Abstract
Breast cancer is a heterogeneous disease, and triple-negative breast cancer (TNBC) continues to be a serious health problem. The potential involvement of lncRNAs in TNBC progression remains unexplored. Here, we demonstrated that LINC01638 is highly expressed in TNBC tissues and cells. LINC01638 maintains the mesenchymal traits of TNBC cells, including an enriched epithelial-mesenchymal transition (EMT) signature and cancer stem cell-like state. LINC01638 knockdown suppresses tumor proliferation and metastasis both in vitro and in vivo. LINC01638 overexpression predicts a poor outcome of breast cancer patients. Mechanistically, LINC01638 interacts with c-Myc to prevent SPOP-mediated c-Myc ubiquitination and degradation. C-Myc transcriptionally enhances MTDH (metadherin) expression and subsequently activates Twist1 expression to induce EMT. Our findings describe LINC01638-mediated signal transduction and highlight the crucial role of LINC01638 in TNBC progression.
Similar content being viewed by others
Introduction
As the most common cancer, breast cancer represents a significant cause of cancer related death for women worldwide [1]. Being a heterogeneous disease, breast cancer has varying immunohistochemical characteristics and genetic alterations, as well as distinct clinical prognoses. Breast cancer can be divided into four major intrinsic subtypes based on gene expression profiling: luminal A, luminal B, ErbB2-enriched, and triple-negative breast cancer (TNBC), which generally includes Basal-like and Claudin-low subtypes [2, 3]. Accounting for 12–17% of breast cancer cases, TNBC is characterized by lacking expression of estrogen receptor (ER), progesterone receptor (PR), and epidermal growth factor receptor (HER2/ErbB2) [4]. Patients diagnosed with TNBC usually have a greater risk of distant metastasis and mortality [5]. TNBC cells display remarkable mesenchymal features, including an enriched EMT (epithelial–mesenchymal transition) signature and cancer stem cell (CSC)-like traits [6]. Due to its aggressive nature, TNBC is a severe breast cancer subtype; thus, it is crucial to investigate newly studied long non-coding RNAs (lncRNAs) as potential genetic markers and therapeutic targets to fight this disease [7, 8].
Long non-coding RNAs (lncRNAs) are an important set of emerging non-coding transcripts that are longer than 200 nucleotides and do not encode proteins [9]. LncRNAs function in a wide range of biological activities, including cell cycle regulation, stem cell pluripotency, lineage differentiation and cancer progression, by regulating gene expression by various mechanisms [10, 11]. Models point out that lncRNAs might function at multiple aspects, such as transcription, mRNA splicing and stability, translation and degradation, and chromatin and protein conformation [12, 13]. Recent evidence has shown abnormal lncRNA expression in a variety of cancers, and lncRNAs have been associated with proliferation, differentiation, survival, EMT, CSCs, metastasis, and prognosis of cancer [14,15,16]. Although several lncRNAs have been revealed to be involved in breast cancer [8, 17], the mechanisms and specific roles of lncRNAs in regulating the mesenchymal phenotype and mediating metastasis remain poorly studied.
A validated lncRNA, LINC01638 (long intergenic non-protein coding RNA 1683), was previously demonstrated to be expressed at a low level in multiple normal human tissues [18] and may be involved in EMT of hepatocellular carcinoma cells [15]. In this study, we report that LINC01638 is selectively upregulated in TNBC and explore the function and mechanism of LINC01638 in maintaining the mesenchymal phenotype of TNBC.
Results
LINC01638 as a lncRNA is upregulated in triple-negative breast cancer subtype
To analyze the expression pattern of LINC01638 in breast cancer, we examined LINC01638 expression in 54 matched, fresh-frozen breast cancer tissues and adjacent non-tumor breast tissues. The results indicated that the LINC01638 transcript is upregulated in TNBC patient cohorts compared with cohorts of luminal A, luminal B and HER2+ patients (Fig. 1a). We further confirmed LINC01638 to be highly expressed in TNBC cells (Fig. 1b). LINC01638 is composed of three exons, with a transcript of 637 nt.LINC01638 is poly (A)-negative (Ensembl:ENSG00000233521). Analysis of the sequences by CPAT online software was unable to predict the protein coding potentiality of LINC01638, althoug there is a predicted ORF among the transcript (Fig. 1c). Moreover, RegRNA 2.0 online software predicted the same ORF, but there is no ribosome binding site in the LINC01638 transcript (Fig. 1d), supporting that LINC01638 no protein-coding potential is discovered. To verify the protein coding ability of LINC01638, full-length LINC01638 was cloned into a eukaryotic expression vector, pCMV-Flag, and the results indicated that LINC01638 does not encode protein (Fig. 1e). Then, to investigate whether the LINC01638 ORF is active, a series of constructs was generated in which the Flag tag was fused to the ORF or the 5′-UTR-ORF of the full LINC01638 transcript. However, no substantial expression of Flag fusion proteins were detected from ORF-Flag and 5′-UTR-ORF-Flag constructs (Fig. 1f). Furthermore, cellular fractionation assays indicated that LINC01638 expression is mainly concentrated in the nuclei of TNBC cells (Fig. 1g). These data show that expression of LINC01638, as an lncRNA, is upregulated in TNBC cells.
LINC01638 is upregulated in the triple-negative breast cancer subtype. a Scatter plots comparing LINC01638 expression in tissue samples from different breast cancer subtypes, including Luminal A (n = 19), Luminal B (n = 5), HER2+ (n = 13) and TNBC (n = 17), as detected by qRT-PCR. b Expression levels of LINC01638 in BC cell lines were detected by qRT-PCR. c, d Schematic of the coding potential and ribosome binding sites of LINC01638 trascript predicted by online analysis. e, f The full-length LINC01638 or ORF was cloned into a eukaryotic expression vector, pCMV-Flag, with or without an N-terminal ATG codon in different expression patterns. MDA-MB-231 cells transfected with related instructs and then western blot was performed. The p21 served as a positive control. An anti-Flag antibody was used to probe transcribed proteins. Black arrowhead indicates p21-Flag protein. g Fractionation of MDA-MB-231 cells followed by qRT-PCR. BCAR4 served as a positive control for nuclear gene expression and GAPDH served as a positive control for cytoplasmic gene expression. Student’s t-test, **p < 0.01, ***p < 0.001, ****p < 0.0001
LINC01638 promotes mesenchymal traits in BC cells
Given that TNBC cells have remarkable mesenchymal features and given that LINC01638 is highly expressed in TNBC tissues and cells, to determine the roles of LINC01638 in TNBC cells, we stably silenced LINC01638 against either exon 1 or exon 3 in MDA-MB-231 and BT549 cells and established cell lines with a stable knockdown of LINC01638 (Fig. 2a). sh-2# (against exon 3) has a more effective knockdown capability. Distinctly, LINC01638 depletion reduced the growth of MDA-MB-231 and BT549 cells (Fig. 2b). Conversely, we overexpressed LINC01638 in T47D cells to establish a cell line that stably overexpressed LINC01638 and found that LINC01638 overexpression dramatically promoted cell proliferation in vitro (Fig. 2c). To directly test whether LINC01638 is involved in mesenchymal features, including EMT and CSC-like traits, we determined the invasion ability of MDA-MB-231 and BT549 cells after LINC01638 knockdown using Matrigel-coated transwell experiments. We observed that the invasion potential of MDA-MB-231 and BT549 cells were inhibited significantly by the knockdown of LINC01638, whereas LINC01638 overexpression enhanced the invasion potential of T47D cells (Fig. 2d). Additionally, LINC01638 knockdown robustly decreased the proliferation of CD44+CD24− cells (CSCs) and the ability of mammosphere formation in MDA-MB-231 and BT549 cells, whereas LINC01638 overexpression enhanced their generation (Fig. 2e, f). To determine whether LINC01638 was involved in regulating EMT, we examined the effect of LINC01638 on EMT marker expression. Analysis of the epithelial marker E-cadherin and mesenchymal markers Vimentin and EMT-associated transcription factor Twist1 revealed that LINC01638 knockdown increased E-cadherin and reduced Vimentin and Twist1 expression at the mRNA (Fig. 2g) and protein (Fig. 2h) levels in MDA-MB-231 and BT549 cells, whereas LINC01638 overexpression showed an inverse effect in T47D cells (Fig. 2g, h). These data suggest that LINC01638 functions by mediating the mesenchymal traits of BC cells.
LINC01638 promotes mesenchymal traits in BC cells. a LINC01638 was knocked down in MDA-MB-231 and BT549 cells by two independent shRNAs targeting either exon 1 or 3. LINC01638-knockdown stable cell lines were established. b LINC01638 knockdown repressed proliferation of MDA-MB-231 and BT549 cell lines in vitro, as determined by the cell number count. c Stable LINC01638 expression in T47D cells was established by transfection with the LINC01638-expressing construct EXP-LV203-LINC01638 (left). LINC01638 overexpression promoted the proliferation potential of T47D cells in vitro, as determined by a cell number count (right). d The indicated cell clones were added to the top of transwells coated with Matrigel. e CD44+CD24− (CSC) subpopulations were detected by FACS analysis in clones with LINC01638 knockdown or overexpression. f The mammosphere forming assays were performed in LINC01638 knockdowned or overexpressed clones. g The transcript levels of EMT were determined in indicated cell clones by qRT-PCR. h The protein levels of EMT markers were detected in indicated cell clones by western blot. Student’s t-test, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
LINC01638 mediates mesenchymal traits via MTDH regulation
Given that metadherin (MTDH) is widely involved in tumor growth, apoptosis inhibition, therapeutic resistance, relapse and metastasis [19] and given that Twist1 acts downstream of MTDH, we next investigated whether LINC01638 modulated MTDH, which would affect Twist1 and mesenchymal traits in BC cells. We first measured the MTDH transcript levels in the 54 matched, fresh-frozen breast cancer tissues and adjacent non-cancerous breast tissues. MTDH mRNA had a high expression in TNBC tissues and significantly correlated with the LINC01638 transcript levels (Fig. 3a). The MTDH mRNA and protein levels were also much higher in TNBC cells (Fig. 3b). LINC01638 knockdown reduced MTDH transcript and protein expression in MDA-MB-231 and BT549 cells, whereas LINC01638 overexpression increased the MTDH transcript and protein levels in T47D cells (Fig. 3c). Next, we determined whether LINC01638 exerts its roles by modulating MTDH. For the rescue experiment, we overexpressed MTDH in LINC01638-knockdown MDA-MB-231 cells. MTDH overexpression abrogated the suppressive effects of LINC01638 knockdown on proliferation, invasion and generation of CD44+CD24- cells in MDA-MB-231 cells (Fig. 3d). Conversely, for the function-loss experiment, we knocked down MTDH in LINC01638-overexpressing T47D cells. MTDH knockdown overcame the promotive effects of LINC01638 overexpression on proliferation, invasion and generation of CD44+CD24− cells in T47D cells (Fig. 3e). At the molecular level, LINC01638 knockdown reduced Twist1 and N-cadherin and increased E-cadherin protein levels in MDA-MB-231 cells, whereas MTDH overexpression abolished these effects (Fig. 3f). By contrast, LINC01638 overexpression downregulated E-cadherin and upregulated the Twist1 and N-cadherin protein levels in T47D cells, whereas MTDH knockdown abrogated these effects (Fig. 3g). These data indicate that LINC01638 promotes the mesenchymal traits of breast cancer cells by positively regulating MTDH expression.
LINC01638 mediates mesenchymal traits by regulating MTDH. a Scatter plots comparing MTDH expression in tissue samples from different breast cancer subtypes, including Luminal A (n = 19), Luminal B (n = 5), HER2+ (n = 13) and TNBC (n = 17), examined using qRT-PCR (left). The correlation between LINC01638 and MTDH transcript levels in tissue samples (right). b MTDH transcript and protein levels in cell lines were examined by qRT-PCR and western blot, respectively. c LINC01638 knockdown downregulated MTDH expression and LINC01638 overexpression upregulated MTDH expression at the transcript and protein levels, as detected by qRT-PCR (upper) and western blot (bottom), respectively. d Ectopic expression of MTDH abrogated the effects of LINC01638 knockdown on proliferation, invasion and generation of CD44+/CD24− (CSC) subpopulations in MDA-MB-231 cells. e MTDH knockdown abolished the effects of LINC01638 overexpression on proliferation, invasion and generation of CD44+/CD24− (CSC) subpopulations in T47D cells. f Ectopic expression of MTDH abrogated the effects of LINC01638 knockdown on the regulation of EMT markers in MDA-MB-231 cells examined by western blot. g MTDH knockdown abolished the effects of LINC01638 overexpression on the regulation of EMT markers in T47D cells examined by western blot. Student’s t-test, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001
MTDH is transcriptionally regulated by c-Myc in BC cells
Because LINC01638 modulates MTDH expression at the transcriptional level, we next sought to identify the transcription factor that is directly regulating MTDH transcription. Previously, c-Myc has been reported to transcriptionally regulate MTDH expression [20]. Here, we wondered whether c-Myc also regulated MTDH expression in BC cells. By using the JASPAR database (http://jaspar.binf.ku.dk), we screened potential transcription factors which located within a two-kb region upstream of the MTDH gene transcription start site and identified two c-Myc binding sites at the potential promoter (Fig. 4a). TNBC cells had higher protein levels of c-Myc than cells from other breast cancer subtypes (Fig. 3b), but no significant difference was revealed at the mRNA level (data not shown). Next, we knocked down c-Myc using shRNAs in MDA-MB-231 and BT549 cells and found that c-Myc knockdown reduced the MTDH mRNA (Fig. 4b) and protein (Fig. 4c) levels. By contrast, we also overexpressed c-Myc in T47D cells and found that c-Myc overexpression notably increased the MTDH mRNA (Fig. 4b) and protein (Fig. 4c) levels. To investigate whether c-Myc directly bound the MTDH promoter, we developed chromatin immunoprecipitation (ChIP) of c-Myc in BC cells followed by quantitative PCR (qPCR) of the MTDH promoter. The experimental results indicated that c-Myc was mainly enriched at site B in MDA-MB-231 and BT549 cells, but not in T47D cells (Fig. 4d). c-Myc knockdown reduced the enrichment of c-Myc to the MTDH promoter, whereas c-Myc overexpression enhanced this enrichment (Fig. 4e). To examine whether c-Myc activates the MTDH promoter, WT or mutant (deletion of the site B binding motif) MTDH promoter reporter constructs were transfected into control and c-Myc-knockdown MDA-MB-231 or BT549 cells. c-Myc knockdown repressed the promoter activity of WT, but not mutant MTDH (Fig. 4f). Additionally, c-Myc overexpression promoted WT MTDH promoter activity in T47D cells (Fig. 4f). These data suggest that MTDH is transcriptionally regulated by c-Myc in BC cells.
MTDH is transcriptionally regulated by c-Myc in BC cells. a Schematic of the predicted c-Myc binding sites in the MTDH promoter. b, c MTDH transcript and protein levels in cell lines following c-Myc knockdown or overexpression were examined by qRT-PCR (b) and western blot (c), respectively. d c-Myc mainly binds the MTDH promoter at site B, as determined by ChIP-qPCR. e The binding level of c-Myc with the MTDH promoter is influenced by c-Myc levels, as determined by ChIP-qPCR. f MTHD promoter-driven luciferase activity positively correlated with c-Myc levels in BC cell lines, as determined by luciferase reporter assays. Data are shown as the means ± SD. Student’s t-test, **p < 0.01, ***p < 0.001, ****p < 0.0001
LINC01638 interacts with c-Myc to prevent SPOP mediated degradation
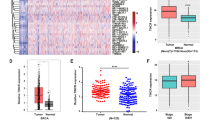
Online analysis based on TCGA data (http://gepia.cancer-pku.cn/) indicates that there are lower c-Myc mRNA levels in BC tissues compared to normal mammary tissues (Fig. 5a). Here, no significant difference was found in the c-Myc mRNA levels between BC cell lines (data not shown). We also measured the c-Myc mRNA levels in the 54 matched, fresh-frozen breast cancer tissues and adjacent non-tumor breast tissues. c-Myc mRNA was slightly lower in BC tissues, with no significant differences detected between BC subtypes (data not shown). However, c-Myc protein was significantly overexpressed in TNBC cell lines (Fig. 3b). Additionally, we found that LINC01638 knockdown resulted in decreased protein levels of c-Myc in MDA-MB-231 and BT549 cells (Fig. 5b). Given these observations, we hypothesized that LINC01638-regulated c-Myc protein in TNBC cells may result from post-transcriptional regulation. To assess this possibility, the proteasome inhibitor MG-132 was applied to MDA-MB-231 and BT549 cells alongside LINC01638 knockdown. MG-132 treatment restored the decrease of c-Myc protein induced by LINC01638 knockdown (Fig. 5b). Studies have reported that the E3 ubiquitin ligase adapter speckle-type POZ (SPOP) can promote c-Myc ubiquitination and degradation [21]. Analysis from TCGA data showed no significant difference in SPOP transcript levels between breast cancer and normal tissues (data not shown). There are also comparable levels of SPOP protein in BC cell lines (Fig. 3b). To examine whether SPOP is involved in LINC01638-mediated changes in c-Myc expression, SPOP was knocked down in LINC01638-knockdown MDA-MB-231 and BT549 cells. SPOP knockdown abolished the decrease in c-Myc protein levels induced by LINC01638 knockdown (Fig. 5c). It is widely recognized that RNA-interacting proteins have a crucial role in enabling lncRNAs to exert their functions; thus, we hypothesized that LINC01638 either interacts with SPOP to inhibit its function or with c-Myc to prevent its degradation. Thus, we performed a RNA immunoprecipitation (RIP) assay which presented that LINC01638 mainly interacts with c-Myc and not SPOP (Fig. 5d). Additionally, the CoIP results indicated that LINC01638 knockdown enhanced the interaction of SPOP with c-Myc in MDA-MB-231 cells (Fig. 5e). Next, a series of LINC01638 truncations were constructed to map its fragment interacting with c-Myc. We found that the exon 3 is responsible for the interacting with c-Myc in 293T cells co-transfected with trunscation and c-Myc overexpressing vectors (Fig. 5f). Additionally, to identify the c-Myc regions that interacts with LINC01638, 293T cells were co-transfected with LINC01638 expressing vector and vectors containing different fragments of the c-Myc protein. Our results indicated that the fragment2 of c-Myc protein mainly interacted with LINC01638 (Fig. 5g), which was also mainly responsible for interaction with SPOP [21]. Moreover, co-transfection with LINC01638 fulllength and LINC01638 exon3 truncation significantly enhaced the nuclear translocation of c-Myc (Fig. 5h). Then, we investigated whehter c-Myc was required for the functions of LINC01638. We found that c-Myc overexpression abrogated the supressive effects of LINC01638 knockdwon on proliferation and invasion in MDA-MB-231 cells (Fig. 5i). Consistently, we found that c-Myc knockdown overcame the promotive effects of LINC01638 overexpression on proliferation and invasion in T47D cells (Fig. 5i). These results show that LINC06138 interacts with c-Myc to prevent SPOP-mediated protein degradation of c-Myc in BC cells.
LINC01638 interacts with c-Myc to prevent SPOP-mediated degradation. a Scatter plots comparing c-Myc expression in breast tumor samples (red box, n = 1085) and normal breast samples (gray box, n = 112) analyzed from TCGA data using online software (http://gepia.cancer-pku.cn/index.html). b The effect of MG-132 treatment on the change in the c-Myc protein level mediated by LINC01638 knockdown as detected by western blot. c The effect of SPOP knockdown on the change of the c-Myc protein level mediated by LINC01638 knockdown as detected by western blot. d The interaction of LINC01638 with c-Myc and SPOP was verified by an RIP assay. e The interaction of c-Myc with SPOP was verified by a CoIP assay. f Mapping analysis of c-Myc interacting domain of LINC01638. Schematic diagram of LINC01638 full-length and truncated fragments (left); The interaction of LINC01638 truncated fragments with c-Myc in 293T cells was verified by an RIP assay (right). g Identify regions of c-Myc protein interacting with LINC01638. The fragments of the c-Myc protein were illustrated (left); The interaction of c-Myc protein regions with LINC01638 in 293T cells was confirmed by an RIP assay (right). h The effect of LINC01638 on nuclear translocation of c-Myc was investigated. i Ectopic expression of c-Myc abrogated the effects of LINC01638 knockdown on proliferation and invasion in MDA-MB-231 cells. c-Myc knockdown abolished the effects of LINC01638 overexpression on proliferation and invasions in T47D cells. The boxes show the median and the interquartile range (a). Student’s t-test, **p < 0.01, ***p < 0.001, ****p < 0.0001
LINC01638 promotes tumor progression of breast cancer cells in vivo
To evaluate the biological functions of LINC01638 in vivo, we next explored the role of LINC01638 in tumor growth. We implemented subcutaneous injection of LINC01638-knockdown MDA-MB-231 and sh-Con cells into nude mice. LINC01638 knockdown repressed tumor growth and tumor weight growth, but restored c-Myc expression disturbed the effect of LINC01638 knockdown on tumor growth in vivo (Fig. 6a). Additionally, to assess the in vivo effect of LINC01638 on breast cancer metastasis, LINC01638-knockdown MDA-MB-231 and sh-Con cells were injected into nude mice intravenously through the tail vein. Hematoxylin and eosin staining of lung tissues revealed the presence of fewer metastatic tumor nodules in the lungs of LINC01638-knockdown MDA-MB-231 cells than those of sh-Con cells, but restored c-Myc expression abrogated the effect of LINC01638 knockdown on metastasis in vivo (Fig. 6b). To further define the role of LINC01638 in human BC, we measured LINC01638 expression via in situ hybridization (ISH) and the protein levels of c-Myc, MTDH and Twist1 using immunohistochemistry (IHC) staining in the 141 BC tissues. Analysis showed that LINC01638 expression level was correlated with BC progression (Table 1). BC tissue samples with high transcript levels of LINC01638 frequently had high protein levels of c-Myc, MTDH and Twist1 (Fig. 6c). The LINC01638 transcript level was correlated with the protein levels of c-Myc, MTDH and Twist1 positively (Fig. 6d). We further examined whether the LINC01638 expression level was correlated with BC outcomes. Kaplan–Meier analysis of the 141 BC patients manifested that high LINC01638 expression in BC tissues significantly associated with a reduction in overall survival (Fig. 6e). These data indicate that a high level of LINC01638 is strongly associated with progression and poor outcome of BC patients.
LINC01638 promotes tumor progression of breast cancer cells in vivo. a LINC01638 knockdown inhibited MDA-MB-231-derived tumor growth in vivo, but restored c-Myc expression disturbed the effect of LINC01638 knockdown (n = 5/group). b Representative images of HE-stained sections derived from metastatic nodules in the lungs of mice injected intravenously through the tail vein with MDA-MB-231 LINC01638-knockdown cells with or without c-Myc overexpression (n = 5/group). Scale bar 50 μm. c Representative ISH staining (red staining: nucleus, blue staining: LINC01638) of the LINC01638 transcript and IHC staining of the c-Myc, MTDH, and Twist1 proteins in breast cancer samples. Scale bar 50 μm. d The correlation between the transcript level of LINC01638 and protein levels of c-Myc, MTDH, and Twist1 was analyzed. e Kaplan–Meier analysis indicated a correlation between high expression of LINC01638 and poor overall survival in breast cancer patients. Student’s t-test, *p < 0.05, **p < 0.01, ***p < 0.001
Discussion
Here, we reported the overexpression of LINC01638 in the TNBC subtype. LINC01638 promoted TNBC cell proliferation in vitro and growth in vivo. LINC01638 promoted TNBC cell invasion in vitro and colonization at the site of metastasis in vivo and was involved in maintaining the mesenchymal state of TNBC cells. Moreover, a higher level of LINC01638 was correlated with poor clinical prognoses. Together, these data support our conclusion that LINC01638 has pleiotropic effects on proliferation, tumor growth, invasion, colonization, and metastasis. Therefore, we conclude that LINC01638 has oncogenic activity in the progression of breast cancer.
lncRNAs play a diversity of roles in regulating gene expression as well as other cellular activities [22, 23]. lncRNAs enact their cellular effects via distinct mechanisms, including regulating co-transcription of gene and pairing with RNAs or interacting with proteins to construct nuclear or cytoplasmic complexes [10, 12, 13]. Here, we demonstrated that LINC01638 exerted its oncogenic role by regulating MTDH expression. MTDH (metadherin) is a crucial oncogene that exerts critical functions in the initiation and progression of most cancers [24]. MTDH was first postulated to be a single-pass transmembrane protein that mediates the adhesion between cancer cell and the lung endothelium [25]. However, later studies revealed that MTDH is mainly localized in the cytoplasm and nucleus rather than the membrane of cells [26]. Recently, accumulated evidence has revealed multifaceted roles of MTDH in conferring tumor growth, therapeutic resistance and metastasis via modulation multiple oncogenic signaling pathways [27,28,29]. Liang et al. reported that MTDH promotes the cancer-stem-cell like traits in breast cancer by epigenetic activation of Twist1 [19]. Consistently, here, we also found that LINC01638 positively regulated Twist1 to maintain the mesenchymal-like traits in TNBC cells via MTDH.
MTDH expression is aberrantly upregulated in a broad range of cancer types and has been correlated with tumor size, regional lymph node metastases, clinical stage, distant metastases and poor outcome [30, 31]. Gene amplification of MTDH contributes to MTDH overexpression in some breast cancers [32]. Whereas, breast cancer is usually regarded as a heterogeneous disease, and whether additional mechanisms exist for MTDH overexpression in breast cancer beyond pure gene amplification remains to be explored. Here, we demonstrated that LINC01638 knockdown decreased the levels of MTDH transcripts and protein. We further demonstrated that c-Myc directly bound the MTDH promoter to transcriptionally regulate MTDH expression in BC cells. C-Myc is one of the most commonly activated oncogenes in a wide variety of cancer types that have critical effects on tumor growth, differentiation and survival [33]. C-Myc has also been associated with invasion, metastasis and EMT-induced breast cancer stem cell (CSC)-like properties [34, 35]. Unexpectedly, online analysis of TCGA data showed that the mRNA levels of c-Myc are reduced in breast cancers, whereas BC cells have higher c-Myc protein levels, indicating the presence of important mechanisms for c-Myc regulation at the protein level. Recently, Geng et al. [21] reported that the E3 ubiquitin ligase adapter speckle-type POZ (SPOP) can promote c-Myc degradation via ubiquitination manner. It is intriguing that there are comparable protein levels of SPOP between breast cancer cells from different subtypes and normal mammary cells. Thus, we speculated that LINC01638 may interact either with SPOP to repress its effect on c-Myc or with c-Myc to prevent SPOP-mediated degradation. Our results indicated that LINC01638 interacts with c-Myc but not SPOP, thus preventing SPOP-induced c-Myc degradation.
In conclusion, our research demonstrates for the first time that LINC01638 plays a crucial part in the progression of TNBC. LINC01638 interacts with c-Myc to prevent SPOP-induced c-Myc ubiquitination and degradation and then activates MTDH-Twist1 signaling to maintain mesenchymal traits with EMT and CSC-like features.
Materials and methods
Tissue samples
The 54 frozen fresh breast tumor and adjacent non-cancerous breast tissues were collected from resected breast cancers from patients at the Affiliated Cancer Hospital and Institute of Guangzhou Medical University and were snap-frozen in liquid nitrogen. Paraffin-embedded human primary breast carcinomas were collected from 141 patients at the Sun Yat-Sen University Cancer Center undergoing breast reduction surgery. Overall survival was calculated from the day of surgery to the last follow-up time or death. Tissue samples and related written informed consents were collected from patients following standard procedures that were carried out under the authorization of the ethics boards of the both indicated hospitals.
Cell culture and transfection
MCF-10A, T47D, MCF7, BT474, SKBR3, ZR-75-30, ZR-75-1, MDA-MB-231, BT549, and HCC1937 cell lines were cultured in the laboratory. Plasmids were transfected uing Lipofectamine 3000 following the manufacturer’s instructions. Containing shRNAs targeting LINC01638, MTDH, SPOP and c-Myc, psi-LVRU6GP vectors were obtained from GeneCopoeiaTM (China, Guangzhou) for knocking down genes. Fifty nanomolar vector with shRNA or negative control was transfected into cells. For gene overexpression, 4 μg of pCMV-Flag, EXP-LV203-LINC01638, pcDNA3.1-MTDH, pcDNA3.1-c-Myc, or control plasmids were transfected into each well, respectively. Follow-up experiments were performed 48 h later.
RNA extraction and quantitative real-time PCR analyses (qRT-PCR)
The TRIzol reagent was used to extracted total RNA. cDNA was generated with the PrimeScriptRT reagent kit. GAPDH was performed as an internal reference for cytoplasmic gene expression, while BCAR4 was performed as a positive control for gene expression in nuclei. The expression change of genes in fold in cancer samples or related cell lines was calculated by the 2−ΔΔCt method using paired adjacent samples or control cell lines as references for normalization. The primers for qRT-PCR: LINC01638: forward primer: 5′-AATACATCAGCACTGTTGCCTTT-3′, RP: 5′-CTCCATACATACATCTCCAAAAAGT-3′; BCAR4: FP: 5′-ACAGCAGCTTGTTGCTCATCT-3′, RP: 5′-TTGCCTTGGGGACAGTTCAC-3′; MTDH: FP: 5′-AAATAGCCA GCCTATCAAGACTC-3′, RP: 5′-TTCAGACTTGGTCTGTGAAGGAG-3′; E-cadherin: FP: 5′-TTGCTACTGGAACAGGGACAC-3′, RP: 5′-GGAGATGTATTGGGAGGAAGG-3′; Twist1: FP: 5′-GAGGCGCCCCGCTCTTCTCCTCTG-3′, RP: 5′-CGTCTGAAGAACGGCGCGAA-3′; N-cadherin: FP: 5′-TGAAGTCCCCAATGTCTCCA-3′, RP: 5′-GCATCATCATCCTGCTTATCC-3′; GAPDH: FP: 5′-AAGGTGAAGGTCGGAGTCAA-3′, RP: 5′-AATGAAGGGGTCATTGATGG-3′.
Cell invasion assay
Cell invasive ability was detected using transwell assay as described previously [6].
Mammosphere forming assay
Mammosphere assays were performed as previously described [6].
Flow cytometry analysis
The flow cytometry analysis was performed as we described previously [36].
Western blotting
The primary antibodies for anti-MTDH (#14065), anti-E-cadherin (#3195), anti-N-cadherin (#13116) were from Cell Signaling Technology. The primary antibody for anti-Twist1 (ab50887) was from Abcam. The primary antibodies for anti-c-Myc (sc-56505), anti-SPOP (sc-377206) and anti-GAPDH (sc-47724) were from Santa Cruz Biotechnology. After protein was extracted, western blotting was performed as described [36]. The ECL detection system (Millipore) was utilized to visualize the target protein.
Chromatin immunoprecipitation (ChIP) assay
The ChIP assay was carried out according to the instructions of the EZ-ChIP™ Chromatin immunoprecipitation kit (Millipore, USA). 1 × 107 cells were fixed with 1% formaldehyde and stopped by glycine, lysed for 10 min on ice and sonicated. Supernatant was prepared for immunoprecipitation with 5 µg of anti-c-Myc antibody, polymerase II, and normal rabbit IgG, respectively. The complexes were incubated overnight at 4 °C. Next, the chromatin/DNA/protein/antibody complexes were digested with Proteinase K at 65 °C, DNA was purified and qRT-PCR was used to test the interaction between c-Myc and MTDH. Primers for the MTDH promoter-with c-Myc binding sites: For site A: forward primer, 5′-ACCTTCCCTGACACGCCTTTGC-3′ and reverse primer, 5′-TCCCTCGGCGGAACAATGG-3′. For site B: Forward primer, 5′-TAGAATGGTCAGAATAGTCAGTGGA-3′ and reverse primer, 5′-AACTTAGCAAATGTTATTGGGATCT-3′. Primers for the human GAPDH gene: forward primer, 5′-AAGGTGAAGGTCGGAGTCAA-3′and reverse primer: 5′-AATGAAGGGGTCATTGATGG-3′.
Luciferase reporter assay
We first constructed wild-type and deletion reporter vectors for the luciferase reporter assay. For the wild-type MTDH promoter, genomic DNA was extracted and the 2000 bp 5′-UTR sequence from the MTDH transcript start site (TSS) was amplified by PCR. This fragment contained a c-Myc binding motif. The pGL4 luciferase vector and MTDH promoter were digested by Bgl II and Xho I then ligated with T4 DNA ligase. The desired clone was sequenced and named pGL4-WT-MTDH. The Site-Directed Mutagenesis kit (Clontech, Japan) was used to delete the c-Myc binding motif in the pGL3-WT-MTDH plasmid. The desired clone was sequenced and named pGL4-MT-MTDH. Cells were seeded in 96-well plates overnight, then transfected with the pcDNA4-MTDH vector, together with either the wild-type or deletion reporter vector, as well as vectors with c-Myc shRNAs or overexpression constructs. pRL-TK was used as an internal control.
RNA immunoprecipitation (RIP)
RIP was performed to detect the interaction of LINC01638 with C-MYC and SPOP via the EZ-Magna RIP Kit (Millipore, USA). LINC01638 truncated fragments were cloned into psi-LVRU6GP vector. C-Myc truncated fragments were cloned into pReceiver-M11 vector with N-terminal Flag-tag. Briefly, cells were lysed in RIP lysis buffer. Magnetic beads and 5 μg antibody of interest (anti-c-Myc, anti-SPOP, anti-Flag, and negative control Normal Rabbit IgG) was added to the beads. The tubes were centrifuged and the supernatant was removed. Hundred microliter RIP lysate and 900 μL RIP Immunoprecipitation Buffer were added to each tube and rotated overnight at 4 °C. Next, the tubes were centrifuged, the beads were washed and each immunoprecipitate was resuspended in 150 μL Proteinase K buffer to digest the protein. Phenol:chloroform:isoamyl alcohol and chloroform were used to extract RNAs. Reverse transcription and qRT-PCR was used to verify the results. The primers for vector, fulllength, and GAPDH were as the same as used for qRT-PCR. The primers for Exon1: FP: 5′-CACACTGTCTCCTGTCAGCTTTC-3′, RP: 5′-CTAACTCAGACCATCCATCGCT-3′; for Exon2: 5′-CATTCCCTCTGGAATACATCAG-3′, RP: 5′-CTGTGAATCCGGAGCTCAGTGG-3′; for Exon3: FP: 5′-GTAGAGATGGGGTTTCACCATG-3′, RP: 5′-TGGCTCACGCCTGTAATCCCA-3′.
Co-immunoprecipitation assay
The Co-immunoprecipitation (CoIP) assay was performed as we described previously [37].
In situ hybridization (ISH)
The LINC01638 transcript level in breast cancer tissues was examined by ISH assay performed with manufacturer’s procedures (Exiqon, Vedbaek, Denmark). After dehydration in the ethanol, the sections were used to hybridize with 40 nM LINC01638 probe tagged with double-DIG at 55 °C for 1 h. After washing in PBST, the AP substrate NBT-BCIP (Roche) was applied for 2 h at 30 °C. The reaction was stopped using KTBT buffer. Next, nuclear counterstaining was performed using Nuclear Fast Red™ (Vector Labs, Burlingame, CA). After ethanol dehydration and mounting, the LINC01638 transcript level was analyzed using microscope.
Immunohistochemistry (IHC)
The protein levels of c-Myc, MTDH, and Twist1 in tissues were detected by IHC performed as we described previously [37]. Two different pathologists evaluated and classified protein levels as high or low based on the scores of the intensity of immunostaining.
Xenograft model in athymic mice
MDA231/sh-Con, MDA231/sh-LINC01638, MDA231/sh-LINC01638 + Vector and MDA231/sh-LINC01638 + cMyc cell lines were injected subcutaneously into the armpit of female Balb/C athymic nude mice to generate xenograft tumors. The tumor growth was monitored and the wet weight of tumors was recorded at the experimental endpoint. For in vivo metastasis assays, 2 × 105 cells for each cell line were injected intravenously via the tail vein into the female Balb/C athymic nude mouse. Six weeks after injection, the mice were euthanized, and the metastatic nodules in the lungs were counted after hematoxylin and eosin staining. There are five mice for each group.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 6. Data are presented as the means ± SD. Pearson’s chi-squared test was used to analyze the clinical variables. The difference between groups for statistical significance was analyzed using Student’s t-test. Pearson correlation analysis was performed to determine the correlation between two variables. Survival curves were plotted using the Kaplan–Meier method and compared using the log-rank test. A p value < 0.05 was considered statistically significant.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. Cancer J Clin. 2016;66:7–30.
Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52.
Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68.
Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–48.
Podo F, Buydens LM, Degani H, Hilhorst R, Klipp E, Gribbestad IS, et al. Triple-negative breast cancer: present challenges and new perspectives. Mol Oncol. 2010;4:209–29.
Shi J, Wang Y, Zeng L, Wu Y, Deng J, Zhang Q, et al. Disrupting the interaction of BRD4 with diacetylated Twist suppresses tumorigenesis in basal-like breast cancer. Cancer Cell. 2014;25:210–25.
Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407.
Lin A, Li C, Xing Z, Hu Q, Liang K, Han L, et al. The LINK-A lncRNA activates normoxic HIF1alpha signalling in triple-negative breast cancer. Nat Cell Biol. 2016;18:213–24.
Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–9.
Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46.
Luo M, Jeong M, Sun D, Park HJ, Rodriguez BA, Xia Z, et al. Long non-coding RNAs control hematopoietic stem cell function. Cell Stem Cell. 2015;16:426–38.
Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–14.
Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–46.
Booy EP, McRae EK, Koul A, Lin F, McKenna SA. The long non-coding RNA BC200 (BCYRN1) is critical for cancer cell survival and proliferation. Mol Cancer. 2017;16:109.
Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–81.
Wang Y, He L, Du Y, Zhu P, Huang G, Luo J, et al. The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell. 2015;16:413–25.
Jiang Z, Slater CM, Zhou Y, Devarajan K, Ruth KJ, Li Y, et al. LincIN, a novel NF90-binding long non-coding RNA, is overexpressed in advanced breast tumors and involved in metastasis. Breast Cancer Res. 2017;19:62.
Fagerberg L, Hallstrom BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteom. 2014;13:397–406.
Liang Y, Hu J, Li J, Liu Y, Yu J, Zhuang X, et al. Epigenetic activation of TWIST1 by MTDH promotes cancer stem-like cell traits in breast cancer. Cancer Res. 2015;75:3672–80.
Lee SG, Su ZZ, Emdad L, Sarkar D, Fisher PB. Astrocyte elevated gene-1 (AEG-1) is a target gene of oncogenic Ha-ras requiring phosphatidylinositol 3-kinase and c-Myc. Proc Natl Acad Sci USA. 2006;103:17390–5.
Geng C, Kaochar S, Li M, Rajapakshe K, Fiskus W, Dong J, et al. SPOP regulates prostate epithelial cell proliferation and promotes ubiquitination and turnover of c-MYC oncoprotein. Oncogene. 2017;36:4767–77.
Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–307.
Flynn RA, Chang HY. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell. 2014;14:752–61.
Sarkar D, Fisher PB. AEG-1/MTDH/LYRIC: clinical significance. Adv Cancer Res. 2013;120:39–74.
Brown DM, Ruoslahti E. Metadherin, a cell surface protein in breast tumors that mediates lung metastasis. Cancer Cell. 2004;5:365–74.
Blanco MA, Aleckovic M, Hua Y, Li T, Wei Y, Xu Z, et al. Identification of staphylococcal nuclease domain-containing 1 (SND1) as a Metadherin-interacting protein with metastasis-promoting functions. J Biol Chem. 2011;286:19982–92.
Yoo BK, Chen D, Su ZZ, Gredler R, Yoo J, Shah K, et al. Molecular mechanism of chemoresistance by astrocyte elevated gene-1. Cancer Res. 2010;70:3249–58.
Emdad L, Das SK, Dasgupta S, Hu B, Sarkar D, Fisher PB. AEG-1/MTDH/LYRIC: signaling pathways, downstream genes, interacting proteins, and regulation of tumor angiogenesis. Adv Cancer Res. 2013;120:75–111.
Wan L, Lu X, Yuan S, Wei Y, Guo F, Shen M, et al. MTDH-SND1 interaction is crucial for expansion and activity of tumor-initiating cells in diverse oncogene- and carcinogen-induced mammary tumors. Cancer Cell. 2014;26:92–105.
Liu L, Wu J, Ying Z, Chen B, Han A, Liang Y, et al. Astrocyte elevated gene-1 upregulates matrix metalloproteinase-9 and induces human glioma invasion. Cancer Res. 2010;70:3750–9.
Hu G, Wei Y, Kang Y. The multifaceted role of MTDH/AEG-1 in cancer progression. Clin Cancer Res. 2009;15:5615–20.
Hu G, Chong RA, Yang Q, Wei Y, Blanco MA, Li F, et al. MTDH activation by 8q22 genomic gain promotes chemoresistance and metastasis of poor-prognosis breast cancer. Cancer Cell. 2009;15:9–20.
Ni M, Chen Y, Fei T, Li D, Lim E, Liu XS, et al. Amplitude modulation of androgen signaling by c-MYC. Genes Dev. 2013;27:734–48.
Cho KB, Cho MK, Lee WY, Kang KW. Overexpression of c-myc induces epithelial mesenchymal transition in mammary epithelial cells. Cancer Lett. 2010;293:230–9.
Cho MH, Park JH, Choi HJ, Park MK, Won HY, Park YJ, et al. DOT1L cooperates with the c-Myc-p300 complex to epigenetically derepress CDH1 transcription factors in breast cancer progression. Nat Commun. 2015;6:7821.
Yin J, Zheng G, Jia X, Zhang Z, Zhang W, Song Y, et al. A Bmi1-miRNAs cross-talk modulates chemotherapy response to 5-fluorouracil in breast cancer cells. PLoS ONE. 2013;8:e73268.
Zheng G, Zhang Z, Liu H, Xiong Y, Luo L, Jia X, et al. HSP27-mediated extracellular and intracellular signaling pathways synergistically confer chemo-resistance in squamous cell carcinoma of tongue. Clin Cancer Res. 2017;24:1163–75.
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China: No. 81672616 (GZ), No. 81402196 (GZ), No. 81772961 (HT), No. 81401989 (NL) and No. 81772825 (HL); supported by grants from Guangdong Natural Science Funds for Distinguished Young Scholars: No. 2016A030306003 (GZ) and Guangdong Natural Science Funds: No. 2017A030313867 (GZ); supported by grants from Science and Technology Program of Guangzhou: No. 201710010100 (GZ) and Guangzhou Municipal University Scientific Research project: 1201610027 (GZ).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Luo, L., Tang, H., Ling, L. et al. LINC01638 lncRNA activates MTDH-Twist1 signaling by preventing SPOP-mediated c-Myc degradation in triple-negative breast cancer. Oncogene 37, 6166–6179 (2018). https://doi.org/10.1038/s41388-018-0396-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-018-0396-8
- Springer Nature Limited
This article is cited by
-
Novel associations between MTDH gene polymorphisms and invasive ductal breast cancer: a case–control study
Discover Oncology (2024)
-
Metformin and long non-coding RNAs in breast cancer
Journal of Translational Medicine (2023)
-
LINC01638 sustains human mesenchymal stem cell self-renewal and competency for osteogenic cell fate
Scientific Reports (2023)
-
LncRNA BCAN-AS1 stabilizes c-Myc via N6-methyladenosine-mediated binding with SNIP1 to promote pancreatic cancer
Cell Death & Differentiation (2023)
-
Functional interplay between long non-coding RNAs and Breast CSCs
Cancer Cell International (2022)