Abstract
The APC/C-Cdh1 ubiquitin-ligase complex targets cell cycle regulators for proteosomal degradation and helps prevent tumor development and accumulation of chromosomal aberrations. Replication stress has been proposed to be the main driver of genomic instability in the absence of Cdh1, but the real contribution of APC/C-Cdh1 to efficient replication, especially in normal cells, remains unclear. Here we show that, in primary MEFs, acute depletion or permanent ablation of Cdh1 slowed down replication fork movement and increased origin activity. Partial inhibition of origin firing does not accelerate replication forks, suggesting that fork progression is intrinsically limited in the absence of Cdh1. Moreover, exogenous supply of nucleotide precursors, or ectopic overexpression of RRM2, the regulatory subunit of Ribonucleotide Reductase, restore replication efficiency, indicating that dNTP availability could be impaired upon Cdh1 loss. Indeed, we found reduced dNTP levels in Cdh1-deficient MEFs. Importantly, DNA breakage is also significantly alleviated by increasing intracellular dNTP pools, strongly suggesting that genomic instability is the result of aberrant replication. These observations highlight the relevance of APC/C-Cdh1 activity during G1 to ensure an adequate supply of dNTPs to the replisome, prevent replication stress and the resulting chromosomal breaks and, ultimately, suppress tumorigenesis.
Similar content being viewed by others
Introduction
The ubiquitin-proteasome system plays a pivotal role in cell cycle control by mediating the precise spatial and temporal proteolysis of critical components of the cell cycle machinery.1, 2 During mitosis and the subsequent G1 phase, this process is initiated by the Anaphase Promoting Complex or Cyclosome (APC/C), a multisubunit E3 ubiquitin ligase whose activity is mostly controlled by the regulated binding of two alternative cofactors, Cdc20 or Cdh1, that recruit specific substrates.3, 4 Cdc20 activates APC/C during early mitosis and towards the end of the mitotic phase, Cdh1 takes over and facilitates exit from mitosis by targeting for degradation protein kinases such as Plk1, Aurora A and Aurora B, mitotic cyclins, and even Cdc20.5 During G1, APC/C-Cdh1 keeps Cdk activity low by promoting proteolysis of Skp2, a component of the SCF complex that, in turn, mediates proteosomal degradation of the Cdk inhibitors p27, p21 and p57. On the other hand, APC/C-Cdh1 ubiquitylates Geminin, the inhibitor of Cdt1, and is thus believed to promote replication origin licensing.6, 7 At the end of G1, Cdh1 is phosphorylated by Cdk2 and dissociates from the APC/C complex that will remain inactive until the following mitosis.
Cdh1 has been shown to be dispensable for the mitotic cell cycle in different organisms.8, 9, 10 However, targeted deletion of the Cdh1 gene in mouse cells, or depletion of the protein in human cells, induce premature entry into S phase and genomic instability.11, 12, 13 Moreover, genetic ablation of Cdh1 in early mouse embryos leads to perinatal lethality of newborn pups, and Cdh1 heterozygous mice show increased susceptibility to epithelial tumor development.11 Cdh1 is thus considered to act as a haploinsufficient tumor suppressor, and loss of genomic integrity has been proposed to facilitate cell transformation in its absence.14, 15
Accurate and complete duplication of the genetic material is essential to preserve genomic stability. However, cells are frequently exposed to endogenous or exogenous insults that compromise the replication process leading to replicative stress.16 The most detrimental consequences of aberrant replication are the generation of DNA double-strand breaks, that can lead to chromosomal rearrangements, and the loss of genomic segments as a result of mitotic entry with under-replicated regions.17 These adverse outcomes can be restricted through activation of the ATR and Chk1-dependent S phase checkpoint, that helps stabilize stalled forks, inhibit late origin firing and arrest cell cycle progression.18 Still, in cases of moderate or low replication stress full activation of the checkpoint may not be achieved, leading to mitotic defects and chromosomal aberrations.
We have previously reported that Cdh1-null mouse embryonic fibroblasts (MEFs) accumulate chromosomal translocations and show non-disjunction figures and chromosome fragments in mitotic spreads.11 However, the underlying molecular dysfunction leading to chromosomal aberrations was not defined. We now show that, in primary MEFs lacking Cdh1 expression, genomic instability is accompanied by altered replication dynamics, and we have identified the resulting replication stress as the main source of DNA breaks in these cells. Moreover, insufficient supply of dNTPs to the replication machinery seems to be behind the replication defects observed upon Cdh1 loss.
Results
Acute depletion of Cdh1 in primary MEFs induces accumulation of DNA breaks
To get further insight into the role of APC/C-Cdh1 in preserving chromosomal integrity, we induced acute depletion of Cdh1 in primary MEFs homozygous for Cdh1 conditional allele (Cdh1(lox/lox)) using adenovirus expressing the Cre recombinase (Adeno-Cre). Efficient downregulation of the protein following infection was confirmed by western blot (Figure 1a). Using flow cytometry, an increased proportion of γH2AX-positive cells was detected in MEFs infected with Adeno-Cre (Figure 1b). Interestingly, this difference was more significant in cells that did not incorporate the thymidine analog EdU (EdU-negative cells), suggesting that DNA damage preferentially occurred in cells that had already completed the bulk of replication. Similar results were obtained when comparing γH2AX expression in control and Cdh1-depleted MEFs by immunofluorescence microscopy (Supplementary Figure S1). Importantly, no change was detected in H2AX phosphorylation upon infection of primary MEFs carrying wild type Cdh1 alleles with the same adenoviruses (Figure 1b). Moreover, equivalent observations were made with Cdh1-deficient primary MEFs (Supplementary Figures S2A, S2B and S3A). Increased detection of 53BP1 foci in Cdh1-depleted or Cdh1-deficient MEFs (Figure 1c and Supplementary Figure S2C) further confirmed the induction of spontaneous DNA lesions upon Cdh1 loss. Finally, to directly visualize and quantify DNA breaks in single cells, we performed alkaline comet assays. As shown in Figure 1d, Adeno-Cre-mediated Cdh1 depletion strongly induced DNA breakage, whereas infection with the same virus had no effect on DNA integrity in wild type MEFs. Likewise, Cdh1-deficient MEFs also showed significantly higher levels of DNA breaks compared to control MEFs (Supplementary Figure S2D). Together, these results indicate that absence, or strong downregulation of the Cdh1 protein, promote accumulation of endogenous DNA damage.
Genomic instability following acute depletion of Cdh1 in primary MEFs. (a) Immunoblot analysis of Cdh1 levels in primary MEFs obtained from embryos with the indicated genotype, either uninfected (Uninf) or infected with empty adenovirus (Cont) or adenovirus expressing Cre recombinase (Cre), and collected four days later. GAPDH was used as loading control. (b) Flow cytometry analysis of γH2AX levels in the same primary MEFs infected with empty adenovirus (Ad-Control) or adenovirus expressing Cre (Ad-Cre). Left, representative density plots showing γH2AX and EdU fluorescence intensities in the indicated primary MEFs. Right, quantification of γH2AX positive cells or γH2AX positive, EdU negative cells in primary cultures of Cdh1(lox/lox) MEFs (top histogram) or wild type MEFs (bottom histogram) infected with the indicated adenoviruses. (c) Left, representative images of 53BP1 and DAPI immunofluorescence staining of the indicated MEFs. Right, quantification of the percentage of cells with 53BP1 foci in each culture. (d) Top, Tail moment values obtained in alkaline comet assays with Cdh1(lox/lox) or wild type MEFs infected with the indicated adenoviruses. Black line indicates median value. Bottom, representative images of cells stained with ethidium bromide showing comet head and tail. *P<0,05, **P<0,01, ****P<0,0001, n.s. not significant. Error bars indicate s.d.; Scale bar : 10 μm.
Signs of replicative stress in the absence of Cdh1
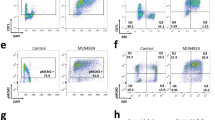
Since replication stress is one of the most common sources of endogenous DNA damage in mammalian cells,19, 20 we looked for signs of replicative stress in Cdh1-mutant MEFs. An increased presence of cells with RPA foci was detected in Cdh1(lox/lox) MEFs following expression of adenoviral Cre recombinase (Figure 2a), as well as in MEFs constitutively deficient for Cdh1 (Supplementary Figure S3B). Phosphorylation of Chk1 kinase and p53 protein were also moderately induced upon acute depletion of Cdh1 (Figure 2b), suggesting a mild activation of the DNA damage response. Finally, we observed that following incubation of asynchronous cultures with EdU, replicating MEFs devoid of Cdh1 incorporated less of the thymidine analog than control MEFs (Figure 2c). We conclude that, upon acute or persistent loss of Cdh1 expression, primary MEFs show signs of inefficient DNA synthesis and replication stress.
Signs of replicative stress in Cdh1-depleted MEFs. (a) Left, representative images of RPA immunofluorescence staining in Cdh1(lox/lox) MEFs infected with the indicated adenoviruses. Right, quantification of the percentage of cells with RPA foci in each culture. (b) Left, the level of the indicated proteins or phospho-proteins (pS15-p53, pS345-Chk1) was analyzed by immunoblotting in whole cell lysates from Cdh1(-/lox) and Cdh1(lox/lox) MEFs infected with Adeno-cre or control adenovirus. GAPDH was used as loading control. Right, band intensity was quantified for the indicated phospho-proteins and normalized to loading control. Relative fold change over samples infected with control adenovirus is shown. (c) Top, representative flow cytometry histogram overlay showing EdU fluorescence intensity in asynchronous control and Cdh1-deficient MEFs. Bottom, quantification of mean EdU fluorescence intensity in different primary MEFs with the indicated genotype. *P<0,05, **P<0,01; Error bars indicate s.d.; Scale bar : 10 μm.
Slow replication fork progression and increased origin activation in the absence of Cdh1
The findings outlined above prompted us to compare replication dynamics in the presence or absence of Cdh1. We first assessed replication fork progression rates by measuring IdU tracks on stretched DNA fibers (Figure 3a). As shown in Figure 3b, replication fork movement in Cdh1(lox/lox) MEFs was notably slower following infection with Adeno-Cre. Similarly, fork speed was lower in Cdh1 null MEFs than in control cells (Figures 3c and d). Moreover, the distance between adjacent replication origins was markedly shorter in Cdh1-depleted and Cdh1-deficient MEFs (Figures 3e and f), indicating increased origin firing in these cells, and suggesting the compensatory activation of dormant origins.21, 22, 23 In contrast, expression of adenoviral Cre in wild type MEFs only marginally reduced fork velocity and had no effect on Inter Origin Distance (IOD) (Figures 3g and h).
Replication defects in Cdh1-deficient and Cdh1-depleted MEFs. (a) Schematic drawing of a DNA fiber after pulse labeling with CldU (red) and IdU (green). Green track measurements allow quantification of fork speed. Origins are identified based on the pattern of green and red staining as shown in the diagram and the distance between two adjacent origins on the same DNA fiber defines IOD. (b,c,g) Scatter plots showing fork rate values measured on stretched DNA fibers obtained from Cdh1(lox/lox) MEFs (b) or wild type MEFs (g) mock infected (Ad-Control) or infected with Adeno-Cre (Ad-Cre) and processed 4 days after infection, and from control and Cdh1-deficient MEFs (c). (d) Representative immunofluorescence images of stretched DNA fibers from the indicated MEFs showing CldU (red) and IdU (green) staining. (e,f,h) Scatter plots showing IOD values for Cdh1(lox/lox) or wild type MEFs (e,h) treated as above, and for control and Cdh1-deficient MEFs (f). Median fork rate and IOD values (indicated by a black line) are : 0.96 and 0.86 Kb/min and 129.5 and 99.4 Kb for control and Cdh1-deficient MEFs, 1.02 and 0.71 Kb/min and 145.23 and 112,02 Kb for Cdh1(lox/lox) MEFs mock infected or infected with Adeno-Cre, and 1.08 and 1.04 Kb/min and 146.06 and 142.17 Kb for wild type MEFs infected with the sane adenoviruses. *P<0,05, ****P<0,0001, n.s. not significant (n=2 in G,H). Scale bar : 10 μm.
To compare the overall duration of the S phase in control and Cdh1-deficient MEFs, we labeled them with two pulses of the thymidine analogs CldU and IdU, separated by a chase period of 0 to 8 h (Figure 4a), and determined what fraction of the cells that had incorporated the first analog (CldU) were also positive for IdU at each time point. As shown in Figure 4c an increasing difference between the two cell cultures became obvious at longer chase times (4 h onward). Based on these data (Figure 4d) we estimated that S phase can be up to 28% longer in cells lacking Cdh1 than in control cells (12,8 h vs 9,9 h). Therefore, the increased origin activity observed upon Cdh1 ablation, does not fully compensate for the slow progression of replication forks. Collectively, these results confirm that the replication process is inefficient in the absence of Cdh1, and that inactivation of the APC/C-Cdh1 complex in cycling cells leads to replication stress.
Extended S phase in the absence of Cdh1. (a) Schematic outline of experiment. Cells were first labeled with CldU for 30 min, allowed to grow for a variable period of chase time (0–8 h) and labeled again with IdU for 30 min before fixation and processing for detection of CldU and IdU. (b) Representative images of immunofluorescence staining of CldU (red), IdU (green) and DAPI (blue) in control and Cdh1-deficient MEFs, corresponding to the 6 hour chase time point. Merged images of CldU and IdU immunostaining are also shown. (c) Top, quantification of CldU/IdU double positive cells within the CldU labeled cell population, at different chase time points, in control and Cdh1-deficient MEF cultures. Bottom, linear plot of the same data shown above. The dotted line corresponds to the linear regression fit that allowed us to estimate the duration of the S phase (time required to have zero double positive cells). *P<0,05, **P<0,01; Error bars indicate s.d.; Scale bar : 10 μm.
Slow replication fork progression in Cdh1-mutant cells is not due to enhanced origin usage
Increased origin firing has been shown to result in reduced fork rate, possibly due to competition for limiting factors.24 To verify if this was the case in the absence of Cdh1, we used the Cdc7 kinase inhibitor, PHA-767491, to partially inhibit replication origin firing and analyze the effect on fork speed.25 Phosphorylation of MCM2 in Ser53, which is mediated by Cdc7, was similarly reduced following PHA-767491 treatment in Cdh1(lox/lox) MEFs infected with Adeno-Cre or with control adenovirus (Figure 5a), as well as in Cdh1-deficient and control MEFs (Supplementary Figure S4A). As expected, PHA-767491 produced a highly significant increase in IOD in both control and mutant cells (Figure 5b and Supplementary Figure S4B). However, whereas treated control cells showed a faster progression of replication forks, presumably because they attempt to compensate for the impaired origin activity, no detectable changes in fork speed were observed in Cdh1-depleted or Cdh1-deficient MEFs after treatment (Figure 5c and Supplementary Figure S4C). Therefore, slow replication fork movement in the absence of Cdh1 is independent of the elevated frequency of origin activation, and fork velocity seems to be intrinsically limited in the absence of Cdh1.
Slow fork progression is not caused by increased origin activity in Cdh1-mutant cells. Cdh1(lox/lox) MEFs were infected with control adenovirus or with Adeno-Cre and four days later, were either mock-treated, or treated with the Cdc7 kinase inhibitor PHA-767491 for four hours. (a) Left, immunoblot analysis of MCM2 phosphorylation (pSer53-MCM2). Right, quantification of the phospho-MCM2 band intensity normalized against loading control (GAPDH) and total MCM2 protein for each sample. (b) Scatter plot showing IOD values obtained with stretched DNA fibers from the indicated, PHA-treated or untreated cell cultures. Median values (indicated by a black line) are 141,7 and 176 Kb for mock infected cultures either untreated or PHA-treated and 107.5 and 158.4 Kb for the corresponding Adeno-Cre infected MEFs. (c) Replication fork speed distribution for the same samples as in b. Median values (shown with a black line) are 1.03 and 1,11 Kb/min for untreated or PHA-treated mock infected cultures and 0.73 and 0.70 Kb/min for the equivalent Adeno-Cre infected cells. ****P<0,0001, n.s. not significant.
Replication dynamics and chromosomal stability are restored in Cdh1-mutant cells by nucleoside supplementation
Adequate supply of nucleotides for DNA synthesis is a major determinant of replication efficiency.26, 27 Therefore, we tested whether addition of nucleotide precursors to the culture medium would improve replication dynamics in cells lacking Cdh1 expression. Indeed, exogenous supply of nucleosides to Cdh1-depleted or Cdh1-deficient MEFs, greatly accelerated replication fork progression in these cells, but had negligible effects on fork speed in the corresponding control MEFs (Figures 6a and c). Notably, nucleoside supplementation also increased IOD in these cells (Figures 6b and d), suggesting that the compensatory use of dormant origins observed upon Cdh1 loss, is no longer required. Alkaline comet assays revealed that nucleoside supplementation substantially reduced DNA breakage in mutant cells while having minimal impact on control cells (Figure 6e). These results strongly suggest that replication stress is the main source of chromosomal instability in the absence of Cdh1. Intriguingly, the same beneficial effects on fork velocity, IOD, and DNA damage were observed with dNTPs (Supplementary Figure S5), possibly implying that, despite their predicted lack of cell-permeability, nucleotides could efficiently traverse the cell membrane (perhaps after partial or complete dephosphorylation).
Replication defects and DNA breakage are corrected with exogenous nucleotide precursors. (a,b). Cdh1(lox/lox) MEFs mock-infected or infected with Adeno-Cre were grown in normal culture medium or in medium supplemented with nucleosides and processed for DNA fiber analysis. The distribution of fork speed values (a) or IOD values (b) is shown. Median fork speed and IOD (black lines) were 1.11 and 1.06 Kb/min and 114,44 and 106,38 Kb respectively for mock-infected cells without or with nucleosides and 0,79 and 1,15 Kb/min and 81,56 and 125,22 Kb for the corresponding Adeno-Cre infected MEFs. (c,d) Control and Cdh1-deficient MEFs were grown in normal culture medium or in medium supplemented with nucleosides and processed for DNA fiber analysis. Scatter plots for fork speed (c) and IOD (d) values are shown. Median values for fork speed and IOD (black lines) are 1,03 and 0,99 Kb/min and 121.39 and 121.92 Kb respectively for control MEFs grown in regular or nucleoside-supplemented medium and 0.88 and 1.06 Kb/min and 103.21 and 130.72 Kb for equivalent mutant MEFs. (e) Top, alkaline comet assays were performed with control and Cdh1-deficient MEFs treated as in c and d and tail moment values were plotted. Black line marks the median value. Bottom, representative images of individual cells from each sample, stained with ethidium bromide after agarose gel electrophoresis. *P<0,05, **P<0,01, ***P<0,001, ****P<0,0001, n.s. not significant. Scale bar : 10 μm.
Intracellular dNTP pools are limiting in Cdh1-deficient MEFs
The fact that replication efficiency could be restored in Cdh1-depleted or -deficient MEFs with exogenous nucleosides, supports the idea that nucleotide pools could be insufficient in these cells. Quantification of intracellular pools of all four dNTPs in control and Cdh1-null cells confirmed a moderate but significant decrease in dCTP and dTTP in mutant cells and a similar tendency in dATP and dGTP (Figure 7a). Whether or not this mild reduction could restrain replication is unclear, but it is possible that dNTP concentrations could be further diminished locally, in the vicinity of replication forks or replication factories, where nucleotide levels have a greater impact on genome duplication efficiency.
Shortage of dNTPs underlies replication defects in the absence of Cdh1. (a) Intracellular dNTP concentrations were measured in asynchronously growing control and Cdh1-deficient MEFs by liquid chromatography coupled to tandem mass spectrometry. Since dNTP pools are maximal during S phase and minimal in G1, and asynchronous cultures of MEFs devoid of Cdh1 show increased percentage of cells in S phase, final values were normalized by the number of cells in S phase in each culture (see Methods) (n=4). (b) Immunoblot analysis of RRM2 levels in control and Cdh1-deficient MEFs infected with empty retrovirus or retrovirus encoding RRM2. GAPDH was used as loading control. (c) Replication fork speed values obtained on stretched DNA fibers from the same MEF cultures as in b. Black line indicates median values (1.01 and 1.06 Kb/min for control MEFs mock infected or infected with RRM2 retroviruses and 0.77 and 0.94 Kb/min for equally treated mutant MEFs). (d) Comet assays were performed with the same MEFs as in b and c, and tail moment was calculated. Median value is shown by black line. ****P<0,0001, n.s. not significant. Error bars indicate s.e.m.
Ribonucleotide Reductase (RNR) is a key enzyme for dNTP synthesis and its regulatory subunit RRM2 is rate-limiting for this process.28, 29 To increase intracellular dNTP pools using a genetic strategy, we ectopically overexpressed RRM2 in control and Cdh1-deficient MEFs (Figure 7b). As shown in Figures 7c and d, RRM2 overexpression significantly improved replication fork progression and restored genomic stability in Cdh1-null MEFs, further confirming our previous observations. These results could suggest that RRM2 might be downregulated in the absence of Cdh1. Yet, RRM2 was reported to be a substrate of APC/C-Cdh1,30 thus predicting higher RRM2 protein levels in cells lacking Cdh1. However, RRM2 expression was only modestly elevated in asynchronous Cdh1-deficient MEFs or even in cultures enriched for S phase cells (20 h after release from G0 arrest) (Supplementary Figure S6A). Therefore, in these mutant cells, even the slightly elevated RRM2 protein levels seem to be insufficient to ensure an adequate supply of dNTPs to the replisome.
Two other enzymes involved in dNTP synthesis, Thymidine Kinase 1 (TK1) and Thymidylate Kinase 1 (TMPK), have also been described as targets of APC/C,31 suggesting that their stabilization could mediate the defective dNTP homeostasis in Cdh1-null cells. Interestingly, overexpression of their non-degradable mutant forms in NIH-3T3 cells was shown to cause a marked increase in the dTTP pool size, growth retardation and higher gene mutation rates.31 However, we did not find either TK1 or TMPK to be upregulated in Cdh1-depleted or Cdh1-deficient MEFs, whereas another known substrate of APC/C-Cdh1 such as Aurora A, was clearly stabilized in the same cells (Supplementary Figure S6B). Consistently, as shown in Figure 7a, dTTP pools were not enlarged but rather decreased in mutant MEFs.
Premature entry into S phase contributes to induce replication stress in the absence of Cdh1
Premature entry into S phase upon Cdh1 deletion or depletion has been attributed to increased Cdk activity during G132 and could lead to replication stress. To test this hypothesis, we first assessed whether limited inhibition of Cdk activity could prevent precocious S phase entry in Cdh1-deficient MEFs. As shown in Figure 8a, following a G0 arrest, cell cycle re-entry in the presence of a low dose of Roscovitine, reduced the percentage of EdU-positive mutant cells to levels comparable to those of control cells at all time points tested, indicating normalization of the timing of S phase onset. DNA fiber analysis on samples collected 15h after release from G0, when most cells have initiated replication, revealed that treatment with the Cdk inhibitor partially improved replication fork progression in Cdh1-deficient cells (Figure 8b). A similar recovery was detected on asynchronous cultures treated with the same dose of Roscovitine, although fork velocity was not fully restored (Figure 8c). In agreement with these observations, DNA breakage was also partly reduced by drug treatment in mutant cells after cell cycle re-entry (Figure 8d) or during asynchronous growth (Figure 8e). Together, these results suggest that increased Cdk activity in G1 and the resulting accelerated entry into S phase, restrain replication fork movement in the absence of Cdh1. However, the fact that this phenotype cannot be fully rescued by Roscovitine indicates that additional factors contribute to the alterations in DNA replication observed in Cdh1-null cells.
Partial inhibition of Cdk activity in Cdh1-deficient MEFs delays S phase entry and improves replication fork progression and genomic DNA integrity. (a) Cdh1-null MEFs were synchronized in G0 by serum starvation (0,5% FBS for 72 h) and re-fed with 20% FBS for the indicated times in the absence or presence of 3 μM Roscovitine. Control cells were cultured in parallel but were left untreated. Edu-positive cells were quantified by flow cytometry in each sample and the combined results of three independent experiments are shown (mean±s.d). (b) Cells treated as in a were labeled with CldU and IdU 15 h after cell cycle re-entry and processed for DNA fiber analysis. The distribution of fork speed values is shown. Median values (black lines) are 1,88 Kb/min for control cells, and 0,93 and 1,02 Kb/min for Cdh1-deficient cells untreated or treated with Roscovitine. (c) Scatter plots showing fork rate values for control and mutant MEFs growing asynchronously and either untreated or treated with Roscovitine for 13 h. Median values (black lines) are 1,11 Kb/min for control cells and 0,94 and 1,01 Kb/min for untreated or treated mutant cells. (d) The indicated cells were grown for 36 h after cell cycle re-entry to allow accumulation of DNA damage after completion of S phase and progression through G2/M. Roscovitine was added at the time of re-feeding and washed out 15 h later. The distribution of tail moment values obtained with alkaline comet assays is shown (n=2). Black line indicates median values (e). Control and Cdh1-deficient MEFs growing asynchronously were treated as in c and processed for alkaline comet assays. Tail moment values were calculated and plotted (n=2) and median values are shown with black line.
Discussion
In this study we have used primary MEFs either deficient in Cdh1 or conditionally depleted of the protein to gain further insight into how APC/C-Cdh1 activity in G1 determines replication dynamics, prevents replicative stress and protects genomic integrity.
Using alkaline comet assays, we were able to directly quantify DNA breaks in Cdh1-proficient or Cdh1-deficient cells, and enhanced DNA breakage in the absence of Cdh1 was substantiated. These findings are in agreement with the previously reported increase in DNA damage markers such as γH2AX in primary mouse and human cells lacking Cdh1 expression.33, 34 Our results showing differential γH2AX staining in proliferating cells that are not replicating their DNA, support the idea that, upon Cdh1 loss, most of the DNA damage is generated in G2, when the bulk of genomic DNA has already been duplicated, or during mitosis. Consistently, Cdh1-mutant MEFs show increased presence of 53BP1 foci, which are typically found in G1 cells suffering from mild replication stress and mark chromosomal lesions generated during G2/M.35 In this regard, aberrant mitosis with lagging chromosomes, anaphase bridges and micronuclei, have been observed in the absence of Cdh1 in MEFs, human fibroblasts and U2Os cells.11, 12, 34 Interestingly, similar mitotic defects have been associated with low but persistent replication stress, that would fail to fully activate the DNA damage response.36, 37 Importantly, Cdh1 mutant MEFs showed mild activation of the DNA damage response.
Several studies have proposed defective replication to be the underlying cause of genomic instability upon Cdh1 loss.11, 33, 34, 38 However, a direct analysis of replication dynamics in primary cells lacking Cdh1 expression was missing. Our results provide the first evidence of slow replication fork movement and altered origin usage in the absence of Cdh1 in non-tumoral cells. A similar reduction in fork velocity was recently found in Cdh1-depleted HeLa cells,32 although no significant changes in origin activity were detected, perhaps reflecting differential sensitivities to Cdh1 loss between normal and transformed cells. Nevertheless, the increased origin activation rate found in Cdh1-mutant MEFs, proved to be secondary to the observed deceleration of replication forks, as origin activity was normalized when fork velocity was restored. Intriguingly, budding yeast Cdh1 was shown to be required for optimal firing efficiency of a subset of origins.39 While analyses of individual origin usage are not yet possible in mammalian cells, we cannot rule out that specific origins could be suboptimally activated in Cdh1-deficient MEFs. On the other hand, replication origin licensing is believed to be restrained in the absence of Cdh1 owing to Cyclin A and Cyclin B stabilization in G1 and increased Cdk1 and Cdk2 activities.6 Accordingly, we previously reported a moderate reduction in MCM4 and MCM5 chromatin loading in Cdh1-null MEFs,11 and similar observations were made in Cdh1-depleted U2Os cells.34 Since a full load of helicase complexes are required when cells are challenged with replicative stress, and dormant origins must be activated,21, 23 the consequences of impaired fork progression in the absence of Cdh1 could be aggravated due to insufficient availability of backup licensed origins.
Shortage of building blocks for DNA synthesis could be a prevalent underlying defect leading to aberrant replication, as nucleoside supplementation has been shown to limit replication stress induced by a variety of triggering factors, including oncogenes.40, 41, 42 Normal replication dynamics were also restored in Cdh1-mutant MEFs by exogenous supply of nucleotide precursors, suggesting that dNTP availability is compromised in the absence of Cdh1. Consistently, we found reduced levels of intracellular dNTP pools in cells devoid of Cdh1. Remarkably, DNA breakage was also significantly alleviated by exogenous nucleosides, further demonstrating that genomic instability in the absence of Cdh1 is mostly caused by replication stress.
Intracellular dNTP levels are determined by the interplay between the synthetic and hydrolytic pathways.43 RNR catalyzes the rate-limiting step for dNTP synthesis, and its cell cycle-dependent activity is dictated by RRM2 levels, which, in turn, are restricted by proteosomal degradation driven by SCF-Cyclin F in G2, and by APC/C-Cdh1 in G1.30, 44 This dual proteolytic control may explain why, in the absence of Cdh1, RRM2 expression is only moderately increased in asynchronous cultures. Moreover, the recent finding that Cyclin F is itself a substrate of APC/C-Cdh1 suggests that, upon Cdh1 loss, RRM2 degradation in G2 could be enhanced.45 TK1 and TMPK could be similarly affected by alternative degradation pathways upregulated in the absence of Cdh1. In this regard, TMPK was shown to be a substrate of both APC/C-Cdh1 and APC/C-Cdc20,31 and Cdc20 is itself targeted for degradation by APC/C-Cdh1. TK1 proteolysis has only been linked to APC/C-Cdh1, and the protein is notably upregulated in HeLa cells following Cdh1 depletion.31 However, additional or alternative control mechanisms could operate in normal primary cells to help balance its steady-state levels, particularly in the absence of Cdh1.
Nevertheless, the fact that replication defects and genomic instability are significantly alleviated in Cdh1-mutant cells by ectopic RRM2 overexpression suggests that, in these cells, endogenous RNR activity could be insufficient to maintain the required concentration of dNTPs or to counterbalance another dysfunction perturbing dNTP homeostasis. More importantly, this observation opens-up the interesting possibility of using genetic strategies to upregulate RRM2 levels in vivo, in mouse models of Cdh1 deficiency, and determine the real contribution of replicative stress to perinatal lethality of Cdh1-deficient pups and to increased tumor susceptibility of Cdh1 heterozygotes. A similar approach has been successfully used to reduce fragile site breakage and improve survival of ATR mutant mice.41
Our results show that limited inhibition of Cdk activity in Cdh1-deficient cells delays entry into S phase, and partially improves replication fork progression and chromosomal stability. Therefore, the precocious S phase onset induced upon Cdh1 deletion contributes to defective replication, perhaps by preventing full activation of the transcriptional program required for efficient replication, which includes nucleotide biosynthesis enzymes. However, full recovery of the phenotype is only achieved by increasing intracellular dNTP pools, suggesting that, in the absence of Cdh1, several mechanisms may contribute to limit dNTP availability or to create a scenario where higher than normal dNTP levels would be required to compensate for additional defects in the replication machinery.
In summary, we have formally established using primary cells, that loss of the APC/C-cofactor Cdh1 disrupts normal replication dynamics, triggers replication stress and, as a consequence, undermines genomic integrity. Moreover, we have identified the primary defect leading to aberrant replication, as APC/C-Cdh1 seems to be required to ensure an adequate supply of dNTPs to the replisome. Understanding the role of APC/C-Cdh1 in preserving nucleotide homeostasis in mammalian cells will require further investigation, but our findings further underscore the relevance of this ubiquitin-ligase complex to prevent replication stress and its pathological consequences, including developmental defects and cancer predisposition.46, 47
Materials and methods
MEFs isolation and treatments
The Cdh1lox/lox and Sox2-Cre mouse strains were previously described.11 Mice were housed under standard conditions in the pathogen-free animal facility of the University of Salamanca following the animal care standards of the institution. Primary MEFs were isolated from E13,5-E14,5 embryos and cultured using routine procedures. To obtain Cdh1-deficient embryos (Cdh1(−/Δ); Sox2-Cre, referred as Cdh1(−/−) for simplicity) crosses were established between Cdh1(lox/lox) female mice and Cdh1(−/+); Sox2-Cre male mice. Control embryos (Cdh1(+/lox), referred as Cdh1(+/+)) were obtained from the same crosses. Wild type embryos (referred as Cdh1(wt/wt)) were obtained from crosses between wild type C57BL/6 mice.
Cells were treated with 20 μM PHA-767491 (Sigma) for 4 h or with 3 μM Roscovitine (Sigma) for the indicated time. Nucleosides (EmbryoMax, Millipore) were used at 30 μM during 72 h and refreshed 24 h before harvesting cells. dNTPs (Sigma) were used at 10 μM.
Viral infections
Adenoviruses expressing the Cre Recombinase (Ad5CMVCre) or control adenoviruses (Ad5CMVEmpty) were supplied by the University of Iowa (Gene Transfer Vector Core Facility) and used at 300 pfu/cell. Cells were processed 4 days after infection. For RRM2 overexpression murine RRM2 cDNA (kindly provided by Dr. Robert S. Weiss, Cornell University, USA) was subcloned in pBABE-puro and MEFs were retrovirally transduced in the presence of 8 μg/ml polybrene (Sigma). Infected cells were selected for three days in 1 μg/ml puromycin (Sigma).
Flow cytometry
For EdU labeling and detection, MEFs were incubated with 10 μM EdU (Invitrogen) for 1 hour before harvesting, and processed according to manufacturer's instructions (Click-iT-EdU Flow Cytometry Assay Kit, Invitrogen). For γH2AX detection, MEFs were fixed in 4% paraformaldehyde, permeabilized in 70% ethanol, blocked in 10% goat serum, and stained with anti-H2AX(pS139)-Alexa Fluor 647 (N1-431, BD). DNA was stained with 7-AAD or Propidium Iodide (Invitrogen). Flow cytometry data were acquired on a FACSCalibur (BD Biosciences) and analyzed using FlowJo software (Tree Star, Ashland, OR).
Immunofluorescence
Cells were grown on glass coverslips, fixed in 4% paraformaldehyde, permeabilized with 0,1% Triton-X100 in PBS, and stained with mouse anti-phospho-H2AX (Ser139) (Millipore, #05-636), rabbit anti-53BP1 (Novus, #NB100-304) or rat anti-RPA32 (Cell Signaling, #2208). For RPA immunostaining, soluble proteins were extracted prior to fixation with 0,5% Triton-X100 in 20mM Tris-HCl pH 8, 50 mM NaCl, 3 mM MgCl2, 300 mM sucrose. For EdU staining cells were processed following manufacturer's instructions (Click-iT-EdU AlexaFluor 594 Imaging Kit, Invitrogen). To estimate S phase duration, MEFs were pulse-labeled with CldU and IdU (10 μg/ml) for 30 min and fixed in cold Methanol for 5 min. Staining was performed essentially as described48 using rat anti-BrdU antibody (Abcam, #ab6326) to detect CldU and mouse anti-BrdU antibody (BD, #347580) to detect IdU.
Alkaline comet assay
Alkaline comet assays were performed as previously described49 with some modifications. Briefly, 30 μl of a cell suspension at a density of 1 × 106 cells/ml were mixed with 65 ul of low melt agarose (0,5 %), cast on pre-coated slides and placed at 4 °C for 20 min. Slides were immersed in lysis buffer (1% Triton-X100, 10% DMSO in 2,5 M NaCl, 100 mM Na2EDTA, 10 mM Tris Base, 250 mM NaOH pH 10), for 1 hour at 4 ºC, then in alkaline buffer (1 mM Na2EDTA, 300 mM NaOH) for 20 min at 4 ºC, and then electrophoresed at 4°C in the same buffer at 0.9 V/cm and 300 mA for 20-30 min. For neutralization, slides were soaked in 400 mM Tris pH 7.5, and then washed, fixed in ethanol for 3 min and allowed to dry at room temperature. DNA was subsequently stained with ethidium bromide (0.5 μg/ml) and visualized by fluorescence microscopy (Nikon Eclipse 90i). Comet parameters were measured using the OpenComet software. At least 100 measurements were made for each sample. Tail moment is defined as the product of the comet tail length and the fraction of total DNA in the tail.
Single-molecule analysis of DNA replication
MEFs were pulse-labeled with 50 μM CldU (20 min) followed by 250 μM IdU (20 min), harvested, and resuspended in 0.2 M Tris pH 7.4, 50 mM EDTA and 0.5% SDS. Stretched DNA fibers were prepared essentially as described.50 For immunodetection of labeled tracks, fibers were incubated with primary antibodies for 1 hour at RT and with the corresponding secondary antibodies for 30 min at RT, in a humidity chamber. DNA was stained with anti-single-stranded DNA antibody (Millipore) to assess fiber integrity. Images were obtained in a Nikon Eclipse 90i microscope with a Plan Apo-Chromat VC 60X objective. The conversion factor used was: 1 μm=2.59 kb. At least 150–200 tracks/sample were measured for fork rate estimation, and at least 40 fibers with two or more origins were scored for IOD estimation.
dNTP pool measurements
1.5–2 × 107 cells were harvested in 0.5 ml mixture of ice-cold 10% trichloroacetic acid (TCA) and 15 mM MgCl2. After centrifugation of the cell debris, the resulting acidic solution was neutralized twice with Freon (1,1,2-trichloro-1,2,2-trifluoroethan/ TOA(Thioctylamine) (5/1.4, v/v) mixture. The upper layer was transferred to an empty tube, and after adjustment of pH to 4.5 with acetic acid, and addition of isotope-labeled internal standards, the samples were loaded onto the Oasis WAX columns (Waters, Milford, USA) for solid phase extraction (SPE). Eluates from SPE were evaporated to dryness under nitrogen at 37 ºC. The residues were reconstituted in 50 μl sample injection solution and analyzed by LC-MS/MS.
Since TCA was added directly to the plates, for normalization, an additional identical plate was cultured in parallel, and incubated with 10μM Edu 1 hour before harvesting. Cells were counted, fixed and processed for EdU staining. The number of S phase cells in each sample was then calculated based on the % of EdU-positive cells determined by flow cytometry.
Immunoblotting
MEFs were lysed in Laemmli buffer and 50 μg of total protein were separated by SDS-PAGE, transferred to nitrocellulose membranes (Bio-Rad) and probed with antibodies against Cdh1 (AR38, a generous gift from Dr J Gannon, Cancer Research UK), phospho-p53 (S15), phospho-Chk1 (S345) (Cell Signaling; #9284, #2348), Aurora A (BD; #610938), phospho-MCM2 (S53), MCM2, RRM2, TK1, TMPK and GAPDH (AbCam: #ab109133, #ab119921, #ab191800, #ab59271, #ab71756, #ab8245). Secondary antibodies were HRP-conjugated (DAKO) and blots were developed with ECL reagents (Western Lightning Plus, Perkin Elmer).
Statistical analysis
In column graphs mean ± s.d. is shown and statistical analysis was performed using Student's t-test. Variance was similar in all groups compared. The distribution of Tail Moment values, Fork Speed and IOD (shown in scatter dot plots) was compared using the Mann-Whitney test (GraphPad Prism 6). Unless otherwise stated, all experiments were replicated (biological replicas) at least three times (n=3).
References
Teixeira LK, Reed SI . Ubiquitin ligases and cell cycle control. Annu Rev Biochem 2013; 82: 387–414.
Craney A, Rape M . Dynamic regulation of ubiquitin-dependent cell cycle control. Curr Opin Cell Biol 2013; 25: 704–710.
Sivakumar S, Gorbsky GJ . Spatiotemporal regulation of the anaphase-promoting complex in mitosis. Nat Rev Mol Cell Biol 2015; 16: 82–94.
Chang L, Barford D . Insights into the anaphase-promoting complex: a molecular machine that regulates mitosis. Curr Opin Struct Biol 2014; 29: 1–9.
Pines J . Cubism and the cell cycle: the many faces of the APC/C. Nat Rev Mol Cell Biol 2011; 12: 427–438.
Sivaprasad U, Machida YJ, Dutta A . APC/C—the master controller of origin licensing? Cell Div 2007; 2: 8.
Eguren M, Manchado E, Malumbres M . Non-mitotic functions of the Anaphase-Promoting Complex. Semin Cell Dev Biol 2011; 22: 572–578.
Sigrist SJ, Lehner CF . Drosophila fizzy-related down-regulates mitotic cyclins and is required for cell proliferation arrest and entry into endocycles. Cell 1997; 90: 671–681.
Kitamura K, Maekawa H, Shimoda C . Fission yeast Ste9, a homolog of Hct1/Cdh1 and Fizzy-related, is a novel negative regulator of cell cycle progression during G1-phase. Mol Biol Cell 1998; 9: 1065–1080.
Fay DS, Keenan S, Han M . fzr-1 and lin-35/Rb function redundantly to control cell proliferation in C. elegans as revealed by a nonbiased synthetic screen. Genes Dev 2002; 16: 503–517.
Garcia-Higuera I, Manchado E, Dubus P, Canamero M, Mendez J, Moreno S et al. Genomic stability and tumour suppression by the APC/C cofactor Cdh1. Nat Cell Biol 2008; 10: 802–811.
Engelbert D, Schnerch D, Baumgarten A, Wasch R . The ubiquitin ligase APC(Cdh1) is required to maintain genome integrity in primary human cells. Oncogene 2008; 27: 907–917.
Sigl R, Wandke C, Rauch V, Kirk J, Hunt T, Geley S . Loss of the mammalian APC/C activator FZR1 shortens G1 and lengthens S phase but has little effect on exit from mitosis. J Cell Sci 2009; 122: 4208–4217.
Wasch R, Robbins JA, Cross FR . The emerging role of APC/CCdh1 in controlling differentiation, genomic stability and tumor suppression. Oncogene 2010; 29: 1–10.
Zhang J, Wan L, Dai X, Sun Y, Wei W . Functional characterization of Anaphase Promoting Complex/Cyclosome (APC/C) E3 ubiquitin ligases in tumorigenesis. Biochim Biophys Acta 2014; 1845: 277–293.
Munoz S, Mendez J . DNA replication stress: from molecular mechanisms to human disease. Chromosoma 2016; 126: 1–15.
Gelot C, Magdalou I, Lopez BS . Replication stress in Mammalian cells and its consequences for mitosis. Genes (Basel) 2015; 6: 267–298.
Labib K, De Piccoli G . Surviving chromosome replication: the many roles of the S-phase checkpoint pathway. Philos Trans R Soc Lond B Biol Sci 2011; 366: 3554–3561.
Ruzankina Y, Asare A, Brown EJ . Replicative stress, stem cells and aging. Mech Ageing Dev 2008; 129: 460–466.
Lopez-Contreras AJ, Fernandez-Capetillo O . The ATR barrier to replication-born DNA damage. DNA Repair (Amst) 2010; 9: 1249–1255.
Woodward AM, Gohler T, Luciani MG, Oehlmann M, Ge X, Gartner A et al. Excess Mcm2-7 license dormant origins of replication that can be used under conditions of replicative stress. J Cell Biol 2006; 173: 673–683.
Ge XQ, Jackson DA, Blow JJ . Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes Dev 2007; 21: 3331–3341.
Ibarra A, Schwob E, Mendez J . Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc Natl Acad Sci U S A 2008; 105: 8956–8961.
Zhong Y, Nellimoottil T, Peace JM, Knott SR, Villwock SK, Yee JM et al. The level of origin firing inversely affects the rate of replication fork progression. J Cell Biol 2013; 201: 373–383.
Montagnoli A, Valsasina B, Croci V, Menichincheri M, Rainoldi S, Marchesi V et al. A Cdc7 kinase inhibitor restricts initiation of DNA replication and has antitumor activity. Nat Chem Biol 2008; 4: 357–365.
Poli J, Tsaponina O, Crabbe L, Keszthelyi A, Pantesco V, Chabes A et al. dNTP pools determine fork progression and origin usage under replication stress. EMBO J 2012; 31: 883–894.
Anglana M, Apiou F, Bensimon A, Debatisse M . Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell 2003; 114: 385–394.
Guarino E, Salguero I, Kearsey SE . Cellular regulation of ribonucleotide reductase in eukaryotes. Semin Cell Dev Biol 2014; 30: 97–103.
Hofer A, Crona M, Logan DT, Sjoberg BM . DNA building blocks: keeping control of manufacture. Crit Rev Biochem Mol Biol 2012; 47: 50–63.
Chabes AL, Pfleger CM, Kirschner MW, Thelander L . Mouse ribonucleotide reductase R2 protein: a new target for anaphase-promoting complex-Cdh1-mediated proteolysis. Proc Natl Acad Sci U S A 2003; 100: 3925–3929.
Ke PY, Kuo YY, Hu CM, Chang ZF . Control of dTTP pool size by anaphase promoting complex/cyclosome is essential for the maintenance of genetic stability. Genes Dev 2005; 19: 1920–1933.
Yuan X, Srividhya J, De Luca T, Lee JH, Pomerening JR . Uncovering the role of APC-Cdh1 in generating the dynamics of S-phase onset. Mol Biol Cell 2014; 25: 441–456.
Eguren M, Porlan E, Manchado E, Garcia-Higuera I, Canamero M, Farinas I et al. The APC/C cofactor Cdh1 prevents replicative stress and p53-dependent cell death in neural progenitors. Nat Commun 2013; 4: 2880.
Greil C, Krohs J, Schnerch D, Follo M, Felthaus J, Engelhardt M et al. The role of APC/C(Cdh1) in replication stress and origin of genomic instability. Oncogene 2016; 35: 3062–3070.
Lukas C, Savic V, Bekker-Jensen S, Doil C, Neumann B, Pedersen RS et al. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol 2011; 13: 243–253.
Wilhelm T, Magdalou I, Barascu A, Techer H, Debatisse M, Lopez BS . Spontaneous slow replication fork progression elicits mitosis alterations in homologous recombination-deficient mammalian cells. Proc Natl Acad Sci U S A 2014; 111: 763–768.
Teixeira LK, Wang X, Li Y, Ekholm-Reed S, Wu X, Wang P et al. Cyclin E deregulation promotes loss of specific genomic regions. Curr Biol 2015; 25: 1327–1333.
Delgado-Esteban M, Garcia-Higuera I, Maestre C, Moreno S, Almeida A . APC/C-Cdh1 coordinates neurogenesis and cortical size during development. Nat Commun 2013; 4: 2879.
Ayuda-Duran P, Devesa F, Gomes F, Sequeira-Mendes J, Avila-Zarza C, Gomez M et al. The CDK regulators Cdh1 and Sic1 promote efficient usage of DNA replication origins to prevent chromosomal instability at a chromosome arm. Nucleic Acids Res 2014; 42: 7057–7068.
Bester AC, Roniger M, Oren YS, Im MM, Sarni D, Chaoat M et al. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell 2011; 145: 435–446.
Lopez-Contreras AJ, Specks J, Barlow JH, Ambrogio C, Desler C, Vikingsson S et al. Increased Rrm2 gene dosage reduces fragile site breakage and prolongs survival of ATR mutant mice. Genes Dev 2015; 29: 690–695.
Ruiz S, Lopez-Contreras AJ, Gabut M, Marion RM, Gutierrez-Martinez P, Bua S et al. Limiting replication stress during somatic cell reprogramming reduces genomic instability in induced pluripotent stem cells. Nat Commun 2015; 6: 8036.
Stillman B . Deoxynucleoside triphosphate (dNTP) synthesis and destruction regulate the replication of both cell and virus genomes. Proc Natl Acad Sci U S A 2013; 110: 14120–14121.
D'Angiolella V, Donato V, Forrester FM, Jeong YT, Pellacani C, Kudo Y et al. Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair. Cell 2012; 149: 1023–1034.
Choudhury R, Bonacci T, Arceci A, Lahiri D, Mills CA, Kernan JL et al. APC/C and SCF(cyclin F) Constitute a Reciprocal Feedback Circuit Controlling S-Phase Entry. Cell Rep 2016; 16: 3359–3372.
Zeman MK, Cimprich KA . Causes and consequences of replication stress. Nat Cell Biol 2014; 16: 2–9.
Gaillard H, Garcia-Muse T, Aguilera A . Replication stress and cancer. Nat Rev Cancer 2015; 15: 276–289.
Yokochi T, Gilbert DM . Replication labeling with halogenated thymidine analogs. Curr Protoc Cell Biol 2007; Chapter 22 (Unit 22): 10.
Pu X, Wang Z, Klaunig JE . Alkaline Comet Assay for Assessing DNA Damage in Individual Cells. Curr Protoc Toxicol 2015; 65: 3 12 11–11.
Mouron S, Rodriguez-Acebes S, Martinez-Jimenez MI, Garcia-Gomez S, Chocron S, Blanco L et al. Repriming of DNA synthesis at stalled replication forks by human PrimPol. Nat Struct Mol Biol 2013; 20: 1383–1389.
Acknowledgements
This research was funded by grants from the Spanish Ministry of Economy and Competitiveness MINECO (CSD2007-0015, BFU2011-28274 and BFU2014-55439) and Junta de Castilla y León (CSI151U13 and CSI084U16), the Swedish Cancer Society, the Knut and Alice Wallenberg Foundation and the Swedish Research Council (AC). IGH is supported by Fundación Científica de la Asociación Española contra el Cáncer (AECC). JG and RR were recipients of CSIC JAE and FPU predoctoral fellowships (MINECO).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Garzón, J., Rodríguez, R., Kong, Z. et al. Shortage of dNTPs underlies altered replication dynamics and DNA breakage in the absence of the APC/C cofactor Cdh1. Oncogene 36, 5808–5818 (2017). https://doi.org/10.1038/onc.2017.186
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2017.186
- Springer Nature Limited
This article is cited by
-
Embryonic stem cells maintain high origin activity and slow forks to coordinate replication with cell cycle progression
EMBO Reports (2024)
-
Efficient terminal erythroid differentiation requires the APC/C cofactor Cdh1 to limit replicative stress in erythroblasts
Scientific Reports (2022)
-
CD98hc (SLC3A2) sustains amino acid and nucleotide availability for cell cycle progression
Scientific Reports (2019)
-
Down-regulation of Cdk1 activity in G1 coordinates the G1/S gene expression programme with genome replication
Current Genetics (2019)
-
Chromatin conformation regulates the coordination between DNA replication and transcription
Nature Communications (2018)