Abstract
Human epithelial cells can be infected by more than 200 types of human papilloma viruses (HPVs), and persistent HPV infections lead to cervical cancer or other deadly cancers. It has been established that mitotic progression is critical for HPV16 infection, but the underlying mechanism remains unknown. Here, we report that oncoprotein E7 of HPV16 but not HPV18 retards mitotic progression in host cell by direct binding to the C terminus of Microtubule-Associated Protein 4 (MAP4). MAP4 is a novel bona fide target of HPV16E7 protein which binds and recruits the latter to spindle microtubule in mitosis. HPV16E7 protein promotes MAP4 stability by inhibiting MAP4 phosphorylation- mediated degradation to increase the stability of microtubule polymerization and cause an extend mitotic progression. We further uncovered that Mps1 is a kinase of MAP4, and E7-MAP4 binding blocks Mps1 phosphorylation of MAP4, thereby interrupting phosphorylation-dependent MAP4 degradation. Mutations of MAP4 at T927ES928E partially abolished E7-binding capacity and rescued mitotic progression in host cells. In conclusion, our study reveals a molecular mechanism by which HPV16E7 perturbs host mitotic progression by interfering Mps1-MAP4 signaling cascade, which results in an extended infection window and may facilitate the persistent HPV16 infection.
Similar content being viewed by others
Introduction
Human papilloma viruses (HPVs) are carcinogenic not only for cervical cancer [1] but also for other deadly cancers in men and women [2,3,4]. Persistent infections by high-risk HPVs are considered as prerequisite for the invasive cancer [5, 6] and persistent HPV16 infection causes half of all cervical cancers [7, 8]. Mitotic progression was shown to be critical for early steps of HPV infection [9], but the exact molecular mechanism on how the virus interact with host cells reminds open at large. A recent study found that viral particle can relocate to the nucleus only when cells progressed into mitosis, during which the nuclear envelope is broken down and the membrane-bound HPV16 particles migrate along with spindle microtubules to the condensed chromosomes [10]. In addition, high-risk HPV-associated lesions showed a significantly higher proportion of cells with mitotic defect [11, 12]. Numerous mitotic proteins were found to be abnormally expressed in cervical cancer and were closely associated with high-grade cervical intraepithelial neoplasia [13, 14].
Previous researches demonstrate that Microtubule-Associated Protein (MAP4) plays a key role in microtubule polymerization and stability during mitosis [15,16,17]. MAP4 is a major protein for spindle microtubule assembly during mitosis, and its mutations contribute to the clinical spectrum of centrosome defects [18]. MAP4 was proven to contribute to tumor genesis and its protein level is positively correlated with bladder cancer grade and progression [19, 20]. Interestingly, MAP4 is necessary in early HIV infection, and MAP4 knockdown inhibits HIV-1 infection [21].
Monopolar spindle kinase 1 (Mps1) is known as a critical mitotic kinase, essential for activation and maintenance of the spindle assembly checkpoint (SAC) [22]. Mps1 plays indispensable roles in centrosome duplication, and its mutation results in incorrect replication of centrosomes with a monopolar spindle [23]. Recent studies have revealed the role of Mps1 in regulating the attachment of spindle microtubules to the kinetochore. Mps1 can destabilize microtubule attachments and dynamically modify kinetochores to correct improper attachments and ensure faithful chromosome segregation [24]. Microtubule polymers disrupt the interaction between Mps1 and HEC1 in vitro, and Mps1 binding to kinetochores is prevented by end-on microtubule attachment [25]. Our previous study also found that Mps1 can localize on spindle microtubules. However, it is unclear on whether Mps1 modulates microtubule dynamics.
Our current study simultaneously linked HPV oncoprotein E7 to MAP4 and Mps1, two of the most critical cell cycle controlling components, for the first time. Here we present the solid evidences that oncoprotein E7 of HPV16 but not HPV18 can localize onto the mitotic spindle of the host cell by direct formation of E7-MAP4 association, which resulted in significant retardation in mitotic progression. Furthermore, we found that Mps1 is actually a kinase for MAP4, responsible for the phosphorylation of MAP4, and E7-MAP4 binding significantly interrupted the phosphorylation-dependent degradation of MAP4. Altogether, our findings revealed a previously unknown molecular mechanism by which the oncoprotein HPV16E7 modulates the mitotic process of host cells by interfering with the Mps1–MAP4 signal pathway.
Results
HPV16E7 binds to MAP4 and induces abnormal MAP4 accumulation
We successfully identified MAP4 as a strong binding factor for HPV16E7 through mass spectrometry analysis of GST-HPV16E7 pulldown products. The binding of HPV16E7 to MAP4 was subsequently confirmed by western blotting, and the binding interactions were weakened by the treatment with Nocodazole, an inhibitor interfering with microtubule polymerization (Fig. 1a). MAP4 has an asymmetric molecular structure consisting of an amino-terminal projection (PJ) domain and a carboxyl-terminal MT-binding (MTB) domain [26]. To narrow down the region of the MAP4-HPV16E7 interaction, MAP4 was divided into two parts: MAP4-N (aa 1–692) and MAP4-C (aa 693–1152) (Fig. S1a). The pulldown assay showed that HPV16E7 associated with MAP4 through its C-terminal domain (Fig. S1b). MAP4-C was further divided into two parts, the proline-rich domain (PRD, aa 693–934) and assembly promoting domain (AP, aa 935–1152) (Fig. S1a), which were expressed and purified in Escherichia coli, and the results showed that HPV16E7 could interact with both the PRD and AP domain directly (Fig. S1c). Taken together, these findings clarified that HPV16E7 associates directly with the MAP4 C-terminus, a microtubule-binding domain.
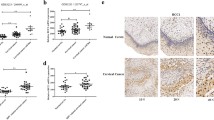
HPV16E7 associates with MAP4 and induces abnormal MAP4 accumulation. a HPV16E7 associates with MAP4. GST-fused HPV16E7 pulled down MAP4 protein in HeLa cells. HeLa cell lysates were treated with Nocodazole or control to demonstrate the effect of cell cycle control on the HPV16E7–MAP4 interaction. See Supplementary Materials for a detailed characterization of the HPV16E7–MAP4 interaction. b MAP4 binds HPV16E7 but not HPV18E7. GST pulldown was executed using purified GST proteins and 293T cell lysate. c HPV16E7 but not HPV18E7 is capable of causing abnormal MAP4 accumulation and increased Ac-tubulin level in cells. 293T cells transiently transfected using HPV16E7 or HPV18E7 plasmid were subjected to a WB assay. d MAP4 was more highly expressed in squamous cell carcinoma than in the control epithelia, similar to HPV16E7. Histochemistry slides were treated following a standard protocol and stained with anti-HPV16E7 and anti-MAP4 antibodies. e Quantitative presentation of d. The expression levels of HPV16E7 and MAP4 proteins were assessed in different tumor tissues and cell types. “16E7” indicates E7 protein encoded by HPV16; “18E7” indicates E7 protein encoded by HPV18
E7 proteins from HPV18 and HPV11, both sharing approximately 40% AA identities with that of HPV16, were also examined for their bindings to MAP4 for comparison (Fig. S1d–f). The results showed that MAP4 binds HPV16E7 but not HPV18E7 (Fig. 1b) and HPV11E7-MAP4-binding capability is significantly weaker than that of HPV16E7–MAP4 (Fig. S1d). We found that only HPV16E7, but not HPV18E7 and HPV11E7, could cause abnormal MAP4 accumulation (Fig. 1c, Fig. S1e). Expression of acetylated α-tubulin (Ac-tubulin), a marker of stabilized microtubules [27], was upregulated when HPV16E7 was overexpressed, hinting that HPV16E7 may act to stabilize the microtubules. Collectively, these data suggested the functional significance of the binding of HPV16E7 with MAP4. In HeLa cells with HPV16E7 overexpressed, the transcription level of MAP4 was not significantly different from that in the control cells (Fig. S2a).
Examination of HPV16E7 and MAP4 expressions in human cervical tissues showed a similar pattern, both showing a significantly higher level of expressions in squamous cell carcinoma (Ib stage) than in other types of cervical carcinoma (Fig. 1d, e). The expression level of MAP4 in stage IIIb squamous cell carcinoma was lower than that in stage Ib, despite a high level of E7 (Fig. 1e). This indicates that their association may occur at an earlier stage of carcinogenesis and contribute positively to HPV infection, integration, or cell transformation.
HPV16E7 retards mitotic progression by inhibiting MAP4 degradation
Overexpression of HPV16E7 in HeLa cells significantly increased the percentage of prometaphase cells at a similar ratio with MAP4 overexpression, and MAP4 knockdown abrogated the HPV16E7-mediated mitotic arrest effect (Fig. 2a, Fig. S2b). To analyze this in detail, mitotic progression in HeLa and HeLa-HPV16E7 cells were monitored using live-cell imaging. The cells were synchronized in the G2/M boundary using RO-3306 (a selective ATP-competitive inhibitor of CDK1) and released into fresh medium. The results showed that the duration of prometaphase in HeLa-HPV16E7 cells was significantly longer than that in control cells (Fig. 2b, c; Fig. S2c) and the mitotic progression was delayed due to HPV16E7 expression. Furthermore, HPV16E7 was co-localized with MAP4 on spindle microtubules during mitosis in HPV16-positive CasKi cells (Fig. 2d). The targeting of HPV16E7 on spindle microtubules depends on MAP4, because HPV16E7 shows diffuse pattern in MAP4 knockdown cells (Fig. 2e).
HPV16E7 retards mitotic progression by inhibiting MAP4 degradation. a The HPV16E7–MAP4 association significantly increased the cell ratio of prometaphase to total mitotic cells. The ratio of prometaphase cells in total mitotic cells was determined (n > 300). b Live-cell imaging showing prolonged duration of prometaphase (red) in HeLa cells expressing HPV16E7. HeLa and HeLa-HPV16E7 cells (n = 50) were synchronized in the G2/M boundary using RO-3306 (9 μM) and submitted to live-cell imaging after release into fresh medium. The duration of prometaphase (red), metaphase (blue), or anaphase/telophase (yellow) was calculated. c Quantitative presentation of b. Significantly prolonged prometaphase times were observed in HeLa cells expressing HPV16E7. d HPV16E7 and MAP4 were co-localized onto spindle microtubules during mitosis. CasKi cells were stained with anti-HPV16E7, anti-MAP4, and anti-β-tubulin antibodies. e HPV16E7 failed to localize onto the spindle in cells with MAP4 knockdown by siRNA. CasKi cells transfected with the specified siRNA were stained with anti-HPV16E7 and anti-MAP4 antibodies and DAPI. f Western blot showing abnormal accumulation of MAP4 protein during mitosis in HeLa cells expressing HPV16E7-EGFP but not in cells expressing EGFP alone. g Western blot showing abnormal accumulation of MAP4 protein in C33A-HPV16E7 cells in which HPV16E7 was stably expressed. h MAP4 protein was also abnormally accumulated in HaCaT-HPV16E7 cells. “MT” indicates microtubule; “Ac-tubulin” indicates acetylated α-tubulin; “RO” indicates RO-3306; “Noco” indicates Nocodazole; “NEBD” indicates nuclear envelope breakdown; “Prometa” indicates prometaphase; “Meta” indicates metaphase; “Ana/Telo” indicates anaphase/telophase
Examination of mitotic progression in synchronized HeLa cells showed that MAP4 was degraded in a mitosis-dependent manner, and HPV16E7 expression significantly inhibited the degradation of MAP4 (Fig. 2f). Especially, the degradation trend of Cyclin B, a critical mitotic marker degraded during M phase, was significantly delayed, and the level of Ac-tubulin was significantly improved in HPV16E7-overexpressing cells (Fig. 2f). Additionally, we examined whether MAP4 was degraded in HPV-negative C33A and HaCaT cells. C33A and C33A-HPV16E7 cells, in which HPV16E7 was stably expressed, were synchronized in prometaphase using Nocodazole and released into fresh medium to assess the protein levels of MAP4 and Cyclin B. The results showed that MAP4 was degraded in a mitosis-dependent manner in C33A cells, similar to Cyclin B; however, under HPV16E7 expression, the degradation of MAP4 and Cyclin B were dramatically inhibited (Fig. 2g). The same results were detected in HaCaT and HaCaT-HPV16E7 cells (Fig. 2h). Thus, our results suggested that HPV16E7 retards mitotic progression by increasing the protein level of MAP4 during mitosis.
Therefore, a new regulatory mechanism of MAP4 was discovered: MAP4 is degraded in a cell-cycle-dependent manner during mitosis. To provide more evidence, HeLa cells were synchronized at the G2/M boundary and released cells into medium with MG132 and degradation of MAP4 in MG132-treated cells was significantly inhibited (Fig. S2d). To test whether MAP4 would be degraded at interphase and whether HPV16E7 could promote re-synthesis of MAP4 protein, Cyclohexane, an inhibitor of protein biosynthesis, was used to treat HeLa and HeLa-HPV16E7 cells. The results showed that MAP4 was not degraded at interphase in HeLa cells and was not increased in HeLa-HPV16E7 cells (Fig. S2e). All these results demonstrate that degradation of MAP4 during mitosis is strictly regulated and that the accumulation of MAP4 in HPV16E7-expressing cells was caused by inhibition of MAP4 degradation.
HPV16E7 disturbs microtubule dynamics during mitosis through association with MAP4
As a microtubule stabilization factor, MAP4 regulates microtubule stability by binding to microtubules and inhibiting their depolymerization [28, 29]. To determine if HPV16E7 affected spindle microtubule polymerization and stability in mitosis through MAP4, HeLa and HeLa-HPV16E7 cells were treated with Nocodazole to breakdown spindle microtubules and then assessed the Ac-tubulin level after release. The results showed that HPV16E7 retarded microtubule depolymerization, causing Ac-tubulin accumulation in HeLa cells (Fig. 3a), which had been found in HeLa cells overexpressing EGFP-16E7 (Fig. 2f). The difference at the 10 min point between the two groups suggested that HPV16E7 might also accelerate spindle microtubule polymerization (Fig. 3a). MAP4 was then knocked down in both HeLa and HeLa-HPV16E7 cells. The results showed that MAP4 downregulation significantly reduced the protein level of Ac-tubulin (Fig. 3b). HPV16E7 and MAP4 were then knocked down in CasKi cells. The results showed that downregulation of HPV16E7 significantly reduced MAP4 and Ac-tubulin protein levels, similar to the results of MAP4 knockdown (Fig. 3c, d). Thus, the association of HPV16E7 with MAP4 promoted Ac-tubulin upregulation in vivo.
HPV16E7 disturbs microtubule dynamics during mitosis by interacting with MAP4. a HPV16E7 expression caused Ac-tubulin accumulation and disturbed the dynamics of microtubules during mitosis. HeLa and HeLa-HPV16E7 cells were treated with Nocodazole for 16 h and then released into fresh medium. Cells were then collected at the time points indicated and subjected to WB. b MAP4 knockdown reduced the Ac-tubulin level in HeLa-HPV16E7 cells. HeLa and HeLa-HPV16E7 cells were separately treated with MAP4 siRNA and then lysed for WB analysis after 72 h. c The Ac-tubulin level decreased under HPV16E7 or MAP4 siRNA treatment in CasKi cells. After treatment with siRNA, the cells were subjected to WB using Ac-tubulin antibody and MAP4 antibody. The protein levels of Ac-tubulin and MAP4 were normalized to those of GAPDH and are displayed as the fold change. d The mRNA levels of HPV16E7 and MAP4 in CasKi cells after HPV16E7 and MAP4 siRNA treatment verified by real-time PCR 72 h after transfection. e In vitro polymerization of tubulin showed that HPV16E7 disturbed the dynamics of tubulin polymerization. Rhodamine-labeled tubulin was incubated with 10% glycerol, 1 mM GTP, and His-MAP4-C in the presence or absence of HPV16E7 (6 μM). The resulting microtubules were fixed, squashed, and sealed under glass coverslips. Scale bar: 10 μm. f The percentage of microtubule area in e was calculated using ImageJ software
HPV16E7 associated with the C terminus of MAP4 (Fig. S1c), and therefore a purified His-MAP4-C was used in vitro assay to explore whether HPV16E7 directly affected the formation of microtubules. In the presence of glycerol and GTP, rhodamine-labeled tubulin can automatically assembly into long microtubules (Fig. 3e). Purified His-MAP4-C can also induce the formation of microtubules in a long polymerization state; however, in the presence of HPV16E7, tubulin polymerization states were significantly changed, with more but shorter polymers in the same area (Fig. 3e). Taking the percentage of microtubule area as a parameter, the quantitative results are shown in Fig. 3f. These in vitro results confirmed that HPV16E7 facilitated excessive tubulin polymerization but destroyed the structure of microtubules. By combining the phenomena observed in vivo and in vitro, our data demonstrated that HPV16E7 could affect the stability and polymerization of spindle microtubules through association with MAP4.
HPV16E7 blocks MAP4 phosphorylation-dependent degradation
HPV16E7 pulled significantly less MAP4 down in Nocodazole-treated cells, suggesting that their binding could be regulated in a cell-cycle-dependent manner (Fig. 1a). For this reason, the binding state by arresting cell cycle at different phases was assessed. With Nocodazole treatment, there was an obvious upward shift of the MAP4 band and the interaction between HPV16E7 and MAP4 was substantially weakened (Fig. 4a). After treatment of the cell lysate with calf intestinal alkaline phosphatase, the shift returned, and the interaction efficiency was rescued (Fig. 4b), which indicated that in Nocodazole-treated cells MAP4 was phosphorylated and the binding of HPV16E7 to low-phosphorylated MAP4 is more efficient than that to high-phosphorylated MAP4.
HPV16E7 downregulates MAP4 phosphorylation and blocks its high-phosphorylation-dependent degradation. a HPV16E7 binds MAP4 in a cell-cycle-dependent manner. HeLa cells were synchronized at the S phase (thymidine, 2 mM, 24 h), G2/M (RO-3306, 9 μM, 19 h), and prometaphase (Nocodazole, 0.1 μg/ml, 16 h, shaken-off) or asynchronized and then lysed and subjected to GST pulldown analysis. b The phosphorylation status of MAP4 affected its association with HPV16E7. Nocodazole-treated cell lysates were treated with calf intestinal alkaline phosphatase (AP). c HPV16E7 inhibited the phosphorylation of MAP4. HeLa and HeLa-HPV16E7 cells were treated with or without Nocodazole and then subjected to WB using 4–20% SDS-PAGE gels. d Change in MAP4 phosphorylation level from c samples determined by mass spectrometry analysis. The data are displayed as fold change and were calculated according to the ratio of intensity. * indicates the sites that were mutated in the next experiments. e HPV16E7 preferred to bind low-phosphorylated MAP4. A GST pulldown assay was conducted using purified GST proteins and 293T cell lysates with MAP4 overexpression, mutant MAP4A5 or MAP4E5 protein. f MAP4 in the high-phosphorylated state (mutant MAP4E5) was degraded during mitosis. HeLa cells transfected with plasmid harboring MAP4, mutant MAP4A5, or MAP4E5 were synchronized with Nocodazole and then released into fresh medium. g High-phosphorylated MAP4 (mutant MAP4E5) was degraded even in the presence of HPV16E7. HeLa-HPV16E7 cells were treated as stated above in f. “AP” indicates alkaline phosphatase; “Thy” indicates thymidine; “MAP4A5” indicates MAP4 T580A/S585A/T620A/T927A/S928A mutant; “MAP4E5” indicates MAP4 T580E/S585E/T620E/ T927E/S928E mutant
The change in MAP4 phosphorylation in HeLa-HPV16E7 cells under Nocodazole treatment was then probed and the data showed that there was not an obvious shift of the MAP4 band (Fig. 4c) compared with control, which suggested that HPV16E7 decreased the phosphorylation level of MAP4. Cell cycle progress was examined using flow cytometry (Fig. S3a, b) to ensure that cells were arrested at prometaphase with Nocodazole treatment.
To explore the MAP4 phosphorylation level and sites, mass spectrometry analysis was performed using four samples from Fig. 4c. As expected, in HeLa cells, the phosphorylation level of MAP4 was significantly upregulated under Nocodazole treatment compared with that in the control at several sites (Fig. 4d, the blue cylinder). Most importantly, with Nocodazole treatment, MAP4 phosphorylation was significantly downregulated at many sites in HeLa-HPV16E7 cells compared with HeLa cells (Fig. 4d, the orange cylinder), especially at the MAP4 C terminus where HPV16E7 binds (Fig. S1a). Then, five sites (580, 585, 620, 927, and 928) that were significantly changed in the presence of HPV16E7 near the C terminus were chosen to make mutants for further study. In one MAP4A5 mutant (T580A/S585A/T620A/T927A/S928A), all five sites were mutated from threonine or serine to alanine residues to mimic the dephosphorylated form, while in the other MAP4E5 mutant (T580E/S585E/T620E/T927E/S928E), all five sites were changed to glutamic acid residues to mimic the phosphorylated form. The binding assay results showed that MAP4A5 interacted with HPV16E7 more strongly than MAP4E5 and the wild-type protein (Fig. 4e), which indicated that HPV16E7 preferred to bind to low-phosphorylated MAP4 to block the phosphorylation of MAP4. Thus, HPV16E7 downregulated the phosphorylation level of MAP4 during mitosis by binding to its low-phosphorylation state.
We further tested whether these MAP4 mutants could retard mitotic progression and if MAP4 mutants would be degraded in mitosis. MAP4A5 and MAP4E5 were overexpressed in HeLa cells, and then, cell samples were collected over a specific time course. MAP4A5, similar to the wild-type MAP4, was not degraded. However, MAP4E5 obviously presented a degradation trend (Fig. 4f). The level of change in cyclin B demonstrated that MAP4A5 delayed mitotic progression and MAP4E5 rescued this delay. The same experiment was performed in HeLa-HPV16E7 cells, and the result indicated that despite HPV16E7 expression exogenous MAP4E5 was still degraded during mitosis and could induce cyclical degradation of cyclin B to restore the delay induced by HPV16E7 (Fig. 4g). Overall, HPV16E7 blocked the high-phosphorylation-dependent degradation of MAP4 to induce mitotic progression arrest.
Mps1 is an upstream kinase of MAP4 and regulates the MAP4 protein level
Given that HPV16E7 slows mitotic progression by blocking the phosphorylation of MAP4, we sought to find the upstream kinase for MAP4 during mitosis. A predictive analysis using NetworKIN software suggested that Mps1 is a potential upstream kinase of MAP4. Consistent with this notion and the observations of other labs, both Mps1 and MAP4 can co-localize onto spindle microtubules in metaphase cells [30] (Fig. 5a). Mps1 formed a complex with MAP4, which was shown by co-immunoprecipitation experiments (Fig. 5b). Furthermore, down-regulation of Mps1 activity by either Mps1-specific siRNA or Reversine can significantly decrease MAP4 phosphorylation levels in mitotic cells, according to the band shift in a Phos-tag gel (Fig. 5c, d). To investigate if Mps1 phosphorylates MAP4 directly, the phosphorylation sites through an in vitro kinase assay were mapped using purified Mps1 and MAP4. Meanwhile, changes in MAP4 phosphorylation sites in response to Mps1 kinase inhibition in mitotic cells were also analyzed. Both assays revealed a common phosphopeptide containing T927S928, which localizes to the C-terminal microtubule-binding domain (Fig. 5e). These two phosphorylation sites (T927S928) are followed by the hydrophobic amino acid alanine, which is a classic phosphorylation pattern targeted by Mps1 [31]. Interestingly, the phosphorylation of T927S928 can also be downregulated by HPV16E7 based on mass-spec data (Fig. 4d), suggesting that these sites are regulated in an opposite way by Mps1 and HPV16E7. Some of phosphorylate sites on MAP4 are not affected by Mps1, which suggests that other kinase(s) are involved in MAP4 phosphorylation.
Mps1 is a potential kinase of MAP4 and regulates MAP4 degradation during mitosis. a MAP4 and Mps1 were co-localized onto spindle microtubules during mitosis. HeLa cells were stained with anti-MAP4, anti-Mps1, and anti-α-tubulin antibodies. b MAP4 associated with Mps1. HeLa cells cotransfected with pEXL-MAP4 and pEXL-Mps1 were submitted to an immunoprecipitation assay using rabbit IgG and anti-MAP4 antibody. c Mps1 inhibition decreased the phosphorylation of MAP4. Forty-eight hours after the HeLa cells were transfected with Mps1 siRNA, the cells were treated with Nocodazole for 16 h. In addition, HeLa cells were treated with Nocodazole for 16 h followed by addition of the Mps1 inhibitor Reversine (1 μM) for 2 h. Another group was released into fresh medium with Reversine and MG132 (10 μM). d Mps1 can phosphorylate MAP4 in vitro. FTH-Mps1 was purified from Mps1-overexpressing 293T cells, and His-MAP4-N and His-MAP4-C were expressed and purified from E. coli. The reaction products were then subjected to western blot analysis using phos-tag gel and anti-His antibody. e Mass-spec analysis of the phosphorylation level of MAP4 induced by Mps1 in vitro and in vivo. The in vitro samples treated as in d were then used for mass spectrometry. For in vivo samples, 293T cells were transfected with MAP4 plasmid followed by treatment with Nocodazole, and another group was treated with Nocodazole, Reversine, and MG132. After 12 h, MAP4 was purified, and a mass spectrometry analysis was conducted to assess the phosphorylation level of MAP4. + indicates the sites that were phosphorylated in the experiments. f Mps1 inhibition increased the protein level of MAP4 during cell cycle progression. HeLa cells were synchronized by RO-3306 and released into fresh medium with Reversine or without Reversine. “Rev” indicates Reversine
Next, we speculated that Mps1 may regulate the degradation of MAP4 during mitosis. To confirm this, the MAP4 level was monitored in late G2 cells, which were arrested by RO-3306 and then released into fresh medium in the presence of Reversine. Consistent with the speculation, inhibition of Mps1 kinase activity increased the protein level of MAP4 in mitosis (Figs. 5f and S3c).
HPV16E7 disrupts MAP4–Mps1 association to inhibit MAP4 phosphorylation
The phosphorylation of T927S928 site can be regulated by Mps1 but is blocked by HPV16E7. To elucidate the relationship between these three proteins, a straightforward hypothesis that HPV16E7 decreases MAP4 phosphorylation by preventing it from associating with Mps1 was proposed. To test this idea, the binding affinities of MAP4 and Mps1 in the presence of HPV16E7 was measured using biolayer interferometry. As shown in Fig. 6a, HPV16E7 disturbed the MAP4–Mps1 interaction. And overexpression of Mps1 in HeLa-HPV16E7 cells rescued HPV16E7-mediated dephosphorylation of MAP4 (Fig. 6b).
HPV16E7 disturbs the MAP4–Mps1 interaction to inhibit MAP4 phosphorylation. a HPV16E7 disturbs the MAP4–Mps1 interaction. Kinetic analysis of the affinity of MAP4-C or the MAP4-C/16E7 complex and Mps1 using Octet Red 96. The protein concentrations of FTH-Mps1, His-MAP4-C, and HPV16E7 were 20 μg/ml, 5 µM, and 2.5 µM, respectively. b Overexpression of Mps1 in HeLa-HPV16E7 cells rescued the inhibition of MAP4 phosphorylation caused by Nocodazole treatment. HeLa-HPV16E7 cells were transfected with plasmid harboring Mps1, followed by Nocodazole treatment and then subjected to WB using a gradient gel. c The phosphorylation status of MAP4 at the T927 and S928 sites was crucial for the degradation of MAP4. MAP4 mutant E2 was degraded during mitosis. HeLa cells transfected with plasmid harboring mutant MAP4A2 or MAP4E2 were synchronized with Nocodazole and then released into fresh medium. d The high-phosphorylated MAP4 mimic (mutant MAP4E2) was degraded even in the presence of HPV16E7. HeLa-HPV16E7 cells were treated as stated above in c. e HPV16E7 preferred to bind MAP4A2 compared with MAP4E2. GST pulldown was conducted using purified GST proteins and 293T cell lysates with MAP4 overexpression, mutant MAP4A2, or MAP4E2 protein. f Overexpression of Mps1 in HeLa-HPV16E7 cells rescued the degradation of MAP4. HeLa-HPV16E7 cells transfected with plasmid harboring Mps1 were synchronized by RO-3306 and released into fresh medium. Cells were then collected at the time intervals indicated and subjected to WB. “MAP4A2” indicates MAP4 T927A/S928A mutant; “MAP4E2” indicates MAP4 T927E/S928E mutant
To further investigate how the phosphorylation of MAP4 by Mps1 affects MAP4 stability under HPV16E7 expression, a phospho-mimic mutant MAP4E2 protein and a phospho-null mutant MAP4A2 protein were made by replacing T927S928 with E927E298 and A927A928, respectively. The results showed that MAP4A2, similar to the wild-type MAP4, was not degraded; however, MAP4E2 obviously presented a degradation trend (Fig. 6c). The level change of Cyclin B demonstrated that MAP4A2 delayed mitotic progression similar to wild-type MAP4 and MAP4E2 rescued this delay. The same results were observed in the presence of HPV16E7 in HeLa-HPV16E7 cells (Fig. 6d). In short, the MAP4 phosphorylation status at sites 927 and 928 plays an important role in both MAP4 degradation and its retarding effect on mitosis. Therefore, our results demonstrated that Mps1 is capable of phosphorylating MAP4 and is involved in regulation of MAP4 degradation during mitosis.
To explore whether the phosphorylation of MAP4 by Mps1 affects the binding of HPV16E7 to MAP4, the binding affinity of HPV16E7 to the MAP4A2 and MAP4E2 mutants were examined. The results showed that MAP4A2 interacted with HPV16E7 more strongly than MAP4E2 and the wild-type protein (Fig. 6e). Moreover, the inhibition of MAP4 degradation caused by HPV16E7 can be rescued to some degree by Mps1 overexpression (Fig. 6f).
Together, our results illustrate that HPV16E7 protein disrupts the Mps1–MAP4 association, resulting in dephosphorylation of MAP4 and extended mitotic progression.
Discussion
In this study, we demonstrated for the first time that HPV16 oncoprotein E7 retards mitotic progression in host cells by binding to MAP4. We also discovered a new mode by which MAP4 activity regulates mitosis: MAP4 is degraded during mitotic progression to promote spindle microtubule movement and its degradation is coupled with its phosphorylation status. An important mitotic kinase, Mps1 is a kinase for MAP4 and is involved in modulating the stability of spindle microtubules together with MAP4. Binding of HPV16E7 to MAP4 blocked the phosphorylation mediated by Mps1, which caused spindle microtubules to be less dynamic and delayed mitotic progression.
During normal mitosis, after nuclear membrane breakdown, chromatin begins to condense, spindle microtubules gradually aggregate and assemble and the MAP4 level dramatically improves, promoting polymerization completion. After metaphase, chromosomes are ready to separate into daughter cells, and spindle microtubules become highly dynamic to facilitate chromosome movement. As a microtubule stability factor, MAP4 is degraded to promote spindle microtubule movement. MAP4 overexpression or a high MAP4 level induced by HPV16E7 inhibits spindle microtubule dynamics, which results in a chromosomal alignment abnormality and delays mitotic progression. Consistent with our results, a previous study found that overexpression of MAP4 causes a potent mitotic block characterized by monoastral spindles [32]. This new regulatory mode presents a new chapter for mechanistic studies of how MAP4 regulates spindle formation.
Mps1 is a critical mitotic kinase, whose function has been well established. Mps1 knockdown causes mitotic slippage from prometaphase because Mps1 is an essential component of Spindle Assembly Checkpoint (SAC) [22]. This means that loss of Mps1 function will accelerate the mitotic process. However, in our study we confirmed that Mps1 modulates MAP4 phosphorylation to enhance the dynamics of spindle microtubule, which illustrates that Mps1 depletion at this time point will delay the mitotic process due to reduced microtubule dynamics. The reason for a conflict here is the different times when the events occur. Mitotic slippage induced by Mps1 depletion occurs before chromosome alignment is completed, but Mps1 promotion of microtubule dynamics occurs after chromosome pairing is completed. Thus, we present another function of Mps1: localizing on the spindle and regulating microtubule dynamics through association with MAP4. Furthermore, the phosphorylation of MAP4 T927S928 site regulated by Mps1 partially abolished E7-binding capacity and rescued mitotic progression in host cells.
HPV cannot enter the nucleus directly until host cell nuclear envelope breakdown, and the viral genome migrates along microtubules to the condensed chromosomes during mitosis [10]. Therefore, the host mitotic progression is critical for the virus to enter the nucleus and initiate viral gene expression. Binding of HPV16E7 with MAP4 blocks mitotic progression and leads to an extended infection window and presents an opportunity for the virus to migrate during mitosis, facilitating integration of the viral genome into the host genome. So, the increased ratio of mitotic cells caused by the HPV16E7–MAP4 interaction is likely to promote persistent HPV infection.
More than 200 HPV types have been identified and 15 of them have been classified as high-risk HPVs, which are carcinogenic for many deadly cancers. Among the high-risk HPVs, HPV16 accounts for nearly half of all cervical cancers. However, the molecular mechanisms under the high case detection rates of HPV16 remains largely unknown. Our study discovered that oncoprotein E7 of HPV16 but not HPV18 retards mitotic progression in host cells by binding to MAP4, which correlates with the high HPV16 prevalence, providing a possibility for understanding this mechanism.
In conclusion, our findings linked HPV16 oncoprotein E7 to MAP4 and Mps1, and uncovered a novel mechanism underlying the regulation of host cell mitosis by HPV16. Our studies take a step forward in understanding of the tumorigenesis mechanism of HPV16 and open a new door for investigating the mechanism of HPV16 prevalence.
Materials and methods
Constructs and stable cell line
HPV16E7, HPV18E7, and HPV11E7 genes were synthesized in Sangon (Shanghai, China) and then cloned into vectors pEGFP-N3 and pGEX4T-1. Human MAP4 constructs were cloned into vector pEXL-Flag for eukaryotic expression and pET30a for prokaryotic expression. Truncated MAP4 proline-rich domain (aa 693–934) and assembly promoting domain (aa 935–1152) were cloned into vector pET30a and the proteins were purified from bacteria. HeLa-HPV16E7, C33A-HPV16E7, and HaCaT-HPV16E7 cell lines, stably expressing HPV16E7, were set up with lenti-virus carrying pLVX-Tight-E7-EGFP (provided by Wuhan Viral Therapy Technologies, Wuhan, China) and selected with puromycin (2 μg/ml). Single colonies were selected by flow cytometry. The expression of HPV16E7 was induced by 2 μg/ml Doxycycline hyclate (Gene Operation, Michigan, USA).
Cell culture, transfection, and drug treatment
HeLa, 293T, CasKi, C33A, and HaCaT cells were obtained from Cell Center of Chinese Academy of Medical Sciences, China. All siRNA duplexes used in this study were purchased from GenePharma (Shanghai, China). The sequences of siRNA duplexes are shown in Supplementary Table S1. Plasmids and siRNA duplexes were transfected with Lipofectamine® 2000 Transfection Reagent (Thermo Fisher Scientific, Waltham, USA) following the manufacturer’s protocol. Thymidine, RO-3306, Nocodazole, Reversine, and MG132 were used as indicated in the main text.
Western blotting, immunofluorescence, and immunoprecipitation
Western blotting and immunofluorescence analyses were performed following standard methods. The antibodies used in this research include: anti-MAP4 (sc-67152; Santa Cruz, Dallas, Texas, USA), anti-HPV16E7 (GTX41516; GeneTex, San Antonio, Texas, USA); anti-Cyclin B (SC-245; Santa Cruz); anti-α tubulin (sc-5286; Santa Cruz), anti-acetylated α tubulin (sc-23950; Santa Cruz), anti-Mps1 (3–472–1; Millipore, Massachusetts, USA), anti-GAPDH (sc-25778; Santa Cruz), anti-GFP (ab290; Abcam, Cambridge, MA, USA), anti-His (sc-57598; Santa Cruz), anti-Flag (F1804; Sigma-Aldrich, USA). Immunofluorescence images were captured using a confocal microscope with a ×63 oil immersion lens (Axio imager. z2; ZEISS) with appropriate filters. For immunohistochemistry, the paraffin-embedded cervix cancer tissues spotted on a chip (T101, Shanxi Chaoyang Biotechnology Co., Xi’an, China) were subjected to incubate with an anti-MAP4 (Santa Cruz, 1:100) antibody and an anti-E7 (GeneTex, 1:20) antibody for 2 h, respectively. Images were acquired with Leica SCN400 slide scanner (Buffalo Grove, IL, USA) and then arranged in Photoshop software. The quantification of E7 and MAP4 levels in cancer tissues and tumor grading was performed manually following the standard methods.
Time-lapse fluorescence imaging
HeLa cells and HeLa-HPV16E7 cells were seeded in a 96-well plate at 50% confluence. Cells were synchronized onto G2/M boundary using RO-3306 for 19 h and then incubated in Hoechst-33342 to stain DNA for a half hour before releasing. Next the plate was transferred into an environmental chamber and images were taken every 5 min using ImageExpress microscope (Thermo Fisher Scientific, USA).
Microtubule assembly assay in vitro
No-tagged HPV16E7 was obtained by thrombin cleavage following a standard procedure from GST beads and Rhodamine-labeled tubulin was brought from Cytoskeleton (TL590M, Denver, USA). The microtubule assembly assays were performed following the procedure as described [15].
Kinetics assay
The FTH-Mps1 fusion protein was biotinylated in PBS buffer with 0.05% tween 20, pH 7.0. The interaction between His-MAP4-C and the FTH-Mps1 fusion protein was determined by biolayer interferometry using an Octet Red96 instrument (Forte Bio Inc., USA). Specifically, the biotinylated FTH-Mps1 at 20 µg/ml was loaded onto streptavidin-coated biosensors and incubated with His-MAP4-C and His-MAP4-C/HPV16E7 mixture. All of the binding data were collected at 30 °C. The experiments comprised five steps: (1) baseline acquisition, (2) FTH-Mps1 protein loading onto the sensor, (3) second baseline acquisition, (4) association of the His-MAP4-C and His-MAP4-C/16E7 proteins, and (5) dissociation of the His-MAP4-C and His-MAP4-C/16E7 proteins. The protein concentrations of His-MAP4-C and HPV16E7 used were 5 and 2.5 μM.
In vitro phosphorylation assay
His-MAP4 fragments were purified from E. coli, and FTH-Mps1 was purified from 293T cells. The in vitro phosphorylation assays were carried out in the absence or presence of 1 μM Reversine (Mps1 inhibitor), and His-MAP4 fragments (3 μg) were mixed on ice with FTH-Mps1 (1 μg) in kinase buffer (50 mM Tris HCl, 0.2 M NaCl, 10 mM MgCl2, 1 mM DTT, 0.2 mM ATP, and 50 ng/μl BSA, pH 7.5). The mixture was incubated for 60 min at 30 °C and then subjected to phos-tagTM WB or mass spectrometry analysis. The group with only His-MAP4 fragments was served as the negative control.
Statistical analysis
Statistical analyses were performed on GraphPad Prism 5.0 software. Results are presented as means ± SD. Statistical significance was determined by Student’s t-test or unpaired two-tailed t-test as indicated in the figure legends. Significance was set at *P < 0.05, **P < 0.01, and ***P < 0.001.
References
Muñoz N, Bosch FX, de Sanjosé S, Herrero R, Castellsagué X, Shah KV, et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348:518–27.
Frisch M, Biggar RJ. Aetiological parallel between tonsillar and anogenital squamous-cell carcinomas. Lancet. 1999;354:1442–3.
Marur S, D’Souza G, Westra WH, Forastiere AA. HPV-associated head and neck cancer: a virus-related cancer epidemic. Lancet Oncol. 2010;11:781–9.
Shaw R, Robinson M. The increasing clinical relevance of human papillomavirus type 16 (HPV-16) infection in oropharyngeal cancer. Br J Oral Maxillofac Surg. 2011;49:423–9.
Bosch FX, Burchell AN, Schiffman M, Giuliano AR, de Sanjose S, Bruni L, et al. Epidemiology and natural history of human papillomavirus infections and type-specific implications in cervical neoplasia. Vaccine. 2008;26(Suppl 10):K1–16.
Koshiol J, Lindsay L, Pimenta JM, Poole C, Jenkins D, Smith JS. Persistent human papillomavirus infection and cervical neoplasia: a systematic review and meta-analysis. Am J Epidemiol. 2008;168:123–37.
Bosch FX, Muñoz N. The viral etiology of cervical cancer. Virus Res. 2002;89:180–90.
Mirabello L, Yeager M, Yu K, Clifford GM, Xiao Y, Zhu B, et al. HPV16 E7 genetic conservation is critical to carcinogenesis. Cell. 2017;170:1164–74.
Pyeon D, Pearce SM, Lank SM, Ahlquist P, Lambert PF. Establishment of human papillomavirus infection requires cell cycle progression. PLoS Pathog. 2009;5:e1000318.
DiGiuseppe S, Luszczek W, Keiffer TR, Bienkowska-Haba M, Guion LGM, Sapp MJ. Incoming human papillomavirus type 16 genome resides in a vesicular compartment throughout mitosis. Proc Natl Acad Sci USA. 2016;113:6289–94.
Burger MP, van Leeuwen AM, Hollema H, Quint WG, Pieters WJ. Human papillomavirus type influences the extent of chromosomal lag during mitosis in cervical intraepithelial neoplasia grade III. Int J Gynecol Pathol. 1997;16:10–4.
Thomas LK, Bermejo JL, Vinokurova S, Jensen K, Bierkens M, Steenbergen R, et al. Chromosomal gains and losses in human papillomavirus-associated neoplasia of the lower genital tract—a systematic review and meta-analysis. Eur J Cancer. 2014;50:85–98.
Espinosa AM, Alfaro A, Roman-Basaure E, Guardado-Estrada M, Palma Í, Serralde C, et al. Mitosis is a source of potential markers for screening and survival and therapeutic targets in cervical cancer. PLoS ONE. 2013;8:e55975.
Steinmann KE, Pei XF, Stoppler H, Schlegel R, Schlegel R. Elevated expression and activity of mitotic regulatory proteins in human papillomavirus-immortalized keratinocytes. Oncogene. 1994;9:387–94.
Kremer BE, Haystead T, Macara IG. Mammalian septins regulate microtubule stability through interaction with the microtubule-binding protein MAP4. Mol Biol Cell. 2005;16:4648–59.
Takahashi M, Shiraishi H, Ishibashi Y, Blade KL, McDermott PJ, Menick DR, et al. Phenotypic consequences of beta1-tubulin expression and MAP4 decoration of microtubules in adult cardiocytes. Am J Physiol Heart Circ Physiol. 2003;285:H2072–83.
Holmfeldt P, Brattsand G, Gullberg M. MAP4 counteracts microtubule catastrophe promotion but not tubulin-sequestering activity in intact cells. Curr Biol. 2002;12:1034–9.
Zahnleiter D, Hauer NN, Kessler K, Uebe S, Sugano Y, Neuhauss SC, et al. MAP4-dependent regulation of microtubule formation affects centrosome, cilia, and Golgi architecture as a central mechanism in growth regulation. Hum Mutat. 2015;36:87–97.
Jiang YY, Shang L, Shi ZZ, Zhang TT, Ma S, Lu CC, et al. Microtubule-associated protein 4 is an important regulator of cell invasion/migration and a potential therapeutic target in esophageal squamous cell carcinoma. Oncogene. 2016;35:4846–56.
Ou Y, Zheng X, Gao Y, Shu M, Leng T, Li Y, et al. Activation of cyclic AMP/PKA pathway inhibits bladder cancer cell invasion by targeting MAP4-dependent microtubule dynamics. Urol Oncol. 2014;32:21–8.
Gallo DE, Hope TJ. Knockdown of MAP4 and DNAL1 produces a post-fusion and pre-nuclear translocation impairment in HIV-1 replication. Virology. 2012;422:13–21.
Abrieu A, Magnaghi-Jaulin L, Kahana JA, Peter M, Castro A, Vigneron S, et al. Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell. 2001;106:83–93.
Lauzé E, Stoelcker B, Luca FC, Weiss E, Schutz AR, Winey M. Yeast spindle pole body duplication gene MPS1 encodes an essential dual specificity protein kinase. EMBO J. 1995;14:1655–63.
Maciejowski J, Drechsler H, Grundner-Culemann K, Ballister ER, Rodriguez-Rodriguez JA, Rodriguez-Bravo V, et al. Mps1 regulates kinetochore-microtubule attachment stability via the ska complex to ensure error-free chromosome segregation. Dev Cell. 2017;41:143–56.
Hiruma Y, Sacristan C, Pachis ST, Adamopoulos A, Kuijt T, Ubbink M, et al. Competition between MPS1 and microtubules at kinetochores regulates spindle checkpoint signaling. Science. 2015;348:1264–7.
West RR, Tenbarge KM, Olmsted JB. A model for microtubule-associated protein 4 structure. Domains defined by comparisons of human, mouse, and bovine sequences. J Biol Chem. 1991;266:21886–96.
Piperno G, LeDizet M, Chang XJ. Microtubules containing acetylated alpha-tubulin in mammalian cells in culture. J Cell Biol. 1987;104:289–302.
Mandelkow E, Mandelkow EM. Microtubules and microtubule-associated proteins. Curr Opin Cell Biol. 1995;7:72–81.
Andersen SS. Spindle assembly and the art of regulating microtubule dynamics by MAPs and Stathmin/Op18. Trends Cell Biol. 2000;10:261–7.
Dou Z, Sawagechi A, Zhang J, Luo H, Brako L, Yao XB. Dynamic distribution of TTK in HeLa cells: insights from an ultrastructural study. Cell Res. 2003;13:443–9.
Xu Q, Zhu S, Wang W, Zhang X, Old W, Ahn N, et al. Regulation of kinetochore recruitment of two essential mitotic spindle checkpoint proteins by Mps1 phosphorylation. Mol Biol Cell. 2009;20:10–20.
Holmfeldt P, Brattsand G, Gullberg M. Interphase and monoastral-mitotic phenotypes of overexpressed MAP4 are modulated by free tubulin concentrations. J Cell Sci. 2003;116:3701–11.
Acknowledgements
This work was supported in part by grants from National Basic Research Program of China (2014CB138505) and National Natural Science Foundation of China (81871130).
Author information
Authors and Affiliations
Contributions
Y.G. and X.Z. performed the majority of experiments, analyzed the data, and prepared the figures. Q.X. performed the live-cell imaging experiment. F.N., W.Z., and J.L. assisted in the experiments and data analysis. F.G. and X.S. performed histological analysis. C.L., R.H., and J.T. constructed the plasmids. R.L., L.Z., and L.C. performed manuscript proofreading. R.Z.M. directed the project. Y.G., X.Z., and R.Z.M wrote the paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Guo, Y., Zhang, X., Xu, Q. et al. Human Papillomavirus 16 oncoprotein E7 retards mitotic progression by blocking Mps1-MAP4 signaling cascade. Oncogene 38, 5959–5970 (2019). https://doi.org/10.1038/s41388-019-0851-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-019-0851-1
- Springer Nature Limited