Abstract
The Tripartite motif-containing 28 (TRIM28) transcriptional cofactor is significantly upregulated in high-grade and metastatic prostate cancers. To study the role of TRIM28 in prostate cancer progression in vivo, we generated a genetically-engineered mouse model, combining prostate-specific inactivation of Trp53, Pten and Trim28. Trim28 inactivated NPp53T mice developed an inflammatory response and necrosis in prostate lumens. By conducting single-cell RNA sequencing, we found that NPp53T prostates had fewer luminal cells resembling proximal luminal lineage cells, which are cells with progenitor activity enriched in proximal prostates and prostate invagination tips in wild-type mice with analogous populations in human prostates. However, despite increased apoptosis and reduction of cells expressing proximal luminal cell markers, we found that NPp53T mouse prostates evolved and progressed to invasive prostate carcinoma with a shortened overall survival. Altogether, our findings suggest that TRIM28 promotes expression of proximal luminal cell markers in prostate tumor cells and provides insights into TRIM28 function in prostate tumor plasticity.

Similar content being viewed by others
Introduction
While potent androgen receptor (AR)-targeting drugs have improved survival for patients with castration-resistant prostate cancer (CRPC), prostate tumor heterogeneity and plasticity remain significant clinical challenges [1]. Recent findings from single-cell RNA sequencing of mouse and human prostate have provided insights into the heterogeneity of luminal cell types in normal prostate cells [2,3,4,5]. Additional knowledge of luminal cell heterogeneity in prostate cancers, which generally display a luminal phenotype, and the key transcriptional regulators that promote prostate cancer progression and plasticity are needed.
The tripartite motif-containing 28 (TRIM28/TIF1b/KAP1) protein is a transcriptional coregulator proposed as a therapeutic target for CRPC [6]. TRIM28 is significantly upregulated in metastatic CRPC and high-grade localized prostate cancers, and knockdown of TRIM28 reduces prostate xenograft growth [6]. Furthermore, overall survival is significantly reduced in prostate cancer patients with high TRIM28 expression [6].
TRIM28 is a corepressor protein for over 300 transcription factor proteins in the Kruppel-associated box (KRAB) zinc finger protein family. KRAB zinc finger proteins are conserved only in tetrapod genomes and function as epigenetic transcriptional regulators by recruiting TRIM28, histone methyltransferase SETDB1, HP1, and histone deacetylases. TRIM28 regulates mouse embryonic development [7], genomic imprinting [8,9,10], self-renewal of embryonic stem (ES) cells [11], differentiation of cells including blood cells [12], muscle [13] and naïve T cells [14], and has been identified as a chromatin reader that sustains oncogenesis [15]. In breast cancers, TRIM28 is upregulated and has been proposed to regulate breast cancer stem cells [16,17,18].
Independent findings from three labs indicate pro-tumorigenic functions for TRIM28 in human prostate cancers [6, 19, 20]. These studies showed that TRIM28 interacts with AR in prostate cancer cell lines and enhances AR signaling, and that TRIM28 also prevents degradation of TRIM24 by SPOP. However, in vivo studies on TRIM28 in prostate cancers have been limited to work using xenograft models using immune-compromised mice. In this study, we investigated the effect of Trim28 inhibition in a genetically-engineered mouse model of prostate cancer and show that TRIM28 promotes the expression of proximal luminal lineage cell markers in prostate tumor cells.
Results
To assess the effect of Trim28 inhibition in prostate cancers, we used the genetically-engineered mouse prostate cancer model NPp53 (Nkx3.1-CreERT2/+; Ptenf/f; p53f/f), which lacks Pten and p53 tumor suppressor proteins in prostate epithelial cells [21]. We selected the NPp53 model for our studies as co-occurrence of PTEN and TP53 mutations are observed in approximately 20–30% of advanced prostate cancer patients [22]. We further inhibited Trim28, generating Nkx3.1-CreERT2/+; Ptenf/f; p53f/f;Trim28f/f (NPp53T) mice.
To determine TRIM28 function in prostate cancers in vivo, we compared NPp53 prostates to NPp53T prostates with Trim28 inhibition (Fig. 1). We induced tumor formation in adult mice and dissected prostates 3 months later (Fig. 1A). Simultaneously, we performed lineage tracing in NPp53 and NPp53T prostates using an R26R-YFP reporter allele, which is expressed upon activation of Nkx3.1-CreERT2. In control NPp53 prostates, all YFP positive (YFP+) cells expressed TRIM28 (Fig. 1B, D). In contrast, in NPp53T prostates, 90% of YFP+ cells were TRIM28 negative (-), indicating a high efficiency of TRIM28 inhibition (Fig. 1B–D, Table S2).
A Genotypes and experimental timeline. B–D Immunofluorescence staining for (B, C) YFP, TRIM28 and DAPI; and (D) quantification (n = 3 mice per genotype). Arrowhead points to a YFP+ cell with TRIM28 expression from an NPp53 prostate. Arrow points to a YFP+ epithelial cell lacking TRIM28 expression from an NPp53T prostate. E, F H&E staining. G, H Immunofluorescence staining for epithelial marker E-cadherin, T cell marker CD3 and DAPI. I–K Immunofluorescence staining for (I, J) YFP, proliferation maker KI67 and DAPI; and (K) quantification (n = 3 mice per genotype). L–O Immunofluorescence staining for YFP, apoptosis maker active caspase 3 (CASP3) and DAPI at (L,M) low and (N,O) high magnification; and (P, Q) YFP, luminal cell marker KRT8, basal cell marker KRT5 and DAPI (R, S) Immunohistochemical staining for NKX3.1. Nuclei were stained with Hematoxylin. *Stromal cells in NPp53T tumors. Scale bars represent 50 mm. Error bars represent standard deviation from the mean. P values were calculated using an unpaired Student’s t test. ****p < 0.0001. See also Fig. S1 and Table S2.
Trim28 inhibition in NPp53 prostates promotes inflammation and apoptosis
By 3 months after tumor induction, NPp53 prostates developed high-grade prostatic intraepithelial neoplasia (PIN) with a cribriform pattern, focal invasion and nuclear atypia (Figs. 1E, S1A, C) [21]. In age-matched NPp53T prostates, in addition to high-grade PIN lesions, we detected necrotic lumens with increased stromal cells surrounding prostate ducts (Figs. 1F, S1B, D). Furthermore, CD3 lymphocyte infiltration was observed in NPp53T prostates suggesting an inflammatory response to Trim28 inhibition (Fig. 1G, H). This finding was unexpected, as prostate cancers are immunologically “cold” with few tumor-infiltrating T cells [23, 24].
To understand how Trim28 inhibition affects the NPp53 tumor phenotype, we analyzed prostates at an earlier stage of tumor progression. We confirmed the high efficiency of TRIM28 inhibition in NPp53T prostates 1 month following tamoxifen treatment (Fig. S1E–G). NPp53 and NPp53T prostate tissues could be distinguished histologically as early as 1 month after tamoxifen treatment, as necrotic lumens were more prominent in NPp53T prostates (Fig. S1H, I; n = 4 prostates). Thus, we focused our analysis on tissues at this stage of tumor progression.
To determine if the increase in lymphocyte infiltration that we observed in NPp53T prostates compared to NPp53 prostates at 3 months after tamoxifen treatment arises during early stages of tumor progression (Fig. 1G, H), we characterized immune cell types in NPp53 and NPp53T whole prostate tissues 1 month after tamoxifen treatment. We compared NPp53 to NPp53T prostates using flow cytometry. In both NPp53 and NPp53T prostates, many CD45+ immune cells were macrophages [25]. The percentage of CD45+ cells that were macrophages (37% ± 6.3% vs. 34% ± 4.4%; n = 5 mice), neutrophils (24% ± 14.3% vs. 10% ± 1.3%; n = 5 mice) and T cells (7% ± 2.4% vs. 10% ± 2.4%; n = 5 mice) did not differ significantly (Fig. S1N–P). These findings suggest that Trim28 inhibition in epithelial cells does not immediately illicit an immune response.
Using flow cytometry analysis, we quantified the percentage of YFP+ cells in NPp53 and NPp53T prostates 1 month after tamoxifen treatment. We observed a slight reduction in the percentage of YFP+ cells in NPp53T prostates compared to NPp53 prostates (Fig. S1K–M).
To determine the effects of Trim28 inhibition on cell proliferation in vivo, we assessed KI67 expression in YFP+ prostate epithelial cells 1 month after tamoxifen treatment (Fig. 1I, J). The percentage of YFP+ KI67+ cells was similar between NPp53 and NPp53T prostates (14 ± 6.5% and 10.7 ± 2.5% respectively; n = 3 mice each; Fig. 1I–K; Table S2). This suggests that at 1 month after tumor induction, Trim28 inhibition in NPp53T prostates does not affect epithelial cell proliferation.
We then assessed for apoptosis by staining for active Caspase 3. Cells with active Caspase 3 were rare in epithelial layers of both NPp53 and NPp63T prostate tissues. However, high expression of active Caspase 3 was detected in lumens of NPp53T prostates (Fig. 1L–O), suggesting that Trim28 inhibition in NPp53T prostates promotes apoptosis.
Prostate tumor cells in hormonally-intact genetically-engineered mouse models with inactivation of Tp53 and Pten in Nkx3.1-expressing cells generally express luminal markers [21]. We confirmed that YFP+ cells in NPp53 and NPp53T prostates expressed high levels of luminal cytokeratin marker KRT8; a few YFP+ cells expressed basal cytokeratin marker KRT5 (Figs. 1P, Q, S1Q, R). Androgen receptor (AR) was expressed in both NPp53 and NPp53T prostates (Fig. S1S, T).
NKX3.1 is an AR target and a prostate luminal cell marker expressed in the wild-type mouse prostate. However, NKX3.1 expression is lost in mouse PIN lesions and cancers in Nkx3.1+/−; Pten+/− [26] and Myc transgenic animals [27]. Unexpectedly, we found differences in NKX3.1 expression in NPp53 and NPp53T prostates, with reduced NKX3.1 expression in control NPp53 prostates but not in NPp53T prostates (Fig. 1R, S).
Single-cell RNA sequencing suggests TRIM28 is required in proximal luminal phenotype cells
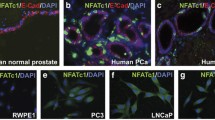
Recent single-cell RNA sequencing studies of the normal mouse prostate have identified a distinct proximal luminal lineage of cells enriched in proximal prostate and distal epithelial invagination tips that are conserved in human and mouse prostates (also known as LumP/L2/Luminal C [2,3,4,5]). In mice, proximal luminal cells are specified early during prostate organogenesis and remain distinct from other luminal cell types [4, 28]. However, in organoid culture and tissue reconstitution assays, proximal luminal lineage cells display enhanced progenitor properties [29]. Furthermore, proximal luminal-like cells are present in human prostate cancers and in mouse models of prostate cancer [2, 30].
To assess the effect of Trim28 inhibition on heterogeneous prostate tumor cell types, we conducted single-cell RNA sequencing of NPp53 and NPp53T prostates 1 month after tamoxifen induction (Figs. 2, S2). Using unsupervised clustering analysis and UMAP visualization, we confirmed the generation of gene expression profiles from distinct cell types and identified luminal cell clusters (Krt8; Fig. S2A–C).
A Experimental timeline. B, C Aggregated UMAP visualization of single-cell RNA sequencing gene expression profiles from YFP+ NPp53 and NPp53T prostate cells. Blue color in (C) indicates expression level of Nkx3.1 and Pbsn, Ppp1r1b and Krt4, Krt5 and Slc12a8 genes. D Bubble plot showing expression of proximal luminal marker genes in YFP+ cells from NPp53 and NPp53T prostates. See also Fig. S2.
To compare lineage-traced cells from NPp53 and NPp53T prostates, we separated YFP+ cells (Fig. S2B, D). We conducted an unsupervised clustering analysis of gene expression profiles from YFP+ lineage-traced cells and identified 6 clusters of cells (Fig. 2B, Supplementary File 1). These clusters included a luminal cell cluster expressing Nkx3.1 and Pbsn resembling luminal cells from anterior or dorsal prostate lobes (Luminal A/D cells), a luminal cell cluster expressing Ppp1r1b and Krt4 resembling proximal luminal cells, two smaller luminal cell clusters expressing luminal ventral or lateral prostate lobe markers (Luminal V/L), and a cluster of basal cells expressing Krt5 (Figs. 2B, C, S2F). We also identified a small cluster of cells expressing goblet cell markers, previously reported in genetically-engineered mouse models of prostate cancer (Figs. 2B, C, S2F) [31]. Surprisingly, we found many YFP+ cells from NPp53 prostates clustered with proximal luminal-like cells, whereas this population was reduced in Trim28 inhibited NPp53T prostates (Fig. 2B). In contrast, a higher percentage of YFP+ cells from NPp53T prostates resembled Luminal A/D cells.
We performed differential gene expression analysis on lineage-labeled YFP+ cells from NPp53 and NPp53T samples (n ≥ 800 cells per sample). We found expression of proximal luminal markers Krt4, Psca, Krt7, Ppp1r1b, Runx1 and Tacstd2 were significantly reduced in YFP+ cells from NPp53T prostates (Fig. 2B–D, Supplementary File 2). Other top downregulated genes in NPp53T YFP+ cells included genes that regulate cancer stem/progenitor cells (Tspan8, Prom1; [32,33,34], Supplementary File 2). Top overexpressed genes in NPp53T YFP+ cells included genes involved in immune cell regulation (Stfa1, Ptpn22; [35, 36]) and apoptosis (Bcl2l15). Notably, the most differentially upregulated gene in Trim28-deleted cells was Bglap3 (Supplementary File 2), which is regulated by TRIM28 and histone methyltransferases including SETDB1 in ES cells [37, 38]. We confirmed the aberrant BGLAP protein overexpression in NPp53T prostate tissues (Fig. S3A, B).
Trim28 promotes expression of proximal luminal cell markers
To further study the reduction in cells expressing proximal luminal markers identified by single-cell RNA sequencing of NPp53T prostates, we conducted immunostaining analysis (Figs. 3, S3). In addition to marking proximal luminal cells, Ppp1r1b expression has been reported in normal prostate neuroendocrine cells [5]. Thus, we evaluated Synaptophysin expression in NPp53 and NPp53T tissues to determine if Trim28 inhibition affects neuroendocrine differentiation. Rare YFP+ cells that expressed Synaptophysin were detected in NPp53 prostates, but not in NPp53T prostates (Fig. S3C, D). Although NPp53 prostates undergo transdifferentiation to neuroendocrine tumors when castrated and treated with abiraterone [21], most cells in hormonally-intact NPp53 and NPp53T tissues did not display characteristics of neuroendocrine cells. This was also confirmed by analysis of the expression of synaptophysin (Syp) and chromogranin A (Chga) neuroendocrine markers in YFP+ cells by single-cell RNA sequencing (Fig. S2F).
A, C–E, G–I Immunofluorescence staining for YFP, proximal luminal marker PPP1R1B and DAPI and (F) quantification of YFP+PPP1R1B+ cells. Arrow in (H) points to a cluster of YFP-negative PPP1R1B+ cells. B Genotypes and experimental timeline. J–M Immunofluorescence staining for YFP, PPP1R1B, KI67 and DAPI and (K, M) quantification (n = 3 mice per genotype for each timepoint). Values were compared only within each timepoint. Error bars represent standard deviation from the mean. P values were calculated using an unpaired Student’s t test. **p < 0.01; ****p < 0.0001. N, O NPp53 and NPp53T prostate epithelial organoids after 10 days of culture. P, Q PPP1R1B immunohistochemical staining of passage 5 organoids. Nuclei were stained with Hematoxylin. Scale bars represent 50 μm. See also Figs. S2, S3, S5 and Table S2.
In the wild-type mouse prostate, NKX3.1-expressing cells are distinct from PPP1R1B expressing proximal luminal lineage cells [2,3,4,5]. Thus, as expected, in Nkx3.1-CreERT2/+; R26R-YFP animals, we did not detect PPP1R1B expression in YFP+ cells (Fig. 3A, Table S2). To investigate the shift of NKX3.1-expressing cells to cells with PPP1R1B expression upon inactivation of Pten and p53, we stained NPp53 prostates at 2 weeks, 1 month and 3 months after tamoxifen treatment. In NPp53 prostates, the percentage of YFP+ PPP1R1B+ cells gradually increased over time (38.2 ± 1.8% of YFP+ cells at 2 weeks following tamoxifen treatment to 65 ± 4.3% of YFP+ cells at 3 months; Fig. 3B–F, Table S2, n = 3 mice). In NPp53 prostates, PPP1R1B+ cells were frequently detected near the lumens of prostate ducts (Fig. S3E). In contrast, in NPp53T prostates with Trim28 inactivation, the percentage of YFP+ PPP1R1B+ cells remained lower, at 28 ± 6.1% of YFP+ cells at 3 months following tamoxifen induction (Fig. 3F–I, Table S2, n = 3 mice). In NPp53T prostates, non-transformed PPP1R1B+ cells without YFP expression were also observed (Figs. 3H, S3F). Expression of additional proximal luminal markers including KRT4 and TACSTD2 was also reduced in live cells of NPp53T prostates (Fig. S3G–J), though cells retained expression of the HOXB13 prostate marker (Fig. S3K, L).
Despite differences in PPP1R1B expression, the overall proliferation of lineage-marked YFP+ cells was similar in NPp53 and NPp53T prostates (Fig. 1I–K, Table S2). We hypothesized that in NPp53 prostates, an increase in proliferation rate in PPP1R1B+ cells could contribute to a shift in tumor phenotype to a higher number of PPP1R1B+ cells over time. Thus, we compared KI67 expression between YFP+ PPP1R1B- and PPP1R1B+ cells but did not detect significant differences in proliferation rates between PPP1R1B+ and PPP1R1B- cells in NPp53 or NPp53T prostates (Fig. 3J–M, Table S2).
To investigate the cell-autonomous effects of Trim28-deletion in YFP+ NPp53 prostate epithelial cells, we isolated EpCAM+ YFP+ cells using fluorescent activated cell sorting (FACS) and generated organoids (Fig. S4A–D) [39]. The growth of NPp53T organoids from sorted YFP+ cells was similar to control NPp53 organoids (Figs. 3N, O, S4E–J). Interestingly, PPP1R1B expression in organoids resembled prostate tissues in vivo, with higher PPP1R1B expression in NPp53 organoids compared to NPp53T organoids (Fig. 3P, Q).
In contrast, when we knocked out Trim28 in NPp53 organoids expressing high levels of PPP1R1B, we could not establish NPp53T organoid lines (Fig. S4K, n = 2 experiments). This finding, along with the increased apoptosis we observed in NPp53T prostate lumens in vivo suggests a differential requirement for TRIM28 in PPP1R1B+ cells that is not present in PPP1R1B- cells.
Altogether, as Nkx3.1-CreERT2 is not expressed in proximal luminal cells before tumor induction (Fig. 3A) these observations suggest that TRIM28 promotes the expression of proximal luminal cell markers in NKX3.1-expressing tumor cells, and that the increase in the percentage of PPP1R1B expressing cells over time is not due to significant differences in proliferation rates between PPP1R1B+ and PPP1R1B− cells. We have further found that this plasticity is not dependent on the genetic inactivation of p53, as the shift to a proximal luminal tumor phenotype occurred in Nkx3.1-CreERT2/+; Ptenf/f; Trim28f/+ (NP), but not Nkx3.1-CreERT2/+;Ptenf/f;Trim28f/f (NPT) prostates (Fig. S5). Our findings are consistent with a recent report demonstrating cellular plasticity in prostates of PSA-CreERT2; Ptenf/f mice [30]. As Hypoxia-inducible factor 1 alpha (HIF1A) signaling in luminal cells promotes luminal cell plasticity [30], we also examined HIF1A expression in NP and Trim28 inhibited NPT prostates and found that HIF1A expression was generally reduced in epithelial cells of NPT tumors (Fig. S5E–H). These findings reveal new functions of TRIM28 in promoting prostate tumor plasticity.
In adult mouse prostates, we confirmed high levels of TRIM28 expression in CK8-expressing luminal prostate epithelial cells, as reported in human prostate luminal cells [19] (Fig. S6A). TRIM28 expression was also detected in CK5-expressing basal prostate epithelial cells (Fig. S6B). Similarly, in normal human prostate tissues, TRIM28 mRNA is ubiquitously expressed across epithelial cell types (Fig. S7A) [3]. To assess TRIM28 function in normal adult prostate luminal cells, we treated adult Nkx3.1-CreERT2/+; Trim28f/f; R26R-YFP mice with tamoxifen (Fig. S6C). This deletion of TRIM28 in adult luminal cells did not affect prostate morphology or weight (Fig. S6C–F), indicating that prostate-specific deletion of TRIM28 appears well-tolerated in normal adult prostate luminal cells expressing NKX3.1.
Trim28 inhibition in NPp53 prostates reduces overall survival
Despite the reduction of proximal luminal-like cells, NPp53T prostates surprisingly progressed to invasive prostate carcinoma by 3 months after tumor induction (Fig. 4). Overall survival in NPp53T mice was significantly reduced compared to control NPp53 mice, and restricted to a median survival of 122 days after tumor induction (Fig. 4B, n = 7 NPp53T mice).
A Experimental timeline. B Kaplan-Meier survival analysis of animals after tamoxifen treatment. P value for the difference in the percent survival of NPp53T compared to NPp53 mice was calculated by log-rank test. C Total prostate weight after tamoxifen induction. Each dot represents a single prostate. n = 26 NPp53, n = 19 NPp53T mice. Exponential growth curves are shown. D, E Whole mount images of NPp53 prostates (D) 3 months and (E) 8 months after tamoxifen induction and (F) immunohistochemical staining for YFP. G, H Whole mount images of NPp53T prostates (G) 3 months and (H) 4 months after tamoxifen induction and (I, J) immunohistochemical staining for YFP in (I) prostate tissue, including in stromal regions (arrowhead), and (J) a metastatic lymph node. Immunofluorescence staining for (K, L) YFP, TRIM28 and DAPI; (M–O) YFP, proliferation marker KI67 and DAPI. Arrows point to proliferating YFP+KI67+ cells in stromal regions of NPp53T prostates. P, Q Immunohistochemical staining for α smooth muscle actin (SMA). R, S Masson’s Trichrome Staining showing blue Aniline Blue collagen staining in NPp53T stroma. SV seminal vesicle. Scale bars represent 500 μm in (D, E, G, H); 50 μm in all other panels.
Comparison of whole prostate weights revealed exponential growth of NPp53T prostates at approximately 100-days after tumor induction (Fig. 4C–E, G, H; n = 26 NPp53, n = 19 NPp53T mice). In late-stage NPp53T mice, lineage-marked YFP+ cells with sarcomatoid features were detected in prostate stromal regions (Fig. 4I). We confirmed that these YFP+ cells remained negative for TRIM28 (Fig. 4K, L). However, when stained for the KI67 proliferation marker, we detected KI67 expression in NPp53T YFP+ stromal cells, suggesting that the proliferation of these cells contributed to the tumor mass (Fig. 4M–O). Enlarged lymph nodes were also common in late-stage NPp53T mice (n = 3 out of 4 NPp53T mice); we confirmed YFP expression, indicating metastasis of prostate cancer cells to lymph nodes (Fig. 4J). Although metastases to distant organs such as lungs or liver were not detected, NPp53T mice succumbed to bladder outlet obstruction. In contrast, age-matched NPp53 mice were viable even at 250 days after tumor induction with only a gradual increase in prostate weight (Fig. 4B, C).
We found that alpha-smooth muscle actin (ACTA2) expression was discontinuous in NPp53T prostates, suggesting remodeling of the basement membrane surrounding prostate ducts and progression to invasive carcinoma (Fig. 4P, Q) [31]. To assess the collagen content of tumors, we conducted Masson’s Trichrome staining. We observed an increase in blue collagen fiber staining in stromal regions of NPp53T prostates compared to age-matched control NPp53 prostates (Fig. 4R, S).
The reduced survival and increase in prostate weights in NPp53T prostates were surprising, given previous work on reduced cell proliferation upon Trim28-deletion in prostate cancer cell lines and cell-line derived xenograft models [6]. Our observations indicate that although Trim28 inhibition reduces the expression of proximal luminal markers and promotes apoptosis and lymphocyte infiltration, NPp53T tumors evolve and rapidly progress to invasive prostate carcinoma. In human prostate cancers, TRIM28 genetic alterations and mRNA expression changes occur in approximately 5–20% of patients, independent of PTEN and TP53 alterations (Fig. S7B–E; no significant correlation or mutual exclusivity). Although TRIM28 overexpression has been associated with prostate tumor progression [6], low TRIM28 expression was also observed in a few advanced prostate cancer cases (Fig. S7C–E).
Long-term Trim28 inhibition promotes a pro-tumorigenic immune microenvironment
To further understand how long-term Trim28 inhibition affects the tumor microenvironment, we characterized immune cells 3 months after tumor induction. Although the percentage of CD45+ immune cells (27.7% ± 3.3% vs. 23% ± 3.6%), myeloid cells (74.8% ± 3.3% vs. 75.4% ± 19%) and neutrophils (37.8% ± 7.7% vs. 30% ± 9.5%) were similar between NPp53 (n = 5 prostates) and NPp53T tumors (n = 4 prostates), the percentage of macrophages was slightly reduced in Trim28-deleted tumors (Fig. 5A; 30.1% ± 5.1% vs. 19.9% ± 6.5%; p = 0.04). Since tumor-associated macrophages influence human prostate cancer progression [40], we further analyzed macrophage markers. Macrophages expressing the M2 macrophage marker CD206 were rare at 1 month after tumor induction (Fig. S1N, O). However, at 3 months after tumor induction, a higher percentage of macrophages in Trim28 inhibited NPp53T prostates expressed the M2 macrophage marker CD206 compared to NPp53 prostates (54.4% ± 7.2% vs. 22.3% ± 3.7%; %; p < 0.0001) (Fig. 5B, C), suggestive of a pro-tumorigenic microenvironment in NPp53T tumors. Interestingly, the CD206+ macrophages also co-expressed the M1 marker CD80 (Fig. 5D).
A–F Immunophenotyping of NPp53 and NPp53T prostates 3 months after tumor induction. A Quantification of immune (CD45+), myeloid (CD11b+), macrophage (F4/80+CD11b), neutrophil (Ly6G+) and T cells (CD3+CD11b+). B, C Representative plot showing gating (B) and quantification (C) of CD206+ macrophages. D CD80 and CD206 expression in F4/80+ macrophages. E, F Representative plots showing gating for CD8 and CD4 T cells (E) and quantification (F). n = 4–5 mice per genotype. Error bars represent standard deviation from the mean. P values were calculated using an unpaired Student’s t test. *p < 0.05; ****p < 0.0001.
Although we frequently observed clusters of CD3+ T cells in NPp53T prostates (Fig. 1E–H), the percentage of CD3+ total T cells (13.7% ± 2.0% vs. 13.7% ± 6.3%) and CD8+ cytotoxic T cells (35.9% ± 6.8% vs. 43.5% ± 5%) were similar between NPp53 and NPp53T tumors (Fig. 5A). However, CD4+ regulatory T cells were significantly more prominent in Trim28 inhibited NPp53T tumors (19.2% ± 2.8% vs. 29.4% ± 7.6%) (Fig. 5E, F). These findings suggest a shift to an immune suppressive and tumor-promoting microenvironment in Trim28 inhibited NPp53T tumors by 3 months after tumor induction leading to an aggressive tumor phenotype.
NPp53T tumors are castration-resistant and remain phenotypically distinct from NPp53 tumors after androgen deprivation
To assess the effects of Trim28 inhibition in CRPC, we castrated NPp53 and NPp53T mice one month after tumor induction (Fig. 6A). We observed no significant differences in whole prostate weights of castrated NPp53 mice compared to NPp53T mice at 2 or 3 months post-induction, though tumor weights were variable in castrated NPp53T mice at 3 months post-induction (Fig. 6B, C). Nuclear AR expression was absent after castration (Fig. 6D, E). Similar to prostates from hormonally-intact NPp53T mice, prostates from castrated NPp53T mice had high-grade PIN lesions with increased stromal cells surrounding prostate ducts and increased lymphocytes (Fig. 6F, G).
A Experimental timeline. B, C Total prostate weight of castrated NPp53 and NPp53T mice at (B) 2 months and (C) 3 months after tumor induction. Each dot represents a single prostate. n = 4 or 5 mice per genotype. ns not significant. D, E Immunofluorescence staining for YFP and AR. Nuclei were stained with DAPI. F, G H&E staining. *indicates stromal cells. H, I Immunohistochemical staining for PPP1R1B. Nuclei were stained with hematoxylin. Scale bars represent 50 μm.
We detected high expression of the proximal luminal marker PPP1R1B in prostates from castrated NPp53 mice, consistent with results in hormonally-intact NPp53 mice (Fig. 6H). In contrast, PPP1R1B expression was reduced in prostates from castrated NPp53T mice at 2 months post-induction (Fig. 6I). The variation observed in weights at 3 months post-induction indicates that further studies are needed to understand how these tumors progress. However, these data suggest androgen-independent functions of TRIM28 in prostate cancer. Additionally, we were unable to grow human androgen-independent PC-3 cells when we knocked out TRIM28 (Fig. S4L), providing further evidence for cell-autonomous androgen-independent functions of TRIM28 in prostate cancer cells.
Discussion
Our study demonstrates that inhibition of Trim28 in prostate tumors reduces the expression of proximal luminal cell markers and promotes cell death. However, NPp53T tumors progress over time to invasive prostate carcinoma with a pro-tumorigenic immune cell phenotype and reduced overall survival. Our finding that Trim28 inhibition resulted in larger tumors with shortened survival was unexpected, as knockdown of Trim28 in organoid culture or using cell-line derived xenograft models leads to reduced tumor weights in immune-compromised mice [6]. However, our findings from immune-competent genetically-engineered mice provide insights into human prostate cancer.
Our findings indicate that TRIM28 promotes a proximal luminal cell phenotype. We demonstrate through lineage-tracing studies that NKX3.1-expressing cells gain proximal luminal marker expression when Pten and p53 are inhibited. In contrast, inhibition of Trim28 led to a significant decrease in PPP1R1B, KRT4 and TACSTD2 proximal luminal marker expression. A previous study showed that proximal luminal cells located in proximal and distal regions of prostate ducts can be cellular origins for prostate cancer and identified analogous cells in human prostate cancers [2]. Using a Tmprss2-CreER; Ptenf/f model with Cre expression in all luminal cells including proximal luminal cells, this study also found that non-proximal luminal cells acquire proximal luminal markers upon Pten deletion [2]. Our study uses genetic lineage-tracing of NKX3.1-expressing cells to definitively demonstrate that tumors arising from non-proximal luminal cells can acquire expression of proximal luminal markers. Additionally, we show that TRIM28 has a key function in promoting these changes.
Recent findings indicate that hypoxia and hypoxia-inducible factor 1 alpha (HIF1A) signaling promote tumor cell plasticity and progression of prostate intraepithelial neoplasia in a Pten inhibited mouse model of prostate cancer [30]. As TRIM28 has been found to interact with HIF-1 and promote HIF-1 transcriptional activity in response to hypoxia in breast cancer cell lines [41], it is possible that TRIM28 may similarly promote HIF1A signaling in prostate cancer. The increased apoptosis we observed in lumens of NPp53T prostates compared to NPp53 prostates resembles the phenotype observed upon Hif1a and Pten deletion in luminal cells [30]. Further studies are needed to characterize the mechanism in which TRIM28 may promote HIF1A signaling, and thus increase cell survival of hypoxic tumors.
Using single-cell RNA sequencing data, we identified Bglap3, which is unique to mouse genomes, as a top overexpressed gene in NPp53T prostates. Since Bglap3 is not conserved in the human genome, its overexpression upon Trim28 inhibition may be specific to the mouse. In mouse ES cells, expression of Bglap3 and an IAP-class endogenous retroviral element (ERV) in the promoter region of Bglap3 are epigenetically regulated by TRIM28 [37]. In our study, the association of increased Bglap3 expression and prostate cancer progression in NPp53T tissues is intriguing, as BGLAP, the human ortholog for mouse Bglap, Bglap2 and Bglap3, encodes for Osteocalcin, a bone remodeling protein that is associated with prostate cancer progression [42, 43].
How Trim28 inhibition alters the tumor microenvironment over time remains unclear. In tumors, the activation of ERVs, which are suppressed by TRIM28 in ES cells, adult cells as well as cancer cells, can induce anti-tumor viral-defense immune responses [44,45,46,47,48]. In prostate cancers, including antiandrogen treatment-resistant prostate cancers, ERV activation has been suggested as an immunotherapeutic strategy [49, 50]. Further studies are needed to determine whether Trim28 inhibition in NPp53T prostates causes overexpression of ERVs that generate an interferon response.
The unexpected tumor evolution and progression in the genetically-engineered NPp53T mouse model was surprising, given the previously reported function of TRIM28 in prostate cancer progression and the promise of TRIM28, SETDB1 and histone methyltransferases as targets for immunotherapy [44, 48, 49, 51]. However, as TRIM28 performs a wide range of functions in the cell, its targeting may result in unexpected downstream effects in immune-competent mice. Our in vivo findings highlight the complexity of tumor cell plasticity, tumor evolution, and interactions of tumor cells with the tumor microenvironment. Although expression of proximal luminal marker TACSTD2 has been associated with human high-grade prostate cancers [2], KLF4, a pluripotency regulator, is also overexpressed in the mouse proximal prostate but marks indolent prostate cancers [52]. Similar to phenotypes we observed with long-term Trim28 inhibition, loss of KLF4 promotes sarcomatoid tumor formation, increasing expression of mesenchymal markers and markers of epithelial-mesenchymal transition [52]. These studies illustrate the complex contributions of cells expressing pluripotency and proximal luminal markers to prostate cancer progression. Additional studies using genetically-engineered mouse models or other models with an intact immune system and further extension of these findings to the context of CRPC and human prostate cancers will be informative in evaluating effective treatment strategies for prostate cancers.
Materials and methods
Mouse models
All animal procedures were performed with prior approval from the Institutional Animal Care and Use Committee at the George Washington University. The Nkx3.1-CreERT2/+;R26rYFP/YFP and Nkx3.1-CreERT2/+;Ptenf/f;p53f/f;R26rYFP/YFP (NPp53) mice were a gift from Dr. Michael Shen, Columbia University Medical Center [21, 53]. Trim28f/f mice were obtained from Jackson laboratory (JAX Strain #018552) [54] and bred to generate Nkx3.1-CreERT2/+;Ptenf/f;p53f/f;Trim28f/f;R26rYFP/YFP (NPp53T), Nkx3.1-CreERT2/+;Ptenf/f;Trim28f/f;R26rYFP/YFP (NPT) and control Nkx3.1-CreERT2/+;Ptenf/f;Trim28f/+; R26rYFP/YFP (NP) and Nkx3.1-CreERT2/+;Trim28f/f;R26rYFP/YFP mice. Animals were maintained on a C57BL/6 mixed genetic background. Genotyping was conducted by Transnetyx or using primers listed in Table S1.
Tamoxifen treatment, castration, tissue dissection and survival curve analysis
Adult male mice of 2–4 months of age were administered tamoxifen (Sigma, St. Louis, MO, USA) dissolved in corn oil by oral gavage for four consecutive days at a dose of 200 mg/kg body weight to activate CreERT2 [39]. For studies of castration-resistance, males were castrated 1 month after tamoxifen treatment. Prostates were dissected in 1X PBS at 2 weeks to 6 months after tamoxifen treatment. Analyses were conducted on anterior prostate lobe tissues unless otherwise noted. For survival curve analysis, mice were euthanized when the body condition score reached ≤2. Investigators were not blinded, and sample sizes were not pre-determined. No randomization was used.
Histopathological and immunostaining analysis
Hematoxylin and Eosin staining was performed on 5 µM paraffin-embedded sections. Masson’s Trichrome staining was performed using the Masson’s 2000 Trichrome Stain Kit (Stat Lab, McKinney, TX, USA). Investigators were blinded when scoring pathological features.
For immunohistochemistry (IHC), formalin-fixed, 5 µM paraffin-embedded tissue sections were subjected to citrate-based or Tris-based heat-induced antigen retrieval (Vector Laboratories, Newark, CA, USA). Sections were blocked with 10% normal goat serum (Vector Laboratories) for 1 h at room temperature (RT) and incubated overnight with primary antibodies at 4 °C. Biotinylated secondary antibodies were used with VECTASTAIN Elite ABC HRP Detection and Vector NovaRed Substrate Kits (Vector Laboratories). Images were captured on a Leica DMi8 microscope (Leica Microsystems, Wetzlar, Germany).
For immunofluorescence (IF) staining, 8 µM sections from 4% paraformaldehyde-fixed, OCT-embedded frozen tissues or 5 µM sections from formalin-fixed, paraffin-embedded tissues were antigen retrieved followed by blocking with 10% normal goat serum (Invitrogen, Waltham, MA, USA) for 1 h at RT. Incubation with primary antibodies (Table S1) was conducted overnight at 4 °C; incubation with Alexa Fluor fluorescent-tagged secondary antibodies (Invitrogen) was performed for 1 h at RT. DAPI was used to stain nuclei. Slides were mounted using glycerol-based mounting medium containing N-propyl gallate. Images were captured on a Zeiss LSM 710 confocal microscope (ZEISS, Jena, Germany) and analyzed using Image J and Adobe Photoshop.
Single-cell RNA sequencing
Prostate tissue was digested with papain [5]. To reduce batch effects, tissues from NPp53 and NPp53T prostates were processed on the same day. 6000 cells with >90% viability were loaded onto a 10X Genomics Chromium Controller (10x Genomics, Pleasanton, CA, USA) using a Chromium Next GEM Single Cell 3ʹ Kit v3.1 (PN-1000269). Libraries were sequenced using a NextSeq High-Output 150 cycle kit (Illumina Inc., San Diego, CA, USA). Each library produced approximately 200 million paired-end reads.
Bioinformatics analysis on raw sequence data was conducted using the 10X Genomics cell-ranger pipeline (v5.0.1), starting with mkfastq for demultiplexing, count for alignment and featurecounting for each of the two samples. The 10X Genomics mouse reference database was used (mm10 July 2020-A version). The Yellow Fluorescent Protein (YFP) gene (JAX Strain #007903) [55] was manually added to the reference database and reindexed with Cell Ranger’s mkref command before using the count command. Additional analysis was conducted using Partek Flow. Cells with less than 5000 detected genes, or greater than 20% mitochondrial genes were removed. Uniform Manifold Approximation and Projection (UMAP) was used to visualize clusters. Samples were split by attribute to identify YFP+ cells. Differential expression analysis was conducted using the Hurdle model. Significantly deregulated genes were identified using a threshold of 1% for False Discovery Rate (FDR ≤ 0.01) together with a fold change of 2 (<−2 or >2).
Flow sorting and organoid culture
Whole prostates from tamoxifen-induced NPp53 or NPp53T mice were collected in cold PBS, 1-month after tamoxifen treatment. The prostates were chopped with scissors and digested with collagenase/hyaluronidase (STEMCELL Technologies, Vancouver, Canada) in Hepatocyte culture media (Corning, Corning, NY, USA) containing 5% FBS for 2 h at 37 °C and briefly with TrypLE and Dispase/DNaseI. After each step, the tissues were washed with cold HBSS containing 1% BSA. Single/near-single cells were obtained by passing through a 40 µM cell strainer (Fisherbrand, Pittsburgh, PA, USA) followed by incubation with anti-EpCAM antibody (1:100) for 25 min on ice. Single cells were enriched by gating from forward and side scatter plots and DAPI was used to exclude dead cells. Lineage-traced epithelial cells were sorted by gating for the YFP+ or YFP+EpCAM+ population (Fig. S4). Sorting was performed on a BD Influx cell sorter. Data were analyzed using FCS Express 7 software.
For organoid culture, YFP+EpCAM+ or YFP+ cells were seeded at 4500–5000 cells/well in a 96-well plate with hepatocyte culture media containing 5% matrigel, 10 µM ROCK inhibitor and 10 nM DHT [39]. Organoids were passaged every 10–12 days using TrypLE, and imaged using a Leica DMi8 microscope.
For CRISPR/Cas9 mediated deletion of Trim28, lentiviral particles were generated in HEK293FT cells (Thermo Scientific, Waltham, MA, USA) using a LentiCRISPRv2 plasmid containing a single guide RNA for mouse Trim28 gRNA [44], human TRIM28 gRNA targeting exon 3 [56], or control empty LentiCRISPR V2 vector (Addgene 52961). The gRNA vectors were sequenced using the hU6-F primer. Lipofectamine 3000 Reagent (Invitrogen) and packaging plasmids pMD2.G (Addgene 12259) and psPAX2 (Addgene 12260) were used. Supernatant containing virus was collected 24 h after transfection of HEK293FT cells and filtered. PC-3 cells (American Type Culture Collection, Manassas, VA, USA) or cells from dissociated NPp53 organoids were spin-infected at 32 °C for 45 min and plated in organoids culture media containing 1 µg/ml puromycin for 3 days (Sigma). Human cell lines were tested for mycoplasma contamination using the MycoSensor PCR assay kit (Agilent, Santa Clara, CA, USA). Organoids were imaged on day 7 (mouse cells) or day 11 (PC-3 cells).
Immunophenotyping
1 month or 3 months after tamoxifen treatment, all prostate lobes were dissected in cold PBS, and dissociated as described above. Single/near-single cells were obtained by passing through a 40 µM cell strainer (Fisherbrand). After washing with cold HBSS containing 1% BSA, cells were resuspended in 1 ml cold PBS followed by a 30 min incubation with a LIVE/DEAD™ Fixable Aqua (Invitrogen) viability dye. Approximately 400,000 cells were stained for myeloid or lymphocyte antibodies (Table S1), fixed in 2% paraformaldehyde for 15 min, and analyzed on a BD Celesta Cell Analyzer (BD, Franklin Lakes, NJ, USA). Fluorescence Minus One (FMO) controls were used for gating. The gating strategy is shown in Fig. S1.
Western Blotting
Mouse prostates were lysed in RIPA buffer (50 mM Tris, 150 mM NaCl, 1% NP-40, 0.5% Sodium deoxycholate, 0.1% SDS) with Complete Protease Inhibitor Cocktail (Roche, Basel, Switzerland). Protein concentrations were determined using the Bradford Assay (BioRad, Hercules, CA, USA). Proteins were resolved by SDS/PAGE and transferred to PVDF membranes (Millipore, Burlington, MA, USA). Membranes were blocked in 3% BSA and incubated with primary antibodies overnight at 4 °C, followed by secondary antibodies for 1 h at RT (Table S1). Pierce ECL Western Blotting Substrate (Thermo Scientific) or SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific) were used for chemiluminescent detection with a LI-COR Odyssey Fc Imaging System.
Human database analysis
TRIM28 expression in human prostate cells was assessed using the Single Cell Portal (singlecell.broadinstitute.org). Publicly available datasets were searched using cBioPortal [57] and datasets from the Cancer Genome Atlas (TCGA) Research Network, Stand Up to Cancer/PCF Dream Team [58], MSK [59] and Fred Hutchinson CRC [60]. Parameters were set to include mutations, structural variants, copy-number alterations, and mRNA expression z-scores relative to diploid samples with a threshold of ±2. Unaltered samples were excluded from visualization.
Statistical analysis
Statistical analyses were conducted with Prism 9 (GraphPad, San Diego, CA, USA) using a two-tailed unpaired Student’s t test unless otherwise noted. Data were assumed to be normally distributed with similar variance between groups. Error bars on graphs represent the standard deviation from the mean. Sample sizes are noted in the figure legends. Immunostaining experiments were replicated on at least two animals of each genotype for each timepoint. For the generation of organoids, n = 2 experiments were conducted for each genotype.
Data availability
Expression data from single-cell RNA sequencing have been deposited in GEO under accession number GEO: GSE199015.
Material availability
All unique reagents generated in the study are available from the lead contact with a completed material transfer agreement.
References
Beltran H, Hruszkewycz A, Scher HI, Hildesheim J, Isaacs J, Yu EY, et al. The role of lineage plasticity in prostate cancer therapy resistance. Clin Cancer Res. 2019.
Guo W, Li L, He J, Liu Z, Han M, Li F, et al. Single-cell transcriptomics identifies a distinct luminal progenitor cell type in distal prostate invagination tips. Nat Genet. 2020;52:908–18.
Karthaus WR, Hofree M, Choi D, Linton EL, Turkekul M, Bejnood A, et al. Regenerative potential of prostate luminal cells revealed by single-cell analysis. Science. 2020;368:497–505.
Mevel R, Steiner I, Mason S, Galbraith LC, Patel R, Fadlullah MZ, et al. RUNX1 marks a luminal castration-resistant lineage established at the onset of prostate development. eLife. 2020;9:e60225.
Crowley L, Cambuli F, Aparicio L, Shibata M, Robinson BD, Xuan S, et al. A single-cell atlas of the mouse and human prostate reveals heterogeneity and conservation of epithelial progenitors. eLife. 2020;9:e59465.
Fong KW, Zhao JC, Song B, Zheng B, Yu J. TRIM28 protects TRIM24 from SPOP-mediated degradation and promotes prostate cancer progression. Nat Commun. 2018;9:5007.
Shibata M, Blauvelt KE, Liem KF Jr, Garcia-Garcia MJ. TRIM28 is required by the mouse KRAB domain protein ZFP568 to control convergent extension and morphogenesis of extra-embryonic tissues. Development. 2011;138:5333–43.
Alexander KA, Wang X, Shibata M, Clark AG, Garcia-Garcia MJ. TRIM28 Controls Genomic Imprinting through Distinct Mechanisms during and after Early Genome-wide Reprogramming. Cell Rep. 2015;13:1194–205.
Messerschmidt DM, de Vries W, Ito M, Solter D, Ferguson-Smith A, Knowles BB. Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science. 2012;335:1499–502.
Li X, Ito M, Zhou F, Youngson N, Zuo X, Leder P, et al. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev Cell. 2008;15:547–57.
Oleksiewicz U, Gladych M, Raman AT, Heyn H, Mereu E, Chlebanowska P, et al. TRIM28 and Interacting KRAB-ZNFs Control Self-Renewal of Human Pluripotent Stem Cells through Epigenetic Repression of Pro-differentiation Genes. Stem cell Rep. 2017;9:2065–80.
Barde I, Rauwel B, Marin-Florez RM, Corsinotti A, Laurenti E, Verp S, et al. A KRAB/KAP1-miRNA cascade regulates erythropoiesis through stage-specific control of mitophagy. Science. 2013;340:350–3.
Singh K, Cassano M, Planet E, Sebastian S, Jang SM, Sohi G, et al. A KAP1 phosphorylation switch controls MyoD function during skeletal muscle differentiation. Genes Dev. 2015;29:513–25.
Gehrmann U, Burbage M, Zueva E, Goudot C, Esnault C, Ye M, et al. Critical role for TRIM28 and HP1beta/gamma in the epigenetic control of T cell metabolic reprograming and effector differentiation. Proc Natl Acad Sci USA. 2019;116:25839–49.
Bacon CW, Challa A, Hyder U, Shukla A, Borkar AN, Bayo J, et al. KAP1 Is a Chromatin Reader that Couples Steps of RNA Polymerase II Transcription to Sustain Oncogenic Programs. Mol Cell. 2020;78:1133–51.e14.
Czerwinska P, Shah PK, Tomczak K, Klimczak M, Mazurek S, Sozanska B, et al. TRIM28 multi-domain protein regulates cancer stem cell population in breast tumor development. Oncotarget. 2017;8:863–82.
Addison JB, Koontz C, Fugett JH, Creighton CJ, Chen D, Farrugia MK, et al. KAP1 promotes proliferation and metastatic progression of breast cancer cells. Cancer Res. 2015;75:344–55.
Hao L, Leng J, Xiao R, Kingsley T, Li X, Tu Z, et al. Bioinformatics analysis of the prognostic value of Tripartite Motif 28 in breast cancer. Oncol Lett. 2017;13:2670–8.
Van Tilborgh N, Spans L, Helsen C, Clinckemalie L, Dubois V, Lerut E, et al. The transcription intermediary factor 1beta coactivates the androgen receptor. J Endocrinol Investig. 2013;36:699–706.
Stelloo S, Nevedomskaya E, Kim Y, Hoekman L, Bleijerveld OB, Mirza T, et al. Endogenous androgen receptor proteomic profiling reveals genomic subcomplex involved in prostate tumorigenesis. Oncogene. 2018;37:313–22.
Zou M, Toivanen R, Mitrofanova A, Floch N, Hayati S, Sun Y, et al. Transdifferentiation as a Mechanism of Treatment Resistance in a Mouse Model of Castration-Resistant Prostate Cancer. Cancer Discov. 2017;7:736–49.
Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–43.
Reimers MA, Slane KE, Pachynski RK. Immunotherapy in Metastatic Castration-Resistant Prostate Cancer: Past and Future Strategies for Optimization. Curr Urol Rep. 2019;20:64.
Pienta KJ, Machiels JP, Schrijvers D, Alekseev B, Shkolnik M, Crabb SJ, et al. Phase 2 study of carlumab (CNTO 888), a human monoclonal antibody against CC-chemokine ligand 2 (CCL2), in metastatic castration-resistant prostate cancer. Investig N Drugs. 2013;31:760–8.
Di Mitri D, Mirenda M, Vasilevska J, Calcinotto A, Delaleu N, Revandkar A, et al. Re-education of Tumor-Associated Macrophages by CXCR2 Blockade Drives Senescence and Tumor Inhibition in Advanced Prostate Cancer. Cell Rep. 2019;28:2156–68e.55.
Kim MJ, Cardiff RD, Desai N, Banach-Petrosky WA, Parsons R, Shen MM, et al. Cooperativity of Nkx3.1 and Pten loss of function in a mouse model of prostate carcinogenesis. Proc Natl Acad Sci USA. 2002;99:2884–9.
Ellwood-Yen K, Graeber TG, Wongvipat J, Iruela-Arispe ML, Zhang J, Matusik R, et al. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell. 2003;4:223–38.
Pletcher A, Shibata M. Prostate organogenesis. Development. 2022;149:dev200394. https://doi.org/10.1242/dev.200394. Epub 2022 Jun 21.
Crowley L, Shen MM. Heterogeneity and complexity of the prostate epithelium: New findings from single-cell RNA sequencing studies. Cancer Lett. 2022;525:108–14.
Abu El Maaty MA, Terzic J, Keime C, Rovito D, Lutzing R, Yanushko D, et al. Hypoxia-mediated stabilization of HIF1A in prostatic intraepithelial neoplasia promotes cell plasticity and malignant progression. Sci Adv. 2022;8:eabo2295.
Ittmann M, Huang J, Radaelli E, Martin P, Signoretti S, Sullivan R, et al. Animal models of human prostate cancer: the consensus report of the New York meeting of the Mouse Models of Human Cancers Consortium Prostate Pathology Committee. Cancer Res. 2013;73:2718–36.
Zhu R, Gires O, Zhu L, Liu J, Li J, Yang H, et al. TSPAN8 promotes cancer cell stemness via activation of sonic Hedgehog signaling. Nat Commun. 2019;10:2863.
Zhu L, Gibson P, Currle DS, Tong Y, Richardson RJ, Bayazitov IT, et al. Prominin 1 marks intestinal stem cells that are susceptible to neoplastic transformation. Nature. 2009;457:603–7.
Agarwal S, Hynes PG, Tillman HS, Lake R, Abou-Kheir WG, Fang L, et al. Identification of Different Classes of Luminal Progenitor Cells within Prostate Tumors. Cell Rep. 2015;13:2147–58.
Hsieh CS, deRoos P, Honey K, Beers C, Rudensky AY. A role for cathepsin L and cathepsin S in peptide generation for MHC class II presentation. J Immunol. 2002;168:2618–25.
Maine CJ, Teijaro JR, Marquardt K, Sherman LA. PTPN22 contributes to exhaustion of T lymphocytes during chronic viral infection. Proc Natl Acad Sci USA. 2016;113:E7231–9.
Ecco G, Cassano M, Kauzlaric A, Duc J, Coluccio A, Offner S, et al. Transposable Elements and Their KRAB-ZFP Controllers Regulate Gene Expression in Adult Tissues. Dev Cell. 2016;36:611–23.
Nicetto D, Donahue G, Jain T, Peng T, Sidoli S, Sheng LH. et al. H3K9me3-heterochromatin loss at protein-coding genes enables developmental lineage specification. Science. 2019;363:294–7.
Chua CW, Shibata M, Lei M, Toivanen R, Barlow LJ, Bergren SK, et al. Single luminal epithelial progenitors can generate prostate organoids in culture. Nat Cell Biol. 2014;16:951–61.
Zarif JC, Baena-Del Valle JA, Hicks JL, Heaphy CM, Vidal I, Luo J, et al. Mannose Receptor-positive Macrophage Infiltration Correlates with Prostate Cancer Onset and Metastatic Castration-resistant Disease. Eur Urol Oncol. 2019;2:429–36.
Yang Y, Lu H, Chen C, Lyu Y, Cole RN, Semenza GL. HIF-1 Interacts with TRIM28 and DNA-PK to release paused RNA polymerase II and activate target gene transcription in response to hypoxia. Nat Commun. 2022;13:316.
Koeneman KS, Yeung F, Chung LW. Osteomimetic properties of prostate cancer cells: a hypothesis supporting the predilection of prostate cancer metastasis and growth in the bone environment. Prostate. 1999;39:246–61.
Desbois C, Hogue DA, Karsenty G. The mouse osteocalcin gene cluster contains three genes with two separate spatial and temporal patterns of expression. J Biol Chem. 1994;269:1183–90.
Lee AK, Pan D, Bao X, Hu M, Li F, Li CY. Endogenous Retrovirus Activation as a Key Mechanism of Anti-Tumor Immune Response in Radiotherapy. Radiat Res. 2020;193:305–17.
Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463:237–40.
Tie CH, Fernandes L, Conde L, Robbez-Masson L, Sumner RP, Peacock T, et al. KAP1 regulates endogenous retroviruses in adult human cells and contributes to innate immune control. EMBO Rep. 2018;19:e45000.
Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell. 2015;162:974–86.
Zhang SM, Cai WL, Liu X, Thakral D, Luo J, Chan LH, et al. KDM5B promotes immune evasion by recruiting SETDB1 to silence retroelements. Nature. 2021;598:682–7.
Baratchian M, Tiwari R, Khalighi S, Chakravarthy A, Yuan W, Berk M, et al. H3K9 methylation drives resistance to androgen receptor-antagonist therapy in prostate cancer. Proc Natl Acad Sci USA. 2022;119:e2114324119.
Morel KL, Sheahan AV, Burkhart DL, Baca SC, Boufaied N, Liu Y, et al. EZH2 inhibition activates a dsRNA-STING-interferon stress axis that potentiates response to PD-1 checkpoint blockade in prostate cancer. Nat Cancer. 2021;2:444–56.
Griffin GK, Wu J, Iracheta-Vellve A, Patti JC, Hsu J, Davis T, et al. Epigenetic silencing by SETDB1 suppresses tumour intrinsic immunogenicity. Nature. 2021;595:309–14.
Xiong X, Schober M, Tassone E, Khodadadi-Jamayran A, Sastre-Perona A, Zhou H, et al. KLF4, A Gene Regulating Prostate Stem Cell Homeostasis, Is a Barrier to Malignant Progression and Predictor of Good Prognosis in Prostate Cancer. Cell Rep. 2018;25:3006–20.e07.
Wang X, Kruithof-de Julio M, Economides KD, Walker D, Yu H, Halili MV, et al. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature. 2009;461:495–500.
Cammas F, Mark M, Dolle P, Dierich A, Chambon P, Losson R. Mice lacking the transcriptional corepressor TIF1beta are defective in early postimplantation development. Development. 2000;127:2955–63.
Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–40.
Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–91.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1.
Abida W, Cyrta J, Heller G, Prandi D, Armenia J, Coleman I, et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci USA. 2019;116:11428–36.
Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22.
Kumar A, Coleman I, Morrissey C, Zhang X, True LD, Gulati R, et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med. 2016;22:369–78.
Acknowledgements
We thank Shibata lab members and Kate Chiappinelli for comments on the paper, Michael Shen for mice, Chuan-Yuan Li for plasmids, Hunain Khawaja for assistance with animal breeding and colony maintenance, Gregory Cresswell and the GW Flow Cytometry Core, Keith Crandall, Caitlin Loeffler and the GW Genomics Core, the GW Nanofabrication and Imaging Center, the McCormick Genomic & Proteomic Center and the GW SMHS Writing Institute. This work was supported by the National Institutes of Health NCI grant R00CA194287 to MS.
Author information
Authors and Affiliations
Contributions
Investigation, ASY, ECW, AP, AHelfand, TMN, MS; Single-cell RNA sequencing analysis, HI, AHorvath, MS; pathology analysis, PSL; writing—original draft, ASY, ECW, MS; writing—review and editing, all authors; conceptualization, funding acquisition, supervision, MS.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yende, A.S., Williams, E.C., Pletcher, A. et al. TRIM28 promotes luminal cell plasticity in a mouse model of prostate cancer. Oncogene 42, 1347–1359 (2023). https://doi.org/10.1038/s41388-023-02655-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-023-02655-0
- Springer Nature Limited