells in the dentate gyrus per 0.3 mm2 in 9.5-month-old mice (E4 PLX: n = 12; TE4 Ctl: n = 5; TE4 PLX: n = 11). *P = 0.0236 for TE4 Ctrl versus TE4 PLX (unpaired two-tailed Student’s t-test). q, Quantification of the number of CD8+ T cells in the dentate gyrus per 0.3 mm2 in 9.5-month-old mice (TE4 Ctrl: n = 5; TE4 PLX: n = 11). **P = 0.002 for TE4 Ctrl versus TE4 PLX. r, Quantification of the area covered by AT8 in the dentate gyrus per slice in 9.5-month-old mice (TE4 Ctrl: n = 5; TE4 PLX: n = 11). ***P = 0.0008 for TE4 Ctrl versus TE4 PLX (unpaired two-tailed Student’s t-test). Data in h–k,m–r are mean ± s.e.m.
IFNγ, a cytokine upregulated in TE4 mice, is a pro-inflammatory cytokine produced by NK cells, NK T cells and T cells that can prime microglia for inflammatory responses to injury as well as promote cytotoxic CD8+ T cell function6. Previous studies identified IFNγ-related transcriptomic signatures in tauopathy and neurodegenerative disease models, although the cell-type expression and functional result of IFNγ on pathology was not described41. Ligand–receptor analysis revealed active interactions between T cells and microglia (Extended Data Fig. 7a). IFNγ receptor was already known to be expressed in both neurons and microglia in the brain42. Notably, we found that in the brain of TE4 mice, IFNγ transcripts were enriched in T cells, especially in CD8+ T cells (Extended Data Fig. 7b). Given that IFNγ can augment antigen-presentation and inflammatory functions of myeloid cells, we further investigated the role of IFNγ in the immune response in tauopathy. To determine whether microglia can present antigen to T cells in vitro, we co-cultured microglia acutely isolated from adult mouse brain with OT-1 T cells, with soluble ovalbumin as the antigen, and found that microglia were capable of weakly stimulating OT-1 T cell proliferation compared to dendritic cells (Extended Data Fig. 8c–f). However, on IFNγ stimulation, OT-1 T cell proliferation was strongly enhanced in the presence of microglia with ovalbumin (Extended Data Fig. 8e,f), nearly to the level observed with dendritic cells, suggesting that microglia in vitro can serve as APCs and that IFNγ can augment this response. Together, these data suggest the possibility that there are active interactions between microglia and infiltrated T cells.
To determine the role of endogenous IFNγ in P301S mice in vivo and to study the interplay between activated microglia and T cells, we blocked IFNγ signalling by peripheral administration intraperitoneally every 5 days with a neutralizing antibody in TE3 mice from 7.5 to 9.5 months of age, right before T cell infiltration into the brain parenchyma. Anti-IFNγ treatment resulted in attenuated brain atrophy as compared to that for the IgG treatment control (Extended Data Fig. 8a–d). CD11c+ microglia were also significantly reduced in number in anti-IFNγ-treated mice (Extended Data Fig. 8e,f), and there was a significant reduction in p-tau staining in anti-IFNγ-treated mice (Extended Data Fig. 8g,h). Together, these results suggest that IFNγ secreted by CD8+ T cells in the brain can augment tau pathology and neurodegeneration, at least in part through promoting inflammatory microglial signalling and antigen-presentation functions.
To further delineate the interrelationship between the activated microglia and infiltrated T cells, we administered PLX3397, a selective inhibitor of CSF1R c-kit and FLT3, in TE4 and E4 control mice from 8.5 months to 9.5 months of age (Extended Data Fig. 9i). PLX3397 treatment resulted in strong depletion of microglia numbers (Fig. 4e–k). PLX3397 treatment also decreased hippocampal atrophy and ameliorated the increase in ventricular volume in TE4 mice (Fig. 4l–o). Notably, CD3+ and CD8+ T cell numbers as well as tau pathology were reduced on microglia depletion (Fig. 4p–r). This suggests a pivotal role for microglia, especially activated microglia, in the setting of the tauopathy-specific immune hubs by recruiting T cells into the brain parenchyma and a detrimental role for this restructured immune hub in facilitating disease progression.
T cell depletion prevents degeneration
To directly investigate whether infiltration of T cells leads to neurodegeneration, we depleted T cells by peripheral administration of neutralizing antibodies in TE4 mice as well as their age-matched non-tau transgenic littermates from 6 months to 9.5 months of age, a critical time window when neurodegeneration develops (Extended Data Fig. 8j). A single dose acute intraperitoneal treatment with anti-CD4 and anti-CD8 antibodies (anti-T) led to strong depletion of CD4+ and CD8+ T cells in brain parenchyma, meninges and peripheral blood, confirming the antibody depletion efficiency (Extended Data Fig. 9a,b). Notably, in TE4 mice given anti-T treatment (intraperitoneally every 5 days) from 6 to 9.5 months, brain atrophy was strongly ameliorated compared to that in the IgG-treated control mice (Fig. 5a–d). T cells were almost completely eliminated in the brain parenchyma in TE4 mice after 3.5 months of anti-T antibody treatment (Fig. 5e,g–i). T cell depletion also reduced overall microglial staining (Fig. 5e–g,j), suggesting that T cells in the brain of TE4 mice can augment microgliosis. To assess the activation status of microglia with and without T cell depletion, we immunohistochemically analysed the parenchyma from TE4 IgG- and TE4 anti-T-treated mice using antibodies to P2RY12, MHC class II proteins and CD11c (Fig. 5f,g). We found significant elevation of P2RY12+ microglia and reduced microglia positive for MHC class II proteins and CD11c+ microglia (Fig. 5k–m) in the anti-T antibody-treated mice, suggesting that microglia shift from activated towards a more homoeostatic state after T cell depletion. scRNA-seq analysis of microglia from anti-T antibody- versus the IgG control-treated mice also revealed strong suppression of different aspects of the disease-related microglia signature and an increase in the homoeostatic signature (Extended Data Fig. 9c–e). To assess tau pathology following T cell depletion, we analysed p-tau immunoreactivity in hippocampus and found a significant reduction in p-tau in anti-T-treated mice (Fig. 5n). Four major p-tau staining patterns, designated as type 1–4, strongly correlated with the level of brain atrophy, with type 1 associated with most preserved brain tissue and type 4 associated with the greatest atrophy. Depletion of T cells resulted in a significant shift of p-tau staining pattern towards the earliest disease stage (Fig. 5o,p). We also assessed plasma protein levels of neurofilament light chain, a marker of neuroaxonal damage and neurodegeneration43. The concentration of neurofilament light chain in T cell-depleted mice was significantly reduced (Fig. 5q). Behavioural performance assessment revealed that after depletion of T cells, nest-building behaviour in 9.5-month-old TE4 mice was significantly improved (Extended Data Fig. 10f). We also assessed an additional cohort of TE4 mice that we treated with anti-T antibody versus the IgG control from 6 months to 8.5 months of age. Assessment of behavioural performance revealed that depletion of T cells resulted in significant improvement in two additional behaviours. Alternation in a Y maze (assessing short-term memory and exploratory behaviour) and freezing in response to an auditory cue (assessing amygdala-dependent memory) were significantly improved (Fig. 5r–u and Extended Data Fig. 9g–i). Freezing behaviour in response to a contextual cue showed a trend towards increased hippocampal-dependent memory after depletion of T cells (Fig. 5t). Both groups showed similar baseline levels of general exploratory behaviour, and locomotor activity levels (Extended Data Fig. 9g,h) and response to tone–shock pairing in the fear conditioning test (Fig. 5s). Together, these data demonstrate that T cell depletion decreases functional decline.
a, Representative images of brain sections from 9.5-month-old mice. Scale bar, 1 mm. b–d, Volumes of brain regions in 9.5-month-old mice (E4 IgG: n = 11; E4 anti-T: n = 10; TE4 IgG: n = 8; TE4 anti-T: n = 12). *P = 0.0112, *P = 0.0397 and *P = 0.0313 for TE4 IgG versus TE4 anti-T for hippocampus, piriform–entorhinal cortex and posterior lateral ventricle, respectively (unpaired two-tailed Student’s t-test). e, IBA1 and CD3 staining in 9.5-month-old mice. Scale bar, 20 μm. f, P2RY12, MHC class II protein and IBA1 staining in 9.5-month-old mice. Scale bar, 50 μm. g, CD11c, CD8 and IBA1 staining in 9.5-month-old mice. Scale bar, 50 μm. h, Quantification of CD3+ T cells in the dentate gyrus per 0.3 mm2 in 9.5-month-old mice (E4 IgG: n = 11; E4 anti-T: n = 11; TE4 IgG: n = 11; TE4 anti-T: n = 11). ***P = 0.0004 (unpaired two-tailed Student’s t-test). i, CD8+ T cells per 0.3 mm2 of the dentate gyrus in 9.5-month-old mice (TE4 IgG: n = 11; TE4 anti-T: n = 11). ***P = 0.0002 (unpaired two-tailed Student’s t-test). j–n, Quantification of the immunostained areas in 9.5-month-old mice (TE4 IgG: n = 11 and TE4 anti-T: n = 11 for j–m; TE4 IgG: n = 8 and TE4 anti-T: n = 12 for n). ***P = 0.0002, *P = 0.0229, ***P = 0.0004, ***P = 0.0002 and ***P = 0.0002 for area of IBA1, P2RY12, MHC class II proteins, CD11c and AT8, respectively (unpaired two-tailed Student’s t-test). o, Distinct p-tau staining patterns. p, Distribution of the four p-tau staining patterns in 9.5-month-old mice. **P = 0.007 for distribution between TE4 IgG and TE4 anti-T (Fisher’s exact test) q, Concentration of neurofilament light chain (NfL) in the plasma of 9.5-month-old mice (E4 IgG: n = 11; E4 anti-T: n = 11; TE4 IgG: n = 11; TE4 anti-T: n = 11). *P = 0.0398 (unpaired two-tailed Student’s t-test). r, Behaviour of 8.5-month-old mice in a Y maze (TE4 IgG: n = 10; TE4 anti-T: n = 15). *P = 0.0239 (unpaired two-tailed Student’s t-test). s, Tone–shock pairing, day 1 (TE4 IgG: n = 10; TE4 anti-T: n = 15). P = 0.2152 (two-way ANOVA, with Bonferroni post hoc comparisons test). t, Freezing in response to a contextual cue, day 2 (TE4 IgG: n = 10; TE4 anti-T: n = 15). P = 0.067 (two-way ANOVA, with Bonferroni post hoc comparisons test) for the treatment with TE4 IgG and TE4 anti-T. u, Freezing in response to an auditory cue, day 3 (TE4 IgG: n = 10; TE4 anti-T: n = 15). **P = 0.0118 (two-way ANOVA, with Bonferroni post hoc comparisons test) for the treatment with TE4 IgG and TE4 anti-T, **P = 0.0099 for 8 min, P = 0.055 for 9 min and **P = 0.007 for 10 min. Data in b–d,h–n,q–u are mean ± s.e.m.
Immune checkpoints are regulatory pathways for maintaining systemic immune homoeostasis and tolerance44. PDCD1 is a checkpoint protein expressed on T cells, which processes inhibitory signals to control the magnitude of adaptive immune responses and tolerance45. Previous reports indicate that PDCD1 immune checkpoint blockade decreases cognitive impairment in mouse models with Alzheimer’s disease pathology46,47. PDCD1 blockade can lead to increased activation of exhausted CD8+ T cells or enhanced immunosuppression through increased activation of PDCD1+CD4+ Treg cells48. To investigate whether a treatment that targets PDCD1–PDL1 blockade could be effective in tauopathy, we administered anti-PDCD1 treatment in TE4 mice from 8 months to 9.5 months of age, a time window in which brain atrophy develops. We found that with 1-week acute anti-PDCD1 treatment increased the percentage of FOXP3+CD4+ Treg cells and PDCD1+FOXP3+CD4+ Treg cells, with no obvious changes on KLRG1+ effector CD8+ T cells and total PDCD1+TOX+CD8+ T cells in the brain (Extended Data Fig. 10a–e). These results suggest that PDCD1-antibody treatment at this age would increase immunosuppressive CD4+ Treg cells. Consistent with this hypothesis, chronic treatment beginning at 8 months significantly decreased tau-mediated neurodegeneration and p-tau staining (Extended Data Fig. 10f–i), further supporting a role for T cells in tau-mediated neurodegeneration.
In this study, a comprehensive map for immune responses at the cellular and molecular level in the brain and meninges during the development of amyloid or tau pathology and neurodegeneration using scRNA-seq and scTCR-seq is presented. We find that an immunological hub involving activated microglia and T cells is overrepresented in brain regions with tauopathy and neuronal loss. Although evidence regarding the pathological changes and the role of microglia in Alzheimer’s disease is emerging, here we expand on the immune microenvironment in the setting of tauopathy and neurodegeneration by assessing a previously less examined adaptive immunological response involving T cells and their interaction with cells in the brain. T cells dynamically shift from activated to exhausted states with unique TCR clonal expansion. We also present direct evidence that breaking the neurodegeneration-associated immune hub between activated microglia and infiltrated T cells effectively prevents neurodegeneration and decreases cognitive decline.
As an innate primary response, microglia seem to have a protective role in the presence of amyloid plaques (restrict plaque growth and local damage) or a pro-inflammatory, damaging role in the presence of tau pathology (response to neuronal damage and aggregated tau, leading to severe neurodegeneration) in Alzheimer’s disease12. Here we found that CD11c and MHC class II protein expression strongly increased in microglia specifically in regions of the brain with atrophy. Genes encoding MHC class I and II proteins were highly upregulated in activated microglia in tauopathy. We also discovered adaptive immune responses in both a mouse model of tauopathy and brain samples from patients with Alzheimer’s disease, finding that T cells are present in the brain parenchyma and also that their enrichment highly correlates with the severity of brain atrophy. Removal and modulation of T cells rescued the brain atrophy and highlighted that T cells have an important role in neurodegeneration.
The complex nature of the central nervous system (CNS) necessitates its own specialized immunological adaptations to detect and respond to environmental changes. Here we found significantly different proportions of T cells in the meninges and brain parenchyma. These results highlight that CNS-border and CNS-resident T cells are functionally different in accordance with their immune niche. The local tauopathy-related microenvironment in the brain parenchyma is likely to be instructive for recruiting and guiding the transformation of T cells. The interaction of T cells with APCs has been well established in peripheral systems49 and CNS borders32. Here our findings raise a fundamental question regarding the interaction of T cells with APCs in the brain parenchyma. We find that T cells actively interact with the disease-related microglia subgroups. Depletion of microglia largely abolishes T cell infiltration and depletion of T cells also remarkably hinders microglia activation, demonstrating communication between the innate and adaptive family of immune cells. In combined scRNA-seq and scTCR-seq analyses, we uncover unique clonal expansion of T cells enriched in the parenchyma with tauopathy and neurodegeneration. Defining which antigens result in T cell activation, such as variously modified forms of tau, other proteins or myelin debris released by damaged neurons that are subsequently presented to adaptive immune cells within tauopathy and Alzheimer’s disease, remains an intriguing question. Sequencing TCRs at the single-cell level combined with high-throughput peptide screening would enable elucidation of the specific antigens, which might in turn yield pathological stage-specific therapeutic strategies. Microglia express many pattern-recognition receptors that bind and internalize foreign misfolded proteins50. We found that T cell infiltration did not increase in the tau mice lacking APOE. Therefore, a previously overlooked immunomodulatory function of APOE may serve as an important mechanism linking both innate and adaptive immunity. Mapping the disease-state-specific interlink between microglia and T cells, including their signalling communications, presented antigens and pathophysiological responses, will be a key nexus to set up unique therapeutic interventions to prevent or reverse brain atrophy and neurodegeneration in tauopathies.
Methods
Animals
Human-APOE-knock-in mice, APOE3 and APOE4 (E3 and E4, respectively), were generated by replacing the mouse genomic sequence from the translation initiation codon in exon 2 to the termination codon in exon 4 with its human counterparts flanked by loxP sites51. Tau(P301S) transgenic mice (Jax, no. 008169) on a C57BL/6 background were crossed to human-APOE-knock-in or Apoe-knockout mice (Jax, no. 002052) to generate P301S/E3 (TE3), P301S/E4 (TE4) and P301S/EKO (TEKO) mice respectively. All tau transgenic mice involved in the final analysis were obtained from the same generation. A/PE4 and 5XFADE4 mice have been described previously8,51. Littermates of the same sex were randomly assigned to experimental groups. All animal procedures and experiments were carried out under guidelines approved by the Institutional Animal Care and Use committee at Washington University School of Medicine.
Human Alzheimer’s disease tissues
All participants gave prospective pre-mortem written consent for their brain to be banked and used for research with information to potentially be published under procedures approved by the human institutional review board at Banner Sun Health Research Institute. Patient demographics are available in Supplementary Table 2.
Volumetric analysis
The left hemi-brain of each mouse was fixed with 4% paraformaldehyde for 24 h at 4 °C and then placed in 30% sucrose at 4 °C overnight. Serial free-floating coronal sections were cut from the rostral crossing of the corpus callosum to the caudal end of the hippocampus at 50 μm on a Leica SM2010 microtome. Brain sections (spaced 300 μm apart) from bregma −1.3 mm to −3.1 mm were mounted for volumetric analysis. All mounted sections were stained with 0.1% Sudan black (Sigma, 199664-25G) in 70% ethanol at room temperature for 20 min, and then washed in 70% ethanol for 50 s, three times. The sections were washed in Milli-Q water three times and covered with Floromount-G (Southern Biotech, 0100-01). Slides were scanned using a Hamamatsu NanoZoomer microscope at ×20 magnification. Hippocampus, piriform–entorhinal cortex and ventricles were traced using NDP viewer. The volume was calculated using the formula: volume = (sum of area) × 0.3 mm × number of sections. The experimenter assessing brain volumes was blinded to experimental groups.
Immunohistochemistry
Two sections from each mouse (300 μm apart), corresponding approximately to bregma coordinates −1.4 mm, −1.7 mm were used for p-tau staining. Brain sections were washed in Tris-buffered saline (TBS) buffer for 3 min, followed by incubation in 0.3% hydrogen peroxide in TBS for 10 min at room temperature. After three washes in TBS, sections were blocked by 3% milk in TBS with 0.25% Triton X-100 (TBSX) for 1 h at room temperature followed by incubation with AT8-biotinylated antibody (Thermo Scientific, MN1020B) overnight at 4 °C. The next day, after three washes in TBS, the slices were developed by VECTASTAIN Elite ABC-HRP kit (Vector Laboratories, PK-6100) following the manufacturer’s instructions. Slides were covered by Cytoseal 60 (Thermo Scientific, 8310-4) and scanned using a Hamamatsu NanoZoomer microscope at ×20 magnification. Images were analysed by ImageJ.
For immunofluorescent staining, two sections (bregma −2.0 mm and −2.3 mm) from each mouse were used. The sections were washed three times in TBS, permeabilized with 0.25% TBSX for 10 min, and then blocked with 3% BSA in 0.25% TBSX for 1 h at room temperature. Sections were incubated in primary antibodies overnight at 4 °C. The next day, sections were washed in TBS and incubated with corresponding fluorescence-labelled secondary antibodies for 1.5 h at room temperature. The slices were washed and mounted in Prolong Gold Antifade mounting medium (Invitrogen, P36930). Primary antibodies were as follows: CD3 (Novus, NB600-1441, 1:200), CD8 (Invitrogen, MA1-145, 1:100), IBA1 (Wako, 019-19741, 1:2,000; Abcam, ab5076, 1:500), AT8 (Invitrogen, MN1020B, 1:500), amyloid-β (made in house, HJ3.4B, 1:1,000), P2RY12 (gift from Butovsky lab, 1:2,000), NeuN (Abcam, ab177487, 1:1,000), myelin basic protein (Abcam, ab7349, 1:500), MHC class II protein (Biolegend, 107650, 1:200), X34 (Sigma, 1954-25MG, 10 mM in dimethylsulfoxide stock, 1:5,000), CD206 (Bio-Rad, MCA2235, 1:300), Hoechst (Sigma, 94403, 1:5,000). Secondary antibodies were as follows: donkey anti-rat 488 (Invitrogen, A21208, 1:500), donkey anti-rabbit 405 (Invitrogen, A48258, 1:500), donkey anti-rabbit 568 (Invitrogen, A10042, 1:500), streptavidin 568 (Invitrogen, S11226, 1:500), donkey anti-goat 647 (Invitrogen, A21447, 1:500). Images were acquired on a Zeiss LSM800 microscope. Areas covered by antibody–fluorophores and their numbers were analysed by ImageJ. Three-dimensional construction was carried out using Imaris 9.7.0 software. CD3, IBA1, CD8 and CD11c were labelled and detected with fluorophores using the surface area function.
PLX3397 formulation and supplement
PLX3397 was purchased from SelleckChem. PLX3397 was formulated in AIN-76A (Research Diet) at a concentration of 400 mg per kilogram of chow. E4 and TE4 mice were treated with PLX3397 for 4 weeks for microglial acute depletion from 8.5 to 9.5 months of age.
IFNγ treatment
For blocking IFNγ signalling, mice were intraperitoneally injected with 100 mg per kilogram of body weight with either control IgG (Leinco, P376) or anti-mouse IFNγ (Leinco, clone H22, I-1190) antibodies52 every 5 days from 7.5 to 9.5 months of age.
Anti-PDCD1 treatment
For blocking PDCD1–PDL1 signalling chronically, mice were intraperitoneally injected with 500 μg anti-PDCD1 antibody (BioXCell, BP0146) every 5 days from 8 to 9.5 months of age. IgG (BioXCell, BP0089) isotype control was administered at the same frequency and dosage. Brains were collected for flow cytometry assessment of T cell populations. To characterize the T cell populations with anti-PDCD1 treatment, mice were acutely treated with 500 μg anti-PDCD1 or IgG every 2 days. At day 7, after perfusion, brains were isolated for single-cell analysis by flow cytometry. Intracellular staining for transcription factors was carried out using eBioscience FOXP3/Transcription Factor Kit (Ref. 00-5523-00) per the manufacturer’s instructions. In brief, cells were stained with LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (Invitrogen, ref. L34966A) for 5 min and then incubated with surface antibody mix and TruStain FcX PLUS (anti-mouse CD16/CD32, Clone S17011E, Biolegend, ref. 156604, 1:200) for 1 h at room temperature. After cell-surface staining, cells were fixed, permeabilized and incubated with intracellular antibody mix overnight at 4 °C. Flow cytometry was carried out on a BD Symphony A3. The following antibodies were used: CD45.2 (Biolegend, 104), CD4 (Biolegend, GK1.5), PDCD1 (Biolegend, 29F.1A12), KLRG1 (Biolegend, 2F1/KLRG1), CD3e (BD, 145-2C11), CD8a (BD, 53-6.7), FOXP3 (Invitrogen, FJK-16s), TOX (Invitrogen, TXRX10).
T cell depletion
For the depletion of CD4+ and CD8+ T cells, mice were intraperitoneally injected with 500 μg anti-CD4 (BioXCell, BP0003-1) and anti-CD8 antibody (BioXCell, BP0061) every 5 days from 6 to 9.5 months of age or for memory-related behavioural experiments from 6 to 8.5 months of age. IgG (BioXCell, BP0090) isotype control was administered at the same frequency and dosage. To characterize the depletion efficiency, mice were acutely treated with 500 μg anti-CD4, or anti-CD8 or IgG. Brain, meninges and blood were extracted for single-cell analysis followed by flow cytometry assessment of CD4+ and CD8+ T cell populations.
Brain extraction
Mouse cortex tissue was weighed and homogenized using a pestle with 10 μl buffer per 1 mg tissue in chilled lysis buffer (Thermo Scientific, 78503). After centrifugation at 20,000g for 10 min at 4 °C, the supernatant was saved and protein concentration was measured by micro BCA protein assay kit (Thermo Scientific, 23235) before multiplex immunoassay (Thermo Scientific).
Nest-building behaviour
Group-housed mice were switched to individual housing in the week of assessment at 9.5 months. A pre-weighed nestlet was provided in each cage. After overnight housing, the remaining nestlet was weighed. A 5-point scale system was assigned on the basis of the percentage of remaining nesting material and shredded conditions. Score 1: nestlet >90% untorn; score 2: nestlet 50–90% untorn; score 3: nestlet 10–50% untorn; score 4: nestlet <10% untorn, but nest is flat and uncompact; score 5: nest is compact and nest wall is higher than the mouse for >50% of its circumference.
General design of behavioural tests
TE4 male mice were treated with IgG or with anti-CD4 and anti-CD8 antibodies for T cell depletion from 6 to 8.5 months of age. They were then tested for behavioural differences. Following 1-week habituation and handling in the Washington University Animal Behavior Core, mice were evaluated on 1 h locomotor activity, spontaneous alternation in a Y maze and fear conditioning. All tests were conducted during the light phase of the light–dark cycle. Behavioural testers were blind to the treatment group.
One-hour locomotor activity and open-field behaviour test
To evaluate general activity levels and possible alterations in emotionality, mice were evaluated over a 1-h period in transparent (47.6 × 25.4 × 20.6 cm high) polystyrene enclosures. Each cage was surrounded by a frame containing a 4 × 8 matrix of photocell pairs, the output of which was fed to an online computer (Hamilton-Kinder, LLC). The system software (Hamilton-Kinder, LLC) was used to define a 33 × 11-cm central zone and a peripheral or surrounding zone that was 5.5 cm wide with the sides of the cage being the outermost boundary. This peripheral area extended along the entire perimeter of the cage. Variables that were analysed included the total number of ambulations and rearing on hindlimbs, as well as the number of entries, the time spent and the distance travelled in the centre area as well as the distance travelled in the periphery surrounding the centre.
Spontaneous alternation in Y maze
Testing was conducted according to our previously published procedures53. In brief, this involved placing a mouse in the centre of a Y maze that contained three arms that were 10.5 cm wide, 40 cm long and 20.5 cm deep with the arms oriented at 120° with respect to each successive other arm. Mice were allowed to explore the maze for 10 min and entry into an arm was scored only when the hindlimbs had completely entered the arm. An alternation was defined as any three consecutive choices of three different arms without re-exploration of a previously visited arm. Dependent variables included the number of alternations and arm entries along with the percentage of alternations, which was determined by dividing the total number of alternations by the total number of entries minus 2, and then multiplying by 100.
Conditioned fear
A previously described protocol54 was used to train and test mice using two clear-plastic conditioning chambers (26 × 18 × 18 cm high; Med-Associates) that were easily distinguished by different olfactory, visual and tactile cues present in each chamber. On day 1, each mouse was placed into the conditioning chamber for 5 min and freezing behaviour was quantified during a 2 min baseline period. Freezing (no movement except that associated with respiration) was quantified using FreezeFrame image analysis software (Actimetrics) that allows for simultaneous visualization of behaviour while adjusting for a ‘freezing threshold’ during 0.75-s intervals. After baseline measurements, a conditioned stimulus consisting of an 80-dB tone (white noise) was presented for 20 s followed by an unconditioned stimulus consisting of a 1-s, 1.0-mA continuous foot shock. This tone–shock (T–S) pairing was repeated each minute over the next 2 min, and freezing was quantified after each of the three tone–shock pairings. Twenty-four hours after training, each mouse was placed back into the original conditioning chamber to test for fear conditioning to the contextual cues in the chamber. This involved quantifying freezing over an 8-min period without the tone or shock being present. Twenty-four hours later, the mice were evaluated on the auditory cue component of the conditioned fear procedure, which included placing each mouse into the other chamber containing distinctly different cues. Freezing was quantified during a 2-min ‘altered context’ baseline period as well as over a subsequent 8-min period during which the auditory cue (conditioned stimulus) was presented. Shock sensitivity was evaluated following completion of the conditioned fear test as previously described55.
Concentration of neurofilament light chain
The concentration of neurofilament light chain in plasma was measured with NF-Light Simoa Assay Advantage kit (Quanterix) by an experimenter blinded to experimental groups.
Single-cell isolation
Mechanical dissociation was carried out as previously described56. In brief, mice were perfused with pre-chilled PBS to fully remove blood contamination. Hippocampus and cortex were dissected followed by Dounce homogenization. Cell suspensions were then passed through Percoll density centrifugation to remove myelin and debris. The cell pellets were washed with 0.5% BSA for analysis or collection. For meninges, meninges were peeled intact from the skullcap using fine forceps and prepared for single-cell analysis as previously described32. In brief, meninges were mashed through a cell strainer, using a sterile syringe plunger, and washed in 0.5% BSA.
Flow cytometry for single cells
All steps were carried out on ice or using a pre-chilled centrifuge set to 4 °C. Single-cell suspensions were incubated with anti-CD16/32 (Fc block; Biolegend) for 5 min and then fluorescently conjugated antibodies were added for 20 min. After washing, samples were collected by 300g followed by a 5 min spin down and suspended in 5% BSA with PI for live–dead selection before sorting. Cells were sorted using a FACS Aria II (BD Bioscience).
Immune scRNA-seq
After quantifying and analysing single-cell integrity, 8,000–16,000 individual single cells per sample were loaded onto a 10x Genomics Chromium platform for Gel Beads-in-emulsion and cDNA generation carrying cell- and transcript-specific barcodes and sequencing libraries constructed using the Chromium Single Cell 5′ library & Gel Bead Kit V2. Libraries were sequenced on the Illumina NovaSeq6000.
Single-cell data processing and TCR analysis
Alignment, barcode assignment and UMI counting with Cell Ranger (v6.1.1) were used for preparation of count matrices for the gene expression library. For alignment, a custom mouse genome (GRCm38) containing human sequences for APOE, PSEN1, APP and MAPT genes was used as a reference.
Barcodes in all samples that were considered to represent noise and low-quality cells were filtered out using the knee-inflection strategy available in default Cell Ranger (v6.1.1) from 10x Genomics.
For downstream analysis, the Seurat package (v4.0.4) was used, and genes expressed in fewer than three cells were also filtered from expression matrices. The fraction of mitochondrial genes was calculated for every cell, and cells with a mitochondrial fraction of more than the highest confidence interval for a scaled mitochondrial percentage were filtered out, which results in removal of cells with a mitochondrial percentage of more than 20%. Additionally, a cutoff of log10[number of unique expressed genes] = 2.5 was used for removing the cells from both CD45hi and CD45total parenchyma cells, and 2 was used as a threshold for the cells from meninges.
Doublets have been excluded on the basis of the co-expression of the canonical cell-type-specific genes.
Each sample was normalized using the SCTransform function with mitochondrial content as a variable to regress out in a second non-regularized linear regression. For integration aims, variable genes across the samples were identified by the SelectIntegrationFeatures function with the number of features equal to 2,000. Then the object was prepared for integration (PrepSCTIntegration function), the anchors were found (FindIntegrationAnchors function) and the samples were integrated into the whole object (IntegrateData function).
Principal component analysis was used for dimensionality reduction, and the first 20 principal components were used further to generate UMAP dimensionality reduction by the RunUMAP function. The clustering procedure was carried out by FindNeighbors and FindClusters with a range of resolutions (from 0.2 to 1.0 with 0.2 as a step) and the first 20 principal components as input.
The object covering all cells was subsetted into T cell-, microglia- and myeloid-specific sub-objects on the basis of expression of canonical gene markers. The T cell object was further split into CD4+ and CD8+ cells. Then, all objects were passed through the iterative process of quality control with doublet removal and exclusion of the cell types that have no relevant markers and contained high mitochondrial content as well as poor coverage (all filters are object-specific).
Cell Ranger’s vdj workflow (v6.1.1) was used for TCR data analysis. Non-canonical T cells (such as γδ T cells and NK T cells) as well as T cells with inappropriate combinations of α- and β-chains were removed. Then, all barcodes were assigned to two populations based on CD4 and CD8 gene expression. The Gini coefficient was calculated using the immunarch package (v0.6.6) to estimate the clonal diversity among samples.
Trajectory analysis was carried out using a slingshot container available at dynverse package with normalized count matrices with barcodes assigned to microglia as input data as well as cells assigned to CD8+ T cells.
Interaction analysis was implemented using the CellChat package (v. 1.1.3) with the Cell-Cell Contact database. As input data, microglia, CD4+ and CD8+ T cells from the E4 genotype and microglia, CD4+ and CD8+ T cells from the TE4 genotype were used. Following the CellChat vignette, CellChat objects were prepared (createCellChat), overexpressed genes and interactions were identified (identifyOverExpressedGenes, identifyOverExpressedInteractions functions), communication probabilities were estimated (computeCommunProb, filterCommunication, computeCommunProbPathway functions) and network analysis (aggregateNet, netAnalysis_computeCentrality functions) was carried out. The genotype-specific as well as genotype-common ligand–receptor pairs were identified (netVisual_bubble function). The number of interactions was evaluated using the netVisual_circle function.
Peripheral immune cell composition assay
Spleens from E4, A/PE4 and TE4 mice were collected and smashed through a 70-μm strainer to prepare single-cell suspensions. After single-cell suspensions were made, the cells were pelleted down and resuspended in 5 ml red blood cell lysis buffer (ACK buffer) at room temperature for 2 min. Cells were blocked in the presence of Fc block (2.4G2; Leinco, C247) in magnetic-activated cell-sorting buffer (0.5% BSA, 2 mM EDTA in PBS) at 4 °C. The following antibodies were used: CD45 (Biolegend, 30-F11), CD19 (Biolegend, 6D5), CD3 (Biolegend, 145-2C11), CD44 (Biolegend IM7), CD4 (Biolegend, PM4-5), CD8 (BD, 53-6.7), FOXP3(Invitrogen, FJK-16s).
Microglia antigen presentation in vitro assay
C57BL/6-Tg (TcraTcrb) 1100 Mb/J (OT-I) (Jax, no. 003831) and B6.SJL-Ptprca Pepcb/BoyJ (B6. CD45.1) (Jax, no. 002014) were from Jackson laboratory. OT-1.CD45.1/2 mice were generated by crossing OT-1 and B6.CD45.1 for one generation. Mice of 8–12 weeks of age were used for the experiment.
To isolate APCs, spleens were chopped into small pieces and digested at 37 °C for 45 min with buffer containing 0.28 U ml−1 Liberase (Roche, 5401119001), 100 U ml−1 hyaluronidase (Sigma, H3506) and 50 U ml−1 DNase I (Roche, 10104159001) in RMPI1640 (Gibco, 11875093). Cells were pelleted down for CD11c microbead (Miltenyi Biotec,130-125-835) enrichment based on the manufacturer’s instructions. Dendritic cells were sorted as CD45+CD11c+MHC-IIhi cells.
To enrich OT-1 CD8+ naive T cells, naive CD8α+ T Cell Isolation Kit (Miltenyi Biotec, 130-096-543) was used for column-based enrichment. OT-1 naive CD8+ T cells were sorted followed by CD45+CD3+CD8+TCRVβ5+TCRVα2+CD62L+CD44low cells.
Microglia were sorted followed by CD45lowCD11b+ cells after single-cell collection from brain parenchyma with cortex and hippocampus. A total of 25,000 T cells labelled with CellTrace Violet (5 μM, Thermo Fisher, C34571) were co-cultured with 20,000 microglia or dendritic cells for 3 days in a U-bottom 96-well plate (Corning, 07-200-720). A serial dilution of ovalbumin (Worthington, LS003049) starting from 1,000 μg ml−1 (2× dilution) was made and added into the wells. For microglia–OT-1 co-culture, two doses of IFNγ (100 ng ml−1 and 1,000 ng ml−1) were added at the same time. After 3 days, cells were analysed by flow cytometry for T cell proliferation.
Flow cytometry and cell sorting were completed on a FACS CantoII or FACS Aria II instrument and analysed using Flowjo (v10). Staining was carried out at 4 °C in the presence of Fc block (2.4G2; Leinco) in magnetic-activated cell-sorting buffer (0.5% BSA, 2 mM EDTA in PBS). The following antibodies were used: CD45 (Biolegend, 30-F11), CD11b (Biolegend, M1/70), I-A/I-E (Biolegend, M5/114.15.2), CD3 (Biolegend, 145-2C11), TCRVβ5 (Biolegend, MR9-4), TCRVα2 (Biolegend, B20.1), CD45.2 (Biolegend, 104), CD45.1 (Biolegend, A20), CD44 (Biolegend, IM7), CD8α (Biolegend, 53-6.7), CD62L (Biolegend, MEL-14).
Statistics
Statistical analysis was carried out using Prism. Differences between groups were evaluated by Student’s t-test, or one-way or two-way ANOVA followed by post hoc tests. For conditioned fear behaviour, two-way ANOVA followed by Bonferroni test was used. Data are expressed as mean ± s.e.m. ***P < 0.0001; **P < 0.001; *P < 0.05; NS, no significant difference.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Immune scRNA-seq sample information, information for samples from patients with Alzheimer’s disease, and immune cell numbers in each cluster in the brain are available in the Supplementary Information. All source data, including sequencing reads and single-cell expression matrices, are available from the Gene Expression Omnibus under accession code GSE221856.
Code availability
Code for preprocessing of immune scRNA-seq bioinformatic analysis is available at https://zenodo.org/record/7566414.
References
Duyckaerts, C., Delatour, B. & Potier, M. C. Classification and basic pathology of Alzheimer disease. Acta Neuropathol. 118, 5–36 (2009).
Masters, C. L. et al. Alzheimer’s disease. Nat. Rev. Dis. Primers 1, 15056 (2015).
Jack, C. R. Jr & Holtzman, D. M. Biomarker modeling of Alzheimer’s disease. Neuron 80, 1347–1358 (2013).
Musiek, E. S. & Holtzman, D. M. Three dimensions of the amyloid hypothesis: time, space and ‘wingmen’. Nat. Neurosci. 18, 800–806 (2015).
Giannakopoulos, P. et al. Tangle and neuron numbers, but not amyloid load, predict cognitive status in Alzheimer’s disease. Neurology 60, 1495–1500 (2003).
Chen, X. & Holtzman, D. M. Emerging roles of innate and adaptive immunity in Alzheimer’s disease. Immunity 55, 2236–2254 (2022).
Ulrich, J. D. et al. Altered microglial response to Aβ plaques in APPPS1-21 mice heterozygous for TREM2. Mol. Neurodegener. 9, 20 (2014).
Xiong, M. et al. APOE immunotherapy reduces cerebral amyloid angiopathy and amyloid plaques while improving cerebrovascular function. Sci. Transl. Med. 13, eabd7522 (2021).
Rupp, N. J., Wegenast-Braun, B. M., Radde, R., Calhoun, M. E. & Jucker, M. Early onset amyloid lesions lead to severe neuritic abnormalities and local, but not global neuron loss in APPPS1 transgenic mice. Neurobiol. Aging 32, 2324 (2011).
Giannoni, P. et al. Cerebrovascular pathology during the progression of experimental Alzheimer’s disease. Neurobiol. Dis. 88, 107–117 (2016).
Yoshiyama, Y. et al. Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron 53, 337–351 (2007).
Long, J. M. & Holtzman, D. M. Alzheimer disease: an update on pathobiology and treatment strategies. Cell 179, 312–339 (2019).
Hammond, T. R., Marsh, S. E. & Stevens, B. Immune signaling in neurodegeneration. Immunity 50, 955–974 (2019).
Heneka, M. T., Kummer, M. P. & Latz, E. Innate immune activation in neurodegenerative disease. Nat. Rev. Immunol. 14, 463–477 (2014).
Krasemann, S. et al. The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity 47, 566–581 (2017).
Mrdjen, D. et al. High-dimensional single-cell mapping of central nervous system immune cells reveals distinct myeloid subsets in health, aging, and disease. Immunity 48, 380–395 (2018).
Prinz, M., Priller, J., Sisodia, S. S. & Ransohoff, R. M. Heterogeneity of CNS myeloid cells and their roles in neurodegeneration. Nat. Neurosci. 14, 1227–1235 (2011).
Fischer, H. G. & Reichmann, G. Brain dendritic cells and macrophages/microglia in central nervous system inflammation. J. Immunol. 166, 2717–2726 (2001).
Gate, D. et al. CD4+ T cells contribute to neurodegeneration in Lewy body dementia. Science 374, 868–874 (2021).
Zenaro, E. et al. Neutrophils promote Alzheimer’s disease-like pathology and cognitive decline via LFA-1 integrin. Nat. Med. 21, 880–886 (2015).
Subbarayan, M. S., Hudson, C., Moss, L. D., Nash, K. R. & Bickford, P. C. T cell infiltration and upregulation of MHCII in microglia leads to accelerated neuronal loss in an alpha-synuclein rat model of Parkinson’s disease. J. Neuroinflammation 17, 242 (2020).
Garber, C. et al. T cells promote microglia-mediated synaptic elimination and cognitive dysfunction during recovery from neuropathogenic flaviviruses. Nat. Neurosci. 22, 1276–1288 (2019).
Gate, D. et al. Clonally expanded CD8 T cells patrol the cerebrospinal fluid in Alzheimer’s disease. Nature 577, 399–404 (2020).
Merlini, M., Kirabali, T., Kulic, L., Nitsch, R. M. & Ferretti, M. T. Extravascular CD3+ T cells in brains of Alzheimer disease patients correlate with Tau but not with amyloid pathology: an immunohistochemical study. Neurodegener. Dis. 18, 49–56 (2018).
Laurent, C. et al. Hippocampal T cell infiltration promotes neuroinflammation and cognitive decline in a mouse model of tauopathy. Brain 140, 184–200 (2017).
Lee, S. H. et al. TREM2-independent oligodendrocyte, astrocyte, and T cell responses to tau and amyloid pathology in mouse models of Alzheimer disease. Cell Rep. 37, 110158 (2021).
Norris, G. T. & Kipnis, J. Immune cells and CNS physiology: microglia and beyond. J. Exp. Med. 216, 60–70 (2019).
Chen, X. et al. Transcriptomic mapping uncovers Purkinje neuron plasticity driving learning. Nature 605, 722–727 (2022).
Da Mesquita, S., Fu, Z. & Kipnis, J. The meningeal lymphatic system: a new player in neurophysiology. Neuron 100, 375–388 (2018).
Da Mesquita, S. et al. Meningeal lymphatics affect microglia responses and anti-Aβ immunotherapy. Nature 593, 255–260 (2021).
Rustenhoven, J. & Kipnis, J. Bypassing the blood-brain barrier. Science 366, 1448–1449 (2019).
Rustenhoven, J. et al. Functional characterization of the dural sinuses as a neuroimmune interface. Cell 184, 1000–1016 (2021).
Korn, T. & Kallies, A. T cell responses in the central nervous system. Nat. Rev. Immunol. 17, 179–194 (2017).
Siller-Farfan, J. A. & Dushek, O. Molecular mechanisms of T cell sensitivity to antigen. Immunol. Rev. 285, 194–205 (2018).
Loetscher, M. et al. Chemokine receptor specific for IP10 and mig: structure, function, and expression in activated T-lymphocytes. J. Exp. Med. 184, 963–969 (1996).
Madore, C., Yin, Z., Leibowitz, J. & Butovsky, O. Microglia, lifestyle stress, and neurodegeneration. Immunity 52, 222–240 (2020).
Gratuze, M. et al. Impact of TREM2R47H variant on tau pathology-induced gliosis and neurodegeneration. J. Clin. Invest. 130, 4954–4968 (2020).
Huang, Y. et al. Microglia use TAM receptors to detect and engulf amyloid beta plaques. Nat. Immunol. 22, 586–594 (2021).
Neefjes, J., Jongsma, M. L., Paul, P. & Bakke, O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 11, 823–836 (2011).
Keren-Shaul, H. et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 169, 1276–1290 (2017).
Mathys, H. et al. Temporal tracking of microglia activation in neurodegeneration at single-cell resolution. Cell Rep. 21, 366–380 (2017).
Filiano, A. J. et al. Unexpected role of interferon-gamma in regulating neuronal connectivity and social behaviour. Nature 535, 425–429 (2016).
Khalil, M. et al. Neurofilaments as biomarkers in neurological disorders. Nat. Rev. Neurol. 14, 577–589 (2018).
Pardoll, D. M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 12, 252–264 (2012).
Sharpe, A. H. & Pauken, K. E. The diverse functions of the PD1 inhibitory pathway. Nat. Rev. Immunol. 18, 153–167 (2018).
Rosenzweig, N. et al. PD-1/PD-L1 checkpoint blockade harnesses monocyte-derived macrophages to combat cognitive impairment in a tauopathy mouse model. Nat. Commun. 10, 465 (2019).
Baruch, K. et al. PD-1 immune checkpoint blockade reduces pathology and improves memory in mouse models of Alzheimer’s disease. Nat. Med. 22, 135–137 (2016).
Kumagai, S. et al. The PD-1 expression balance between effector and regulatory T cells predicts the clinical efficacy of PD-1 blockade therapies. Nat. Immunol. 21, 1346–1358 (2020).
Turley, S. J., Fletcher, A. L. & Elpek, K. G. The stromal and haematopoietic antigen-presenting cells that reside in secondary lymphoid organs. Nat. Rev. Immunol. 10, 813–825 (2010).
Kigerl, K. A., de Rivero Vaccari, J. P., Dietrich, W. D., Popovich, P. G. & Keane, R. W. Pattern recognition receptors and central nervous system repair. Exp. Neurol. 258, 5–16 (2014).
Huynh, T. V. et al. Lack of hepatic apoE does not influence early Aβ deposition: observations from a new APOE knock-in model. Mol. Neurodegener. 14, 37 (2019).
Noguchi, T. et al. Temporally distinct PD-L1 expression by tumor and host cells contributes to immune escape. Cancer Immunol. Res. 5, 106–117 (2017).
Wozniak, D. F. et al. Motivational disturbances and effects of L-dopa administration in neurofibromatosis-1 model mice. PLoS ONE 8, e66024 (2013).
Yuede, C. M. et al. Behavioral consequences of NMDA antagonist-induced neuroapoptosis in the infant mouse brain. PLoS ONE 5, e11374 (2010).
Khuchua, Z. et al. Deletion of the N-terminus of murine map2 by gene targeting disrupts hippocampal ca1 neuron architecture and alters contextual memory. Neuroscience 119, 101–111 (2003).
Hammond, T. R. et al. Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity 50, 253–271 (2019).
Acknowledgements
We thank X. Zhang and S. Li for advice on scRNA-seq analysis; J. Rustenhoven, B. Korin and A. Rolls for advice on meninges isolation; D. Bender for assistance with multiplex immune monitoring; D. Gate for advice on T cell immunohistochemistry on human samples; N. Saligrama for advice on TCR and antigen analysis; and M. Gratuze for PLX3397 drug formulation. We thank the Department of Pathology and Immunology Flow Cytometry and Fluorescence Activated Cell Sorting Core for help with cell sorting. This work was supported by a Carol and Gene Ludwig Award for Neurodegeneration Research (D.M.H.), National Institute of Health grant NS090934 (D.M.H.), the JPB Foundation (D.M.H.), Cure Alzheimer’s Fund (D.M.H.) and Rainwater Charitable Foundation (D.M.H.). M.F. was supported by the Ministry of Science and Higher Education of the Russian Federation (agreement no. 075-15-2022-301). Single-nucleus sequencing was carried out at the McDonnell Genome Institute. Confocal microscopic analyses were carried out at the Washington University Center for Cellular Imaging supported by Washington University School of Medicine, The Children’s Discovery Institute of Washington University and St Louis Children’s Hospital (CDI-CORE-2015-505 and CDI-CORE-2019-813) and the Foundation for Barnes-Jewish Hospital (3770 and 4642). We thank E. Reiman, G. Serrano and T. Beach for human brain tissue. The schematic representations of the fear conditioning behavioural paradigms in Extended Data Fig. 9i were created with BioRender.com.
Author information
Authors and Affiliations
Contributions
X.C. and D.M.H. conceived the study. X.C. designed the study, carried out the experiments, analysed the data and wrote the draft of the manuscript. M.M., M.B.F., J.H., J.K., X.B., I. Smirnov. and C.W. were blinded to sample IDs for brain volume and neurofilament light chain measurement. M.F., E.A., X.C., M.N.A. and J.D.U. analysed scRNA-seq data. X.C., H.H. and M.W.K. carried out microglia in vitro antigen presentation assay. X.C., and I. Shchukina carried out anti-PDCD1 acute treatment flow cytometry assay. C.M.Y. was blinded to sample IDs, and carried out and analysed the memory behavioural assay. D.M.H. supervised the overall research. X.C. and D.M.H. wrote the manuscript with comments from all authors.
Corresponding author
Ethics declarations
Competing interests
D.M.H. is an inventor on a patent licensed by Washington University to C2N Diagnostics on the therapeutic use of anti-tau antibodies. D.M.H. co-founded and is on the scientific advisory board of C2N Diagnostics. D.M.H. is on the scientific advisory board of Denali and Cajal Neuroscience and consults for Genentech and Alector. J.K. is a member of a scientific advisory group for PureTech. All other authors declare no competing interests.
Peer review
Peer review information
Nature thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 ApoE4 exacerbates tau-mediated neurodegeneration.
(a) Representative image of 9.5-month A/PE4 mouse brain sections stained with an anti-amyloid-Aβ (Aβ) antibody and X-34. Scale bar = 500 μm. (b) Representative images of 6 and 9.5-month TE4 mouse brain sections stained with AT8 antibody. Scale bar = 500 μm. (c) Quantification of brain regional volumes of 9.5-month mice normalized to 6-month in b. TE4-6 months: n = 7, TE4-9.5 months: n = 13. ***p < 0.0001 for hippocampus (Hip) vs. cortex dorsal to the hippocampus (Ctx); amygdala (Amyg) vs. Ctx and entorhinal/piriform cortex (Ent) vs. Ctx. One-way ANOVA with Tukey’s post hoc test. (d) Quantification of the area covered by AT8 of 9.5-month TE4 mouse brain sections in b. TE4-6 months: n = 7, TE4-9.5months: n = 13. ***p < 0.0001 for Hip vs. Ctx; Amyg vs. Ctx and Ent vs. Ctx. One-way ANOVA with Tukey’s post hoc test. (e) Representative images of 6-month TE4, 9.5-month E4, and 9.5-month TE4 mouse brain sections stained with NeuN. Scale bar = 50 μm. (f) Thickness of granule cell layer of the DG in 9.5-month E3, TE3, E4, TE4 mice. (E3: n = 5, TE3: n = 6, E4: n = 15 and TE4: n = 13). *p = 0.0130 for TE3 vs. TE4. Two-way ANOVA with Tukey’s post hoc test. (g) Correlation between DG neuronal layer thickness and hippocampal volume. n = 39 biologically independent animals from f. Pearson correlation analysis. R2 = 0.8335, p < 0.0001. (h, i) Representative images of 9.5-month E4 and TE4 mouse brain sections stained with MBP. Scale bar = 500 μm in h. Scale bar = 100 μm in i. (j-l) Volumes of hippocampus, entorhinal/piriform cortex and posterior lateral ventricle in 9.5-month E3, TE3, E4, TE4 mice. (E3: n = 5, TE3: n = 6, E4: n = 15 and TE4 = 13). p = 0.0505 for TE3 vs. TE4 in comparing the volume of the hippocampus, *p = 0.0207 for TE3 vs. TE4 in comparing the volume of the posterior lateral ventricle. Two-way ANOVA with Tukey’s post hoc test. (m—o) Volumes of hippocampus, entorhinal/ piriform cortex and posterior lateral ventricle in 9.5-month TE3, TE4, A/PE4, 5xE4 male and female mice. (TE3-M: n = 6, TE3-F: n = 11, TE4-M: n = 13, TE4-F: n = 17, A/PE4-M: n = 7, A/PE4-F: n = 6, 5xE4-M: n = 6, 5xE4-F: n = 7). **p = 0.0087 for TE4 male vs. female entorhinal/piriform cortex volume. Two-way ANOVA with Tukey’s post hoc test. Data are mean ± s.e.m.
Extended Data Fig. 2 Immune cell composition in brain parenchyma and meninges.
(a) FACS sorting of CD45Total and /or CD45high cells from brain parenchyma and meninges from A/PE4 mice for single cell immune RNA-seq. (b) Analysis of the CD4 and CD8 positive T cells present in the brain of E4 and TE4 mice by flow cytometry. (E4: n = 8, TE4: n = 14) Data are mean ± s.e.m., ***p < 0.0001, Unpaired two-tailed Student’s t test. (c) Representative cell type specific makers in brain parenchyma (CD45Total) clusters. (d) CD45high immune cells from parenchyma assigned into 12 cell types as visualized by UMAP plot. (e) Representative cell type specific makers in meninges (CD45Total).
Extended Data Fig. 3 T cell infiltration in the brain parenchyma with significant tauopathy.
(a) Representative flow cytometry gating plot of splenic lymphocytes. (b) Quantification of the proportion of indicated lymphocytes and their subsets among 9.5-month E4, A/PE4, and TE4 mice. (E4: n = 4, A/PE4: n = 4, TE4: n = 8). p = 0.648, 0.492, 0.992 for E4 vs. A/PE4; E4 vs. TE4; A/PE4 vs. TE4 in T cells; p = 0.614, 0.518, 0.089 for E4 vs. A/PE4; E4 vs. TE4; A/PE4 vs. TE4 in CD4+ T cells; p = 0.665, 0.719, 0.196 for E4 vs. A/PE4; E4 vs. TE4; A/PE4 vs. TE4 in CD8+ T cells. Two-way ANOVA with Tukey’s post hoc test. p = 0.629, 0.472, 0.095 for E4 vs. A/PE4; E4 vs. TE4; A/PE4 vs. TE4 in Treg. One-way ANOVA with Tukey’s post hoc test. (c) Quantification of CD3+ T cell number per DG area with 0.3 mm2 in 9.5 month E3, TE3, E4, TE4 mice. (E3: n = 5, TE3: n = 6, E4: n = 15 and TE4: n = 13). Two-way ANOVA with Tukey’s post hoc test. p = 0.1342 for TE3 vs. TE4. (d) Representative images of 9.5-month old E4, TE4, A/PE4 and 5xE4 mouse brain sections stained with CD3, Iba1, Aβ and X-34. Scale bar = 20 μm. Images are representative of results from n = 4 in E4, A/PE4 and TE4 respectively. (e) TEM image demonstrating presence of a cell with T cell like features in brain parenchyma of 9.5 month of TE4 mouse. Scale bar = 2 μm. Images are representative of results from n = 3 in TE4 mice.
Extended Data Fig. 4 Characterization of T cell populations within the parenchyma and meninges of mice with amyloid or tau pathology.
(a) Heatmap showing identified marker genes in each of the categorized cell types in Total T cells, CD4+ and CD8+ T cells. (b) Total T cells from brain parenchyma and meninges with TCR assigned into 13 cell types as visualized by UMAP plot. (c) TRAV and TRBV enrichment in CD8+ and CD4+ T cells in TE4 and A/PE4 mice. (d) Representative TCR-TRBV projection in E4, A/PE4 and TE4 mice.
Extended Data Fig. 5 Expression of cytokines, chemokines, growth factors and soluble receptors in brain lysates.
Quantification of cytokines, chemokines, growth factors and soluble receptors in brain lysates in 9.5 month old E4, TE4, and TEKO mice. (E4: n = 10, TE4: n = 10 and TEKO: n = 10). One-way ANOVA with Tukey’s post hoc test. With Q=0.1% identify outlier function, n = 1 E4 and n = 1 TEKO samples for IFN-γ measurements were removed; n = 1 E4 sample for IL-1β measurements was removed. ***p = 0.0001, ***p < 0.0001, ***p < 0.0001, ***p < 0.0001, p = 0.1261, ***p < 0.0001, **p = 0.0038, ***p = 0.0005, ***p < 0.0001, *p = 0.0144, **p = 0.0022, ***p < 0.0001, *p = 0.0126, ***p = 0.0002, ***p = 0.0001, **p = 0.0074, **p = 0.0059, *p = 0.0434, ***p < 0.0001, **p = 0.007, *p = 0.038, ***p < 0.0001 for E4 vs. TE4 following the panel order. p = 0.2757, ***p < 0.0001, ***p < 0.0001, ***p < 0.0001, *p = 0.489, ***p < 0.0001, p = 0.2539, p = 0.9952, **p = 0.0011, p = 0.053, n = 0.625, ***p < 0.0001, n = 0.7867, p = 0.1013, p = 0.087, p = 0.9905, p = 0.1107, p = 0.7159, p = 0.9193, p = 0.8426, p = 0.83, **p = 0.0076 for TE4 vs. TEKO following the panel order. Data are mean ± s.e.m.; One-way ANOVA with Tukey’s post hoc test.
Extended Data Fig. 6 Changes in microglia and T cells with Tau-meditated neurodegeneration require ApoE.
(a) MHCII, CD206, Iba1 and GFAP staining in 9.5-month TE4 mice. Scale bar = 100 μm. Images are representative of results from n = 13 in TE4 mice. (b) Iba1, MHCII and Aβ staining in 9.5 month E4, A/PE4, TE4 and TEKO mice in Cortex dorsal to Hippocampus, Hippocampus and Ent/Piri cortex. Scale bar = 50 μm. (c) Quantification of the area covered by MHCII in Ctx, Hip and Ent in 9.5-month TE4 and TEKO mice. (TE4: n = 13 and TEKO: n = 15). ***p < 0.0001 for TE4-Hip vs. TEKO-Hip. One-way ANOVA with Tukey’s post hoc test. (d) Iba1, CD11c and Aβ staining in 9.5-month E4, A/PE4, TE4 and TEKO mice in Prefrontal cortex, Hippocampus and Ent/Piri cortex. Scale bar = 50 μm. (e) Quantification of the area covered by CD11c in Ctx, Hip and Ent in 9.5-month TE4 and TEKO mice. (TE4: n = 13 and TEKO: n = 15). ***p < 0.0001 for TE4-Hip vs. TEKO-Hip. One-way ANOVA with Tukey’s post hoc test. (f) Volume of hippocampus in 9.5-month TE4 and TEKO mice. (TE4: n = 13 and TEKO: n = 15). ***p < 0.0001. Unpaired two-tailed Student’s t test. (g) Quantification of numbers of CD3+ T cells in DG per 0.3 mm2. (TE4: n = 13 and TEKO: n = 15). ***p < 0.0001. Data are mean ± s.e.m.; Unpaired two-tailed Student’s t test.
Extended Data Fig. 7 IFN-γ in the T cell population and microglia can directly present antigen to CD8+ T cells in vitro.
(a) Ligand-receptor analysis in T cells and microglia. (b) IFN-γ expression in brain parenchyma (CD45Total) 12 cell types and T cells from brain parenchyma 15 clusters of T cells as visualized by UMAP plot. (c) The gating strategy for sorting naïve OT-1 CD8+ T cells, dendritic cells (DCs), and microglia. (d) Representative flow cytometry plot to assess the proliferation of OT-1 T cells by cell tracer violet (CTV) dilution after 3 days of co-culture with APCs in the presence of OVA. (e) Representative flow cytometry plot showing dose dependent OVA antigen presentation by DCs, microglia, or microglia in the presence of IFNγ assayed by OT-1 proliferation. (f) Percent of proliferating OT-1 T cells under the indicated conditions. Data are from one representative experiment. Two independent experiments were done showing similar results.
Extended Data Fig. 8 Blocking IFNγ signaling reduces tau-mediated neurodegeneration and tau pathology.
(a) Representative images of 9.5-month TE3-IgG and TE3-αIFN-γ treated mouse brain sections stained with Sudan black. Scale bar = 1 mm. (b–d) Volumes of hippocampus, entorhinal/piriform cortex and posterior lateral ventricle in 9.5-month TE3-IgG and TE3-αIFNγ treated mice. (TE3-IgG: n = 12 and TE3-αIFNγ: n = 12). p = 0.0479, 0.0398, 0.265 for TE3-IgG vs. TE3-αIFNγ in comparing the volumes of hippocampus, entorhinal/piriform cortex and posterior lateral ventricle, respectively. Unpaired two-tailed Student’s t test. (e) CD11c, CD8, Iba1 staining in 9.5-month TE3-IgG and TE3-αIFNγ treated mice. Scale bar = 50 μm. (f) Quantification of area covered by CD11c in 9.5-month TE3-IgG and TE3-αIFNγ mice. (TE3-IgG: n = 12 and TE3-αIFNγ: n = 12). *p = 0.0344. Unpaired two-tailed Student’s t test. (g) Representative images of 9.5-month TE3-IgG and TE3-αIFNγ treated mouse brain sections stained with AT8 antibody. Scale bar = 250 μm. (h) p-Tau (AT8) covered area in 9.5-month TE3-IgG and TE3-αIFNγ treated mice. (TE3-IgG: n = 12 and TE3-αIFNγ: n = 12). *p = 0.0122. Unpaired two-tailed Student’s t test. (i) Schematic representation of the timeline of PLX3397 treatment for microglia depletion. (j) Schematic representation of the timeline of anti-CD4 and anti-CD8 antibody treatment for T cell depletion.
Extended Data Fig. 9 T cell depletion in tauopathy mice.
(a) Representative flow cytometry gating plot and quantification of CD4+, CD8+ T cells in brain parenchyma, meninges and blood in IgG control or α-CD4 and α-CD8 (αT) treated mice. (b) Bar plot showing the number of CD8+ T and CD4+ T cells in IgG and αT treated mice. (IgG: n = 2 and αT: n = 4). Data are mean ± s.e.m. (c) Microglia from brain parenchyma of TE4-IgG and TE4-αT treated mice assigned into 3 categories as visualized by UMAP plot. (d) Bar plot showing the percentage of the 3 categories of microglia in TE4-IgG and TE4-αT mice. (e) Heat map showing representative functional genes specifically expressed in active microglia clusters in TE4-IgG and TE4-αT mice. (f) Quantification of nest-building behavior at 9.5 months age. (TE4-IgG: n = 15 and TE4-αT: n = 21). *p = 0.02 for IgG vs. αT. Fisher’s exact test. (g) Quantification of total rearing at baseline levels of general exploratory behavior in 1 h. (TE4-IgG: n = 10 and TE4-αT: n = 15). p = 0.4363 for TE4-IgG vs. TE4-αT. Unpaired two-tailed Student’s t test. (h) Quantification of total ambulations at baseline levels of locomotor activity levels in 1 h. (TE4-IgG: n = 10 and TE4-αT: n = 15). p = 0.0709. Two-tailed Mann-Whitney test. (i) Schematic representation of fear conditioning behavioral paradigms. Schematics in i were created with BioRender.com.
Extended Data Fig. 10 Blocking PD-1 immune checkpoint increases Foxp3+CD4+ Tregs and reduces tau-mediated neurodegeneration.
(a) The gating strategy for sorting Foxp3+ CD4+ Treg, Pd1+ Foxp3+ CD4+ Treg, Klrg1+ effector CD8+ T cells, PD-1+ Tox1+ CD8+ exhausted T cells in the brain parenchyma. (b–e) Quantification of T cell populations in the brain parenchyma of mice acutely treated with IgG control and αPD-1 antibodies. (TE4-IgG: n = 12 and TE4-αPD-1: n = 13 for b, d, e; TE4-IgG: n = 10 and TE4-αPD-1: n = 11 for c). *p = 0.041, **p = 0.0075, p = 0.52, p = 0.945 for Foxp3+CD4+ Treg, Pd1+ Foxp3+ CD4+ Treg, Klrg1+ effector CD8+ T cells, PD-1+ Tox1+ CD8+ exhausted T cells. Unpaired two-tailed Student’s t test. (f) Representative images of 9.5-month TE4-IgG and TE4-αPD-1 treated mouse brain sections stained with Sudan black. Scale bar = 1 mm. (g-h) Volumes of hippocampus, entorhinal/piriform cortex in 9.5-month TE4-IgG and TE4-αPD-1 treated mice. (TE4-IgG: n = 14 and TE4-αPD-1: n = 14). *p = 0.018 and 0.015 for TE4-IgG vs. TE4-αPD-1 in comparing the volumes of hippocampus, entorhinal/piriform cortex, respectively. Unpaired two-tailed Student’s t test. (i) Quantification of the area covered by AT8 in DG per slice in 9.5-month TE4-IgG vs. TE4-αPD-1 mice (TE4-IgG: n = 14 and TE4-αPD: n = 14). **p = 0.0009. Unpaired two-tailed Student’s t test.
Supplementary information
Supplementary Video 1
CD3 and IBA1 staining in hippocampus of TE4 mice. IBA1 (red) and CD3 (green) staining in 9.5-month-old TE4 mice with tau pathology in DG. Scale bar, 10 μm.
Supplementary Video 2
CD11c, CD8 and IBA1 staining in hippocampus of TE4 mice. IBA1 (blue), CD11c (red) and CD8 (green) staining in 9.5-month-old TE4 mice with tau pathology in DG. Scale bar, 15 μm.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, X., Firulyova, M., Manis, M. et al. Microglia-mediated T cell infiltration drives neurodegeneration in tauopathy. Nature 615, 668–677 (2023). https://doi.org/10.1038/s41586-023-05788-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-023-05788-0
- Springer Nature Limited
This article is cited by
-
Therapeutics for neurodegenerative diseases by targeting the gut microbiome: from bench to bedside
Translational Neurodegeneration (2024)
-
Unraveling the dual nature of brain CD8+ T cells in Alzheimer’s disease
Molecular Neurodegeneration (2024)
-
ANU-ADRI scores, tau pathology, and cognition in non-demented adults: the CABLE study
Alzheimer's Research & Therapy (2024)
-
LRRK2 kinase inhibition reverses G2019S mutation-dependent effects on tau pathology progression
Translational Neurodegeneration (2024)
-
CD4 T-cell aging exacerbates neuroinflammation in a late-onset mouse model of amyotrophic lateral sclerosis
Journal of Neuroinflammation (2024)