Abstract
N6-methyladenosine (m6A) is the most abundant modification in eukaryotic messenger RNAs (mRNAs), and plays important roles in many bioprocesses. However, its functions in bladder cancer (BCa) remain elusive. Here, we discovered that methyltransferase-like 3 (METTL3), a major RNA N6-adenosine methyltransferase, was significantly up-regulated in human BCa. Knockdown of METTL3 drastically reduced BCa cell proliferation, invasion, and survival in vitro and tumorigenicity in vivo. On the other hand, overexpression of METTL3 significantly promoted BCa cell growth and invasion. Through transcriptome sequencing, m6A sequencing and m6A methylated RNA immuno-precipitation quantitative reverse-transcription polymerase chain reaction, we revealed the profile of METTL3-mediated m6A modification in BCa cells for the first time. AF4/FMR2 family member 4 (AFF4), two key regulators of NF-κB pathway (IKBKB and RELA) and MYC were further identified as direct targets of METTL3-mediated m6A modification. In addition, we showed that besides NF-κB, AFF4 binds to the promoter of MYC and promotes its expression, implying a novel multilevel regulatory network downstream of METTL3. Our results uncovered an AFF4/NF-κB/MYC signaling network operated by METTL3-mediated m6A modification and provided insight into the mechanisms of BCa progression.
Similar content being viewed by others
Introduction
Urothelial carcinoma of the bladder is the fourth most common malignancy in men, with ~81,190 new cases and 17,240 deaths estimated in 2018 in the United States [1]. Most cases are diagnosed in advanced stages and patients cannot undergo curative surgery. Chemotherapy has been the main treatment option with agents such as gemcitabine, cisplatin, oxaliplatin, capecitabine, and 5-fluorouracil (5-FU) [2]. However, despite the introduction of target molecules and immunotherapy into clinical application in recent years, the response to these treatments are still limited with modest impact in overall survival [3,4,5], mainly because of the innate and adaptive drug resistance. Thus, understanding the mechanism of BCa progression and identifying new therapeutic approaches for this disease is an urgent and unmet need.
N6-Methyladenosine (m6A) is the most prevalent internal chemical modification of mRNAs in eukaryotes [6,7,8]. In mammalian cells, this modification is catalyzed by a methyltransferase complex consisting of the proteins methyltransferase-like 3 (METTL3), METTL14, Wilms tumor 1 associated protein (WTAP), VIRMA (KIAA1429), and RBM15 [9,10,11,12], and “erased” by two mammalian RNA demethylases, the fat mass and obesity-associated protein (FTO) [13] and alkylation repair homolog protein 5 (ALKBH5) [14], which demonstrated a dynamic nature of m6A methylation. In human cells, thousands of mRNAs are subject to m6A modification [7, 15], which has been implicated in mRNA splicing, nuclear export, mRNA stability, translation [16] as well as microRNA processing [17]. Recent studies demonstrate that m6A regulation play important roles in many biological processes, such as development, metabolism, fertility [18], osteoporosis [19], stemness maintenance and differentiation [20]. It also contribute to synaptic function [21], axon regeneration of neuron [22], and facilitates hippocampus-dependent learning and memory [23]. Besides, Treg suppressive functions [24] and innate immunity [25] of the immune system were regulated by mRNA m6A modification as well. As the critical methyltransferase of RNA m6A modification, METTL3 was reported to promote lung cancer [26], liver cancer [27], breast cancer [28] and myeloid leukemia progression [29], as well as the chemo- and radioresistance of pancreatic cancer cells [30] by some most recent studies. Additionally, METTL3 mediated m6A modification is also involved in the maintenance of embryonic stem cells [31] and, not surprisingly, cancer stem cells [32]. Nevertheless, there is also evidence about METTL3 plays a tumor suppressor role in glioblastoma (GBM) [33], suggesting its context-dependent roles in cancer progression. However, the definite role of METTL3 in BCa has never been studied. In the present study, we sought to investigate the biological function of METTL3 in the pathogenesis of BCa and also explored the underlying molecular mechanism through identification of its critical mRNA targets.
Results
METTL3 expression is elevated in BCa tissue
To gain insight into the role of m6A modification in BCa progression in vivo, we detected mRNA m6A level in two human BCa samples. Compared with the corresponding Para-tumor bladder urothelial tissues (PT), the m6A level in both tumor tissues were significantly elevated (Fig. 1a). We then checked the expression of m6A writer and eraser in the Cancer Genome Atlas (TCGA) datasets (http://cancergenome.nih.gov) to determine which subunit may play the crucial role of m6A deregulation in BCa, and found that METTL3 mRNA expression is significantly elevated in BCa compared with the normal tissues (Fig. 1b). In contrast, expression of METTL14, WTAP, FTO or ALKBH5 were not significantly changed in these patient samples (Fig. 1b). We then examined the protein level of METTL3 in the samples from a cohort of 22 BCa patients by immunohistochemistry (n = 22). In line with the TCGA data, METTL3 expression was significantly higher in tumor samples than in the para-tumor samples (n = 7, Fig. 1c). Besides, we also examined the section samples from previously established N-butyl-N-4-hydroxybutyl Nitrosamine (BBN)-induced bladder carcinogenesis mouse model [34], and found that the expression of METTL3 in mice with invasive BCa (n = 12) was dramatically elevated compared with normal bladder tissue (n = 12, Fig. 1d). With the above evidence, we concluded that METTL3 is frequently up-regulated in BCa and may be implicated in pathogenesis and progression of BCa.
METTL3 expression in normal bladder tissue and BCa samples. a mRNA m6A level in two human BCa samples determined by LC-MS/MS analysis (*p < 0.05; **p < 0.01, student t-test). b METTL3 was up-regulated in BCa (TCGA data, Red box for tumor tissue, n = 404; gray box for normal tissue, n = 19). Representative image of immunohistochemical staining by METTL3 antibody and quantitative measurement in both human (c, n = 22) and mouse BCa samples (d, n = 12) compare with human para-tumor tissue (PT, n = 7) and mouse normal bladder tissues (Normal, n = 12). The intensities of immunostaining were quantitatively measured using Image-Pro Plus 6.0 image analysis software (**p < 0.01, student t-test). Scale bar. 50 μm
METTL3 Promotes BCa Cell Proliferation, Migration and Suppresses Apoptosis in vitro
Considering our findings that METTL3 is aberrantly expressed in BCa tissue, we’re motivated to determine if METTL3 regulated m6A modification plays a role in BCa tumorigenesis. We knocked down METTL3 using two distinct siRNAs (si-METTL3-1, si-METTL3-2) in the BCa cell line 5637. Knockdown of METTL3 expression by both siRNAs was confirmed by qRT-PCR (Fig. 2a) and western blot (Fig. 2b). As expected, METTL3 depletion in BCa cells decreased the global mRNA m6A level (Fig. 2c), and also results in strong inhibition of cancer cell growth (by MTS assay, Fig. 2d), increases cell apoptosis (by Flow Cytometry, Fig. 2e), and decreases the invasive ability of BCa cells (by the Boyden Chamber Assay, Fig. 2f). The effects of METTL3 were also confirmed in another BCa cell line UM-UC-3. Similarly, cell growth and invasion were decreased, while cell apoptosis were elevated upon METTL3 knockdown (Supplementary Fig. 1a-1c). We next investigated how overexpressed METTL3 affect the growth and invasion of bladder cells. Immortalized human uroepithelial cells SV-HUC-1 were transduced with lentivirus expressing control vector or METTL3. mRNAs and proteins were isolated from both cells before RT-PCR analysis and western blot confirmed the overexpression of METTL3 with METTL3-expressing lentivirus (Fig. 2g, h). In contrast to the knockdown assays, ectopic expression of METTL3 promote the cell growth and invasion (Fig. 2i, j). Taken together, our data demonstrated the oncogenic role of METTL3 in BCa cell growth, survival, and invasion.
Effect of METTL3 on BCa cell proliferation, invasion and cell apoptosis. The knockdown effect of specific siRNAs (si-METTL3-1 and -2) in 5637 cells was verified at both the mRNA (a, by qRT-PCR) and protein levels (b, by western blot). c, mRNA m6A level in METTL3 knockdown 5637 cells determined by LC-MS/MS analysis (*p < 0.05 relative to scramble, One Way ANOVA). d MTS assay of cellular proliferation in 5637 cells (**p < 0.01 Two Way ANOVA). e Annexin V/PI staining of METTL3 knockdown and control 5637 cells analyzed by FACS. Quantification of apoptotic cells were plotted, numbers represent the sum of early and late apoptotic cells (**p < 0.01 ***p < 0.001 relative to scramble, One Way ANOVA). f In vitro cell invasion assay, Quantification of invasive 5637 cells (***p < 0.001 relative to scramble, One-way ANOVA). Overexpression of METTL3 in SV-HUC-1 cells transfected with METTL3 expressing plasmid (METTL3) and control vector (control) respectively were confirmed by qRT-PCR g and Western Blot h. i MTS assay of cellular proliferation in SV-HUC-1 cells (**p < 0.01 Two Way ANOVA). j In vitro cell invasion assay, Quantification of invasive SV-HUC-1 cells. Bar graphs: mean ± S.D., n = 3
Identification of METTL3 targets by high-throughput RNA-Seq and m6A-Seq
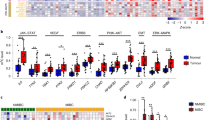
To identify potential mRNA targets of METTL3 the m6A levels of which are induced by METTL3 in BCa cells, we performed transcriptome sequencing and m6A-sequencing to interrogate the expression changes and map the m6A modification in METTL3 knockdown 5637 cells. RNA-seq revealed that 1759 genes were significantly downregulated, while 1793 genes were significantly upregulated (fold change > 2.0) upon METTL3 knockdown. Gene set enrichment analysis (GSEA) revealed there are multiple signaling pathways positively/negatively correlated with METTL3 depletion (Supplementary Table S1), among which the MYC target genes as well as TNF-α/NF-κB pathway target genes showed significant negative correlation with METTL3 knockdown.
Global profiling of m6A target genes using meRIP-seq identified 3,588 m6A peaks (in 2,537 genes) in 5637 cells. Consistent with published studies, when mapped the m6A methylomes in 5637 cells, we found the GGAC motif was identified to be highly enriched in the immunopurified RNA (Fig. 3b), and a metagene analysis revealed that m6A peaks were predominantly localized near stop codons, with a subset of m6A peaks located in the 5′-UTR and internal exons (Supplementary Figure.S2). When searching for m6A peaks in the key regulators of pathways listed in Supplementary Table S1,, we found that significant m6A abundances near the stop codon and downstream 3′-UTR region of MYC, AFF4, RELA(p65) and IKBKB(IKK-β) (Fig. 3c), which involved in MYC and NF-κB pathways, respectively. qRT-PCR using primers to amplify either the m6A peak region or a control (non-peak) region was used to validate the meRIP-seq data (Fig. 3d-g).
Identification of potential targets of METTL3 in BCa via transcriptome-wide m6A seq and RNA-Seq assays. a GSEA showing MYC and NF-κB targets are significantly enriched and correlated with METTL3 knockdown in 5637 cells. b Global profiling of m6A in 5637 cells, sequence motif identified from the top 1,000 m6A peaks. c The m6A abundances on MYC, AFF4, IKBKB and RELA mRNA transcripts in 5637 cells as detected by m6A-seq are plotted using Integrative genomics viewer (IGV). The y axis shows sequence read number, blue boxes represent exons, and blue lines represent introns. Reduction of m6A modification in specific regions of MYC d, AFF4 e, RELA f and IKBKB g transcripts upon METTL3 knockdown as tested by gene-specific m6A-qPCR assay in 5637 cells. h qPCR showing mRNA level of indicated genes after knockdown of METTL3. i Western blot assays of MYC, AFF4, IKBKB (IKK-β), and RELA (p65) in 5637 cells with METTL3 siRNAs or Scramble RNA transfection. α-Tubulin was used as the endogenous control protein for loading control. METTL3 targets’ protein level determined by immunohistochemical staining using indicated antibodies in both human j and mouse BCa samples k compare with human para-tumor tissue (PT) and mouse normal bladder tissues (Normal). (*p < 0.05; **p < 0.01; ***p < 0.001 by student t-test). *p < 0.05; **p < 0.01; ***p < 0.001 relative to the scramble group, One-way ANOVA followed by Tukey’s test
Further qRT-PCR analysis confirmed that knockdown of METTL3 using siRNA resulted in significantly reduced levels of MYC and AFF4 mRNAs, while had little effect on the abundance of RELA or IKBKB mRNA (Fig. 3h). As for the protein levels determined by western blot, expression of all 4 genes were substantially reduced upon METTL3 knockdown (Fig. 3i). Additionally, protein levels of MYC, AFF4 and RELA were also elevated in both human (Fig. 3j and Supplementary Fig. S3a) and mouse (Fig. 3k and Supplementary Fig. S3b) BCa samples. These data strongly indicate that MYC, AFF4, RELA and IKBKB as bona fide targets of METTL3.
MYC, AFF4, RELA and IKBKB are functionally important target genes of METTL3 in BCa
When we transfected human BCa cells with specific siRNAs targeting METTL3, a significant inhibition on activities of MYC (Fig. 4a) and NF-κB (Fig. 4b) pathways determined by dual luciferase assay were observed. To explore the mechanism of how METTL3 regulated cell survival, proliferation and invasion of BCa cells through NF-κB signaling, we further investigated the expression of some NF-κB target genes that are associated with these phenotypes in BCa [35]. Specifically, the mRNA levels of BCL2A1, BIRC3, KITLG, MMP9 and PLAU were reduced after knockdown of METTL3 (Fig. 4c). Furthermore, similar to METTL3 knockdown, when 5637 cells were transfected with siRNAs targeting MYC, AFF4, RELA and IKBKB, respectively, their growth/proliferation (Fig. 4d) and invasion ability (Fig. 4e) were inhibited and apoptosis were induced (Fig. 4f). Taken together, our results demonstrated that MYC, AFF4, RELA and IKBKB are functionally important targets of METTL3 and are important for METTL3-mediated promotion of BCa cell proliferation, invasion and survival.
MYC, AFF4, RELA and IKBKB are critical targets of METTL3 that mediate BCa cell growth, survival, and invasion. a and b, The relative activities (mean ± S.D) of the Myc a and NF-κB b pathways in 5637 cells transfected with indicated siRNAs. c mRNA levels of NF-κB downstream targets determined by qRT-PCR. d MTS assay of cellular proliferation in 5637 cells transfected with indicated siRNAs. e, Quantification of invasive 5637 cells by in vitro cell invasion assay. f AnnexinV/PI staining 5637 cells transfected with indicated siRNAs analyzed by FACS. Quantification of apoptotic cells were plotted, numbers represent the sum of early and late apoptotic cells. **p < 0.01; ***p < 0.001 relative to the scramble control group. One way ANOVA. Error bars represent the S.D. of three independent experiments
AFF4 directly regulates MYC gene expression in BCa cells
The roles of both MYC [36, 37] and NF-κB [35, 38, 39] pathways have been well defined in BCa. However, how AFF4 is implicated in BCa initiation and progression has not been investigated yet. AFF4 is an essential component of super elongation complex (SEC), which is involved in regulation of transcription elongation of many oncogenic genes including MYC [40]. To identify the potential targets of AFF4 (and SEC) in BCa cells, we therefore performed transcriptome sequencing in AFF4 knockdown 5637 cells. Interestingly, GSEA suggested an enrichment of MYC target genes downregulated in AFF4 knockdown cells (Fig. 5a), which agrees with the results of METTL3 knockdown in BCa cells. To investigate whether MYC is directly regulated by AFF4 in BCa cells, we further confirmed expression of MYC in response to AFF4 knockdown by qRT-PCR and western blot. The results showed knockdown of AFF4 reduced MYC expression at both mRNA level and protein level (Fig. 5b, c). Furthermore, we found AFF4 directly bound to MYC promoter as determined by ChIP assay, which is significantly decreased after AFF4 knockdown (Fig. 5d). The above results suggest that AFF4 may involve in the BCa tumorigenesis by acting as a direct upstream regulator of the MYC.
MYC expression in BCa cells is regulated by AFF4. a GSEA showing MYC targets are significantly enriched and correlated with AFF4 knockdown in 5637 cells. Expression of MYC and knockdown of AFF43 in 5637 cells by siRNAs (si-AFF4-1 and -2) was verified at both the mRNA (b, by qRT-PCR) and protein levels (c, by western blot). d ChIP assay showed the recruitment of AFF4 at MYC promoter in 5637 cells transfected with indicated siRNAs at 48 h post transfection. n = 3. **p < 0.01 relative to the scramble group, Two-way ANOVA followed by Tukey’s test
Oncogenic role of METTL3 relies on its methyltransferase activity
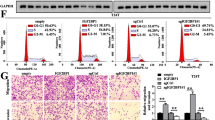
METTL3 has been reported to function without METTL14 and promotes translation of specific mRNAs independently of its catalytic activity in vitro [26]. To investigate if the m6A catalytic activity is responsible for the oncogenic role of METTL3, we established stable METTL3 knockdown model in 5637 cells with specific shRNA (Fig. 6a), and try to rescue its function by transfecting METTL3 cDNA containing nucleotide substitutions that make it resistant to shRNA for expression of Flag tagged wild-type (METTL3R WT) or catalytic mutant METTL3 (METTL3R Mut) protein into the stable knockdown cells. As expected, western blotting and qRT-PCR showed strongly reduced endogenous expression of MYC, AFF4, RELA and IKBKB upon knockdown of METTL3 without affecting the mRNA abundance of RELA and IKBKB (Fig. 6a, b). Expression of METTL3R WT rescued the expression of all four targets but METTL3R Mut failed to recover target gene expression (Fig. 6b). Moreover, phenotypes of cell growth (Fig. 6c), invasion (Fig. 6d) and apoptosis (Fig. 6e) can also be rescued by METTL3R WT but not the catalytic mutant METTL3. These data indicate that the oncogenic role of METTL3 is dependent on its methyltransferase activity in BCa cells.
METTL3 regulates BCa progression and target gene expression via its methyltransferase activity. a METTL3 stable knock down 5637 cell was generated by lentiviral-based shRNA expression and further transfected with METTL3R WT (Rwt) or METTL3R Mut (Rmut), Expression of indicated genes were determined by qRT-PCR a and Western Blot b. c MTS assay of cellular proliferation (**p < 0.01 Two Way ANOVA). d Annexin V/PI staining of the cells analyzed by FACS. Quantification of apoptotic cells were plotted, numbers represent the sum of early and late apoptotic cells (***p < 0.001 relative to control, One Way ANOVA). e In vitro cell invasion assay, Quantification of invasive cells (***p < 0.001 relative to control, One-way ANOVA)
Knock down of METTL3 diminish tumorigenicity of BCa cells in vivo
To investigate the functional roles of METTL3 in vivo, we performed subcutaneous implantation experiment in nude mice. To minimize the inter-mouse bias, 5637 control or 5637 sh-METTL3 cells (1.5 × 107 cells/site) were subcutaneously injected at two back sites of eight mice each (Fig. 7a). Tumor volume were monitored each week and tumor mass was weighed on day 35th at the end of this study. Stable knockdown of METTL3 effectively suppressed tumor growth as reflected by the significant reduction of tumor size and weight when compared with the nontarget shRNA control (Fig. 7b, c). Further confirmation of the METTL3’s role in BCa came from the immuno-histological analysis of Ki67 (an indicator for cell proliferation), active Caspase-3 (an indicator for cell apoptosis), MYC, AFF4 and RELA (targets of METTL3) in the tumor sessions. Knockdown of METTL3 reduced cell proliferation but induced cell apoptosis in BCa tumor (Fig.7d and Supplementary Fig. S3c), and also led the expected changes of MYC, AFF4 and RELA’s protein level in tumor slides (Fig. 7e and Supplementary Fig. S3c). All these results consolidate the conclusion that suggest that METTL3 and its targets play a pivotal role in promoting BCa progression in vivo.
Silencing METTL3 inhibited BCa progression in vivo. a The experimental scheme of the in vivo assay, METTL3 stable knock down or control cells were subcutaneously injected at two points at back of each nude mice, respectively. b Tumor growth curve of xenografts generated by METTL3 stable knock down BCa cells and control cells (***p < 0.001 using an equivalent of ANCOVA). c The image of the tumors on the 35th day. d The mean + S.D. of tumor weight of the tumor from each group. e Quantitative measurement of Ki67, Active Caspase-3(AcCasp-3), MYC, AFF4 and RELA in xenografts generated by METTL3 stable knock down BCa cells and control cells. (*p < 0.05; **p < 0.01; ***p < 0.001 by student t-test)
Discussion
M6A is the most prevalent chemical modification for human mRNA, it has been reported to be essential for cancer progression. Due to the differences of regions that m6A distributed on mRNAs, different readers responding to these m6A modifications, cancer content and target genes regulating different cellular processes, functions of m6A in cancer progression may sometimes be controversial. For instance, both the m6A writer (METTL3 and METTL14) and eraser (FTO and ALKBH5) were reported to play oncogenic roles in different types of cancer such like acute myeloid leukemia and GBM [41], whereas METTL3 and METTL14 can also act as tumor suppressor in GBM [33] and hepatocellular carcinoma [42], respectively. Thus, studying the detailed mechanisms underlying m6A modification is needed to illuminate its role in certain types of cancer. Here, we first unveiled that the major m6A writer METTL3 rather than METTL14 or m6a erasers, was aberrantly expressed in human and mouse BCa. We then functionally demonstrated the essential role of METTL3 in promoting BCa growth and survival through both in vitro and in vivo models. More importantly, we identified the key regulators in NF-κB pathway (IKBKB and RELA), Myc pathway (MYC) and RNA elongation (AFF4) as direct downstream targets of METTL3-mediated m6A modification in BCa. Collectively, our study demonstrates the functional importance of the m6A methylation and the corresponding proteins in BCa, which identifies the m6A mRNA methylation machinery as promising therapeutic targets for BCa.
METTL3-mediated m6A modification was recently discovered to regulate numerous genes in various types of cancer, yet its targets in BCa remain unknown. In this study, we utilized a combination of m6A sequencing and transcriptome mRNA sequencing to provide the first evidence for METTL3–mediate m6A modification profiling of BCa. Consistent with the reported m6A modification of MYC mRNA in normal hematopoietic cells, leukemia cells [43, 44] and glioma [45], expression of MYC was also regulated by METTL3–mediated m6A modification in BCa. However, different with METTL14 and FTO, both of which promote stability of MYC mRNA [44, 45], METTL3 mediated m6A modification was thought to promote the translational efficiency of MYC mRNA through affecting m6A abundance on different sites [43]. Our results suggested that depletion of METTL3 in BCa cells decreased the stability of MYC transcripts through affecting m6A abundance mainly around the stop codon and 3′-UTR regions. It has been reported that there is a ~250- nucleotide cis-acting element termed as coding region instability determinant (CRD) in the 3′-region of MYC, which is required for regulating the stability of MYC mRNA [46], and m6A reader IGF2BP preferentially recognize and bind to the m6A-modified CRD region of MYC mRNA, thereby stabilizing MYC mRNA and promoting translation [47]. Meanwhile, AFF4 might share the similar regulatory mechanism employed by METTL3 –mediated m6A modification with MYC, since both of them have reduced mRNA and protein expression upon METTL3 depletion. On the other hand, although the protein levels of IKBKB and RELA were efficiently reduced upon METTL3 knockdown, little effect has been seen on their mRNA levels. Indeed, it has been reported that m6A reader YTHDF1 could selectively recognize m6A-modified mRNAs and promote translation efficiency, and m6A-modified mRNAs of both IKBKB and RELA were characterized to be direct targets of YTHDF1 via a photoactivatable ribonucleoside crosslinking and immunoprecipitation (PAR-CLIP) assay [48], which indicate METTL3 might promote the expression of IKBKB and RELA mainly by regulating the translational efficiency.
Oncogene MYC is known to be aberrantly expressed in BCa and the antitumor effect of MYC inhibitor has already been tested [36, 49]. Indeed, MYC is upregulated in the majority of all human cancers. The broad oncogenic effects of MYC rely on triggering the expression of target genes to benefit cell proliferation, cell survival as well as stemness maintenance [50, 51]. Mechanisms of MYC deregulation in BCa include DNA mutation [52, 53], signal transduction/transcriptional regulation [37, 54] and microRNA mediated post-transcriptional regulation [55, 56]. NF-κB is a well-known upstream regulator of MYC expression [57], by which NF-κB signaling enhances the proliferation and survival of cancer cells during the development and recurrence of BCa [35]. Besides, previous studies have provided evidence that MYC is one of the direct targets of AFF4/SEC, and SEC recruitment to the MYC gene regulates its expression in different cancer cells [40]. Inspired by the close correlation between AFF4 and cancer progression in acute lymphoblastic leukemia [58] and head and neck squamous cell carcinoma (HNSCC) [59], we supposed that AFF4 may also be involved in BCa progression. In the current study, expression AFF4 was found to be elevated in BCa samples from both mouse model and clinical samples. Furthermore, AFF4 directly bound to the promoter of MYC to facilitate transcription elongation. Hence, signal diverging from METTL3-mediated m6A modification finally converges into MYC expression. Collectively, our observations demonstrated that METTL3 mediated m6A modification upregulate MYC expression at multiple levels, which include activating NF-κB signaling (through IKBKB and RELA) to induce MYC transcription [60], MYC mRNA elongation (through AFF4) [40] and m6A modification abundance of MYC mRNA. This malignant regulatory network coordinated by METTL3-mediated m6A modification efficiently raised MYC protein level in BCa and probably makes MYC difficult to be decreased by blocking a single signaling.
It is well known that cancer is driven by hundreds or even thousands of dysregulated genes. AFF4/SEC and NF-κB also have a broad effect on gene expression of various cancer related genes. Therefore, the oncogenic role of METTL3 might not merely rely on MYC. Other potential target genes of METTL3 involved in BCa progression still await further investigation. Among these candidates, SOX2 was also identified as a functionally important target of METTL3 in GBM, and METTL3-mediated m6A modification of SOX2 mRNA transcripts makes them more stable [61]. Intriguingly, recent study found AFF4 could also be recruited to SOX2 promoter and promote SOX2 expression in HNSCC [59]. In our previous study, we have found SOX2 is a marker of stemness BCa stem cells both in vivo and in vitro [34]. Taken together, these clues hint the possibility that METTL3 might involve in maintaining the stemness of BCa stem cells through inducing m6A modification of SOX2.
In sum, our studies demonstrate the critical role of METTL3 in BCa progression, as featured by promoting cancer cell growth, survival and invasion. Importantly, we uncovered that METTL3 operated a regulatory network which involves AFF4, NF-κB and MYC signaling. Thus, we present the first insight of METTL3-mediated cancer progression and speculate that targeting METTL3 might be an effective therapeutic strategy to treat BCa.
Methods and materials
Samples, plasmids, and cell lines
We used human BCa samples obtained from Department of Urology, Huadong Hospital, Fudan University with patients’ informed consent. The pathological condition was determined by experienced urologists at Huadong Hospital. Patient characteristics for the samples used for m6A level analysis are described in the supplementary Table S2. The study was approved by the local ethics committee (20160059). METTL3 expressing lentivirus vector were purchased from Hanbio (Shanghai). Plasmids for expression of Flag tagged wild-type (METTL3R WT) and catalytic mutant METTL3 (METTL3R Mut) were kindly provided by Dr. Shuibin Lin (Sun yet-sen University) [62]. Mouse BCa samples were prepared in our previous study [34]. BCa cell line 5637 (ATCC NO. HTB-9), UM-UC-3 (ATCC NO.CRL-1749) and immortalized uroepithelial cell SV-HUC-1 (ATCC NO. CRL-9520) were obtained from the Chinese Academy of Cell Resource Center (Shanghai, China) and maintained as previously described [63]. Cell lines were routinely tested for mycoplasma and not cultured for longer than 20 passages.
LC-MS/MS analysis of m6A level
RNA from patient samples and 5637 cell lines were extracted with TRIzol (Invitrogen). The integrity and quantity of each sample was examined using agarose gel electrophoresis and Nanodrop™ instrument. mRNA was isolated and purified from total RNA using NEBNext® Poly(A) mRNA Magnetic Isolation Module (For next generation sequencing using) and then hydrolysed to single nucleosides. Nucleosides were further dephosphorylated by enzyme mix. Pretreated nucleosides solution was deproteinized using Satorius 10,000-Da MWCO spin filter.
Analysis of nucleoside mixtures was performed on Agilent 6460 QQQ mass spectrometer with an Agilent 1290 HPLC system. Multi reaction monitoring (MRM) mode was performed because of its high selectivity and sensitivity attained working with parent-to-product ion transitions. LC-MS/MS data was acquired using Agilent Qualitative Analysis software. MRM peaks of each modified nucleoside were extracted and normalized to peak areas of normal adenosine in each sample. Samples were run in duplicate, and m6A/A ratios were calculated.
Detect gene expression
For mRNA level examination, total RNA of BCa cells was extracted by using Trizol reagent (Invitrogen). Complementary DNA (cDNA) synthesis was performed with GeneAmp RNA PCR kit (Invitrogen) using 1 µg RNA per sample. qPCR reactions were performed using Power SYBR Green Master Mix to determine mRNA transcript level. Primers for qRT-PCR are listed in Supplementary Table S3.
For Western blotting, 5637 cells were lysed with RIPA buffer as standard protocol. Cell lysate were then mixed with loading buffer and incubated at 100 ℃ for 5 min and subject to conventional Western analysis. Antibodies are listed in Supplementary Table S4.
For paraffin sections of bladder cancer tissue samples from mice and human patient were antigen retrieved, blocked and processed as described before [64]. Hematoxylin–eosin stains were performed using standard histology procedures. The intensity of immunostaining was measured by Image-Pro Plus 6.0 image analysis software (Media Cybernetics). The intensity of each image was calculated by normalizing the average integrated optical density with the total selected area of interest.
In vitro cell proliferation, invasion, and apoptosis assay
For cell proliferation assay, 5 × 103 cells per well were seeded onto a 96-well plate on day 0. Absorbances at 490 nm were measured using CellTiter 96 AQueous One Solution Cell Proliferation Assay kit (Promega) for 4 consecutive days. Triplicated samples were counted.
For cell invasion assay, BioCoat™ Matrigel invasion chamber was used according to the manufacturer’s instruction (BD Biosciences). Briefly, 4 × 104 cells transfected with siRNAs were resuspended in 100 μl of DMEM medium, and seeded in the upper portion of the invasion chamber. The lower portion of the chamber contained 500 μl of medium supplemented with 2% FBS and glutamine, which served as a chemoattractant. After 24 h, non-invasive cells were removed from the upper surface of the membrane with a cotton swab. The invasive cells on the lower surface of the membrane were stained with crystal violet, and counted in four separate areas with an inverted microscope.
The numbers of apoptotic cells were quantified by flow cytometric assays using FITC Annexin V Apoptosis Detection Kit I (BD biosciences) following the manufacturer’s instructions.
m6A meRIP-Seq, mRNA sequencing and gene set enrichment analysis
meRIP-seq and data analysis were performed as previously described [26]. Briefly, mRNA was purified from total RNA of 5637 cells using PolyATtract mRNA Isolation Systems (Promega). 5 mg mRNA was then fragmented and immunoprecipitated with anti-m6A antibody (Synaptic Systems, 202003). The immunoprecipitated RNA was washed and eluted by competition with N6-methyladenosine (Sigma-Aldrich, M2780), and analyzed either through quantitative reverse-transcription polymerase chain reaction (qRTPCR) or by high-throughput sequencing. For highthroughput sequencing, purified RNA fragments from m6A-MeRIP were used for library construction using TruSeq Stranded mRNA Sample Prep Kits (Illumina RS-122-2101). Samples were then sequenced with Illumina HiSeq 2000. Reads mapping, m6A peak calling, motif search and following analysis were performed as described [65].
For RNA-sequencing, RNA from the 5637 cells transfected with different siRNAs were extracted with TRIzol (Invitrogen). RNA libraries were prepared using the KAPA RNA-Seq Library Preparation Kit (KAPA Biosystems Cat#07960140001). RNAs were single-end sequenced on Illumina HiSeq 3000 machines. Alignment of reads was done using Tophat with the Hg38 build of the human genome. Transcript assembly and differential expression was determined using Cufflinks with Refseq mRNAs to guide assembly. Analysis of RNA-seq data was done using the cummeRbund package in R (http://cole-trapnell-lab.github.io/cufflinks/). Transcripts with extremely low expression in both cell lines were filtered out and all rest genes were preranked by expression fold change and subject to a Geneset enrichment analysis to find enriched functional annotations.
GSEA were performed with GSEA software (http://www.broad.mit.edu/GSEA), which is a computational method that determines whether there is any difference of a priori defined set of genes between two biological states. P values were computed using a bootstrap distribution created by resampling gene sets of the same cardinality. Details of gene lists that were used in the analysis HALLMARK_MYC_TARGETS_V1 (n = 200) and HALLMARK_TNFA_SIGNALING_VIA_NFKB (n = 200) are described by the following link: http://software.broadinstitute.org/gsea/msigdb/.
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed using a Simple ChIP Assay Kit (Cell Signaling Technology, Danvers, MA) according to the manufacturer’s instruction. The precipitated DNA samples were purified and measured by Q-PCR. Results were shown as the percentage of input controls. Antibodies and primers used for CHIP assay were listed in the supplementary table S2 and Table S3.
The pathway luciferase reporter assay
Activities of the NF-κB and MYC signaling pathways were determined using the Qiagen’s pathway reporter systems The analysis was carried out according to the manufacturer’s instruction (Qiagen) [66]. Briefly, the cells were transfected in triplet with each firefly luciferase reporter construct in combination with the Renilla luciferase construct, and both luciferase activities in cell extracts at 24 h after transfection were measured by a Promega Dual-Luciferase Reporter assay (Promega) using a Promega GloMax 20/20 luminometer. Firefly luciferase activities from each set were normalized to the activity of Renilla luciferase to control the inter-transfection bias. The relative luciferase activities (luciferase unit) of the pathway reporter over the negative control in the transfected cells were calculated as a measurement of the pathway activity.
In vivo xenografts model
The pLKO.1 lentiviral shRNA constructs targeting the human METTL3 (shMETTL3: RHS3979-201764032 and GFP: RHS4459) were purchased from Dharmacon. BALB/c male nude mice of 8 weeks of age were used for this study, no randomization or blinding of groups was done. METTL3 stable knockdown or control (sh-GFP) cells were embedded in BD Matrigel™ Matrix and subcutaneously injected into the left or right flank of mice back, respectively (overall 8 mice). Tumor volume were assessed weekly (Length × Width2 × 0.52). Mice were humanely sacrificed on day 35, and the tumors were weighed and photographed. The tumor weight was described as the mean ± S.D. Tumor samples were paraffin-embedded, sectioned for further IHC staining. All animal procedures were performed under a protocol approved by the Laboratory Animal Center of Anhui Medical University and in accordance with National Institutes of Health guide for the care and use of Laboratory animals (NIH Publications No. 8023, revised 1978).
Statistics
Data are presented as the mean ± standard deviation (S.D.) or standard error (S.E.). All of the statistical analyses were performed using Excel (Microsoft, Redmond, WA) or Prism (GraphPad Software Inc., La Jolla, CA). The two-tailed Student’s t-test, one-way analysis of variance was used to calculate statistical significance. A p-value of <0.05 was considered significant.
Data availability
The authors declare that all relevant data are available within the article and its Supplementary information files or from the corresponding author upon reasonable request. The m6A-sequencing and RNA-sequencing datasets have been submitted to the NCBI database under the accession number PRJNA498900, SAMN10337857 and SAMN10337858.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA. 2018;68:7–30.
Felsenstein KM, Theodorescu D. Precision medicine for urothelial bladder cancer: update on tumour genomics and immunotherapy. Nat Rev Urol. 2018;15:92–111.
Ribas A, Tumeh PC. The future of cancer therapy: selecting patients likely to respond to PD1/L1 blockade. Clinical cancer research: an official journal of the American Association for. Cancer Res. 2014;20:4982–4.
Oing C, Rink M, Oechsle K, Seidel C, von Amsberg G, Bokemeyer C. Second line chemotherapy for advanced and metastatic urothelial carcinoma: vinflunine and beyond-a comprehensive review of the current literature. J Urol. 2016;195:254–63.
van Kessel KE, Zuiverloon TC, Alberts AR, Boormans JL, Zwarthoff EC. Targeted therapies in bladder cancer: an overview of in vivo research. Nat Rev Urol. 2015;12:681–94.
Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci USA. 1974;71:3971–5.
Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–6.
Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O. et al. 5′ UTR m(6)A promotes cap-independent translation. Cell. 2015;163:999–1010.
Bokar JA, Rath-Shambaugh ME, Ludwiczak R, Narayan P, Rottman F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J Biol Chem. 1994;269:17697–704.
Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L. et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93–95.
Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ. et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–89.
Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, Zhao JC. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16:191–8.
Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–7.
Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49:18–29.
Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L. et al. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15:707–19.
Meyer KD, Jaffrey SR. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol. 2014;15:313–26.
Alarcon CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF. N6-methyladenosine marks primary microRNAs for processing. Nature. 2015;519:482–5.
Niu Y, Zhao X, Wu YS, Li MM, Wang XJ, Yang YG. N6-methyl-adenosine (m6A) in RNA: an old modification with a novel epigenetic function. Genom Proteom Bioinform. 2013;11:8–17.
Wu Y, Xie L, Wang M, Xiong Q, Guo Y, Liang Y. et al. Mettl3-mediated m(6)A RNA methylation regulates the fate of bone marrow mesenchymal stem cells and osteoporosis. Nat Commun. 2018;9:4772
Wu Y, Zhou C, Yuan Q. Role of DNA and RNA N6-adenine methylation in regulating stem cell fate. Curr Stem Cell Res Ther. 2018;13:31–38.
Merkurjev D, Hong WT, Iida K, Oomoto I, Goldie BJ, Yamaguti H. et al. Synaptic N(6)-methyladenosine (m(6)A) epitranscriptome reveals functional partitioning of localized transcripts. Nat Neurosci. 2018;21:1004–14.
Weng YL, Wang X, An R, Cassin J, Vissers C, Liu Y. et al. Epitranscriptomic m(6)A regulation of axon regeneration in the adult mammalian nervous system. Neuron. 2018;97:313–25.e316.
Shi H, Zhang X, Weng YL, Lu Z, Liu Y, Lu Z. et al. m(6)A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature. 2018;563:249–53.
Tong J, Cao G, Zhang T, Sefik E, Amezcua Vesely MC, Broughton JP. et al. m(6)A mRNA methylation sustains Treg suppressive functions. Cell Res. 2018;28:253–6.
Zheng Q, Hou J, Zhou Y, Li Z, Cao X. The RNA helicase DDX46 inhibits innate immunity by entrapping m(6)A-demethylated antiviral transcripts in the nucleus. Nat Immunol. 2017;18:1094–103.
Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell. 2016;62:335–45.
Chen M, Wei L, Law CT, Tsang FH, Shen J, Cheng CL. et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67:2254–70.
Cai X, Wang X, Cao C, Gao Y, Zhang S, Yang Z. et al. HBXIP-elevated methyltransferase METTL3 promotes the progression of breast cancer via inhibiting tumor suppressor let-7g. Cancer Lett. 2018;415:11–19.
Barbieri I, Tzelepis K, Pandolfini L, Shi J, Millan-Zambrano G, Robson SC. et al. Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature. 2017;552:126–31.
Taketo K, Konno M, Asai A, Koseki J, Toratani M, Satoh T. et al. The epitranscriptome m6A writer METTL3 promotes chemo- and radioresistance in pancreatic cancer cells. Int J Oncol. 2018;52:621–9.
Cao G, Li HB, Yin Z, Flavell RA. Recent advances in dynamic m6A RNA modification. Open Biol. 2016;6:160003.
Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I. et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA. Proc Natl Acad Sci USA. 2016;113:E2047–2056.
Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G. et al. m(6)A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18:2622–34.
Zhu F, Qian W, Zhang H, Liang Y, Wu M, Zhang Y. et al. SOX2 is a marker for stem-like tumor cells in bladder cancer. Stem Cell Rep. 2017;9:429–37.
Mukherjee N, Houston TJ, Cardenas E, Ghosh R. To be an ally or an adversary in bladder cancer: the NF-kappaB story has not unfolded. Carcinogenesis. 2015;36:299–306.
Jeong KC, Kim KT, Seo HH, Shin SP, Ahn KO, Ji MJ. et al. Intravesical instillation of c-MYC inhibitor KSI-3716 suppresses orthotopic bladder tumor growth. J Urol. 2014;191:510–8.
Mahe M, Dufour F, Neyret-Kahn H, Moreno-Vega A, Beraud C, Shi M. et al. An FGFR3/MYC positive feedback loop provides new opportunities for targeted therapies in bladder cancers. EMBO Mol Med. 2018; 10:pii: e8163.
Zhu J, Li Y, Chen C, Ma J, Sun W, Tian Z. et al. NF-kappaB p65 overexpression promotes bladder cancer cell migration via FBW7-mediated degradation of RhoGDIalpha protein. Neoplasia. 2017;19:672–83.
Zheng J, Kong C, Yang X, Cui X, Lin X, Zhang Z. Protein kinase C-alpha (PKCalpha) modulates cell apoptosis by stimulating nuclear translocation of NF-kappa-B p65 in urothelial cell carcinoma of the bladder. BMC Cancer. 2017;17:432.
Luo Z, Lin C, Guest E, Garrett AS, Mohaghegh N, Swanson S. et al. The super elongation complex family of RNA polymerase II elongation factors: gene target specificity and transcriptional output. Mol Cell Biol. 2012;32:2608–17.
Deng X, Su R, Weng H, Huang H, Li Z, Chen J. RNA N(6)-methyladenosine modification in cancers: current status and perspectives. Cell Res. 2018;28:507–17.
Ma JZ, Yang F, Zhou CC, Liu F, Yuan JH, Wang F. et al. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6)-methyladenosine-dependent primary MicroRNA processing. Hepatology. 2017;65:529–43.
Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G. et al. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med. 2017;23:1369–76.
Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L. et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell. 2018;22:191–205. e199.
Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y. et al. R-2HG exhibits anti-tumor activity by targeting FTO/m(6)A/MYC/CEBPA signaling. Cell. 2018;172:90–105. e123.
Doyle GA, Betz NA, Leeds PF, Fleisig AJ, Prokipcak RD, Ross J. The c-myc coding region determinant-binding protein: a member of a family of KH domain RNA-binding proteins. Nucleic Acids Res. 1998;26:5036–44.
Huang H, Weng H, Sun W, Qin X, Shi H, Wu H. et al. Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20:285–95.
Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H. et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–99.
Seo HK, Ahn KO, Jung NR, Shin JS, Park WS, Lee KH. et al. Antitumor activity of the c-Myc inhibitor KSI-3716 in gemcitabine-resistant bladder cancer. Oncotarget. 2014;5:326–37.
Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV. MYC, metabolism, and cancer. Cancer Discov. 2015;5:1024–39.
Gabay M, Li Y, Felsher DW. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harbor Perspect Med. 2014; 4:pii: a014241.
Watters AD, Latif Z, Forsyth A, Dunn I, Underwood MA, Grigor KM. et al. Genetic aberrations of c-myc and CCND1 in the development of invasive bladder cancer. Br J Cancer. 2002;87:654–8.
Tabach Y, Kogan-Sakin I, Buganim Y, Solomon H, Goldfinger N, Hovland R. et al. Amplification of the 20q chromosomal arm occurs early in tumorigenic transformation and may initiate cancer. PLoS One. 2011;6:e14632
Shiina H, Igawa M, Shigeno K, Terashima M, Deguchi M, Yamanaka M. et al. Beta-catenin mutations correlate with over expression of C-myc and cyclin D1 Genes in bladder cancer. J Urol. 2002;168:2220–6.
Li Y, Liu H, Lai C, Du X, Su Z, Gao S. The Lin28/let-7a/c-Myc pathway plays a role in non-muscle invasive bladder cancer. Cell Tissue Res. 2013;354:533–41.
Li Y, Xu Z, Wang K, Wang N, Zhu M. Network analysis of microRNAs, genes and their regulation in human bladder cancer. Biomed Rep. 2013;1:918–24.
Duyao MP, Buckler AJ, Sonenshein GE. Interaction of an NF-kappa B-like factor with a site upstream of the c-myc promoter. Proc Natl Acad Sci USA. 1990;87:4727–31.
Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L. et al. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell. 2010;37:429–37.
Deng P, Wang J, Zhang X, Wu X, Ji N, Li J. et al. AFF4 promotes tumorigenesis and tumor-initiation capacity of head and neck squamous cell carcinoma cells by regulating SOX2. Carcinogenesis. 2018;39:937–47.
Kamat AM, Tharakan ST, Sung B, Aggarwal BB. Curcumin potentiates the antitumor effects of Bacillus Calmette-Guerin against bladder cancer through the downregulation of NF-kappaB and upregulation of TRAIL receptors. Cancer Res. 2009;69:8958–66.
Visvanathan A, Patil V, Arora A, Hegde AS, Arivazhagan A, Santosh V. et al. Essential role of METTL3-mediated m(6)A modification in glioma stem-like cells maintenance and radioresistance. Oncogene. 2018;37:522–33.
Choe J, Lin S, Zhang W, Liu Q, Wang L, Ramirez-Moya J. et al. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature. 2018;561:556–60.
Li Y, Deng H, Lv L, Zhang C, Qian L, Xiao J. et al. The miR-193a-3p-regulated ING5 gene activates the DNA damage response pathway and inhibits multi-chemoresistance in bladder cancer. Oncotarget. 2015;6:10195–206.
Liang Y, Zhu F, Zhang H, Chen D, Zhang X, Gao Q. et al. Conditional ablation of TGF-beta signaling inhibits tumor progression and invasion in an induced mouse bladder cancer model. Sci Rep. 2016;6:29479
Dominissini D, Moshitch-Moshkovitz S, Salmon-Divon M, Amariglio N, Rechavi G. Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nat Protoc. 2013;8:176–89.
El‐Sheikh A, Fan R, Birks D, Donson A, Foreman NK, Vibhakar R. Inhibition of Aurora Kinase A enhances chemosensitivity of medulloblastoma cell lines. Pediatr Blood Cancer. 2010;55:35–41.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81872313 and 81672776 to YL, 81802391 to QG, 31501838 to XH Z), and Anhui Provincial Natural Science Foundation (1808085QH266 to QG)
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Cheng, M., Sheng, L., Gao, Q. et al. The m6A methyltransferase METTL3 promotes bladder cancer progression via AFF4/NF-κB/MYC signaling network. Oncogene 38, 3667–3680 (2019). https://doi.org/10.1038/s41388-019-0683-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-019-0683-z
- Springer Nature Limited
This article is cited by
-
PIGT promotes cell growth, glycolysis, and metastasis in bladder cancer by modulating GLUT1 glycosylation and membrane trafficking
Journal of Translational Medicine (2024)
-
The relationship between the network of non-coding RNAs-molecular targets and N6-methyladenosine modification in tumors of urinary system
Cell Death & Disease (2024)
-
RNA N6-methyladenosine modifications in urological cancers: from mechanism to application
Nature Reviews Urology (2024)
-
METTL3 facilitates renal cell carcinoma progression by PLOD2 m6A-methylation under prolonged hypoxia
Cell Death & Disease (2024)
-
ZFHX3 acts as a tumor suppressor in prostate cancer by targeting FTO-mediated m6A demethylation
Cell Death Discovery (2024)