Abstract
Krüppel-like transcription factor 10 (KLF10), also named as TIEG1, plays essential roles in mediating transforming growth factor beta (TGFβ) signaling and has been shown to function as a tumor suppressor in multiple cancer types. However, its roles in mediating cancer progression in vivo have yet to be fully characterized. Here, we have employed two well-characterized Pdx-1CreLSL-KrasG12D and Pdx-1CreLSL-KrasG12Dp53L/L pancreatic cancer models to ablate KLF10 expression and determine the impact of KLF10 deletion on tumor development and progression. We show that loss of KLF10 cooperates with KrasG12D leading to an invasive and widely metastatic phenotype of pancreatic ductal adenocarcinoma (PDAC). Mechanistically, loss of KLF10 in PDAC is shown to increase distant metastases and cancer stemness through activation of SDF-1/CXCR4 and AP-1 pathways. Furthermore, we demonstrate that targeting the SDF-1/CXCR4 pathway in the context of KLF10 deletion substantially suppresses PDAC progression suggesting that inhibition of this pathway represents a novel therapeutic strategy for PDAC treatment.
Similar content being viewed by others
Introduction
Pancreatic cancer is the deadliest malignancy of all gastrointestinal cancers, with a 5-year survival rate of only 7% and a median survival of less than 6 months.1 More than 90% of these cancers are pancreatic ductal adenocarcinomas (PDAC), an aggressive malignancy with late presentation and resistance to conventional and targeted therapies.2, 3 In contrast to other major cancers, decades of clinical trials with diverse novel and traditional agents have failed to provide appreciable survival benefit. In the area of targeted therapeutics, it was universally recognized that many PDAC genetic lesions (that is, potential therapeutic targets) remain to be discovered and characterized.4 Additionally, elucidation of the biological roles of genes which are frequently mutated in PDAC (KRAS, p16INK4a, p19ARF, P53 and SMAD4) in refined animal models are lacking.5, 6, 7
Over the past decade, a series of transgenic and gene knockout mouse models have been generated that develop pancreatic cancers with features reflective of ductal adenocarcinoma in humans. The most well-studied model has been the engineering of a Cre-mediated activation of mutant KRAS allele (Lox-Stop-Lox-KrasG12D, designated hereafter as LSL-KrasG12D) that, upon induction of expression with a Pdx1-Cre transgene during pancreatic development, is expressed, eliciting progressive development of pancreatic intraepithelial neoplasia-1 (PanIN-1), -2 and -3 lesions, with sporadic progression to PDAC in older mice.8 If, however, a Cre-conditional knockout of the tumor suppressor p53L/L is overlaid, pancreatic carcinogenesis is fully penetrant and extremely rapid, producing aggressive ductal adenocarcinomas 8–10 weeks after birth.9 This latter model, and second-generation derivatives, collectively represents tumor phenotypes with striking parallels to human PDAC.
Transforming growth factor (TGFβ1) signaling is potentiated or inhibited by intersection with numerous pathways through regulating Smad transcription activity, protein stability and subcellular localization.10 TGFβ is a potent suppressor of epithelial cell growth and survival, although these effects are highly dependent on the cell context. In numerous epithelial cell lines and in epithelial tissue in vivo, TGFβ exerts a growth inhibitory program that involves modulation of expression of cell cycle regulators and activation of apoptosis.11, 12 The tumor suppressor role of TGFβ signaling is underscored by the presence of inactivating TGFβ receptor mutations in a number of cancer.13, 14, 15 On the other hand, TGFβ can enhance the malignant growth of some established epithelial tumors, promoting tumor cell proliferation, migration and the epithelial-to-mesenchymal transition (EMT)—a process by which advanced carcinomas acquire a highly invasive and undifferentiated phenotype and become metastatic.16, 17 One of the tumor suppressors, Krüpple- like factor 10 (KLF10), also known as TIEG1, was originally identified as the product of a TGF-β inducible early-response gene (TIEG) in osteoblastic cells using differential display PCR.18, 19 KLF10 also has been shown to be induced by estrogen, TGF-βs, bone morphogenetic protein (BMP), nerve growth factor and epidermal growth factor (EGF), all with diversified functional roles depending on cellular and environmental contexts.18, 20, 21, 22, 23, 24 Further studies on TGF-β1’s regulation of KLF10 transcription have revealed that increased intracellular levels of KLF10 mimic the anti-proliferative and apoptotic effects of TGF-β1 on epithelial cells, suggesting that KLF10 is an important factor for mediating TGF-β1 signaling.25, 26, 27 However, KLF10’s targets and the related downstream signal transduction pathways remain largely unknown. We have recently succeeded in generating animal models of PDAC that recapitulate the human disease with great accuracy. Using these PDAC models, we sought to elucidate the role of KLF10 in normal pancreas development and its involvement in cancer progression and metastasis. We demonstrate that concomitant KrasG12D expression and loss of KLF10 results in rapid development of an invasive and malignant PDAC. This phenotype is shown to be mediated by the SDF-1/CXCR4 pathway and inhibition of this pathway offers substantial therapeutic benefit in these mouse models. SDF-1 (CXCL12) is a CXC-type chemokine that binds to and signals through the seven transmembrane receptor CXCR4, which has been shown to be necessary for bone marrow neutrophil mobilization.28 CXCL12 and CXCR4 knockout mice both die in the perinatal period, and CXCL12/CXCR4 signaling has been shown to be involved in organogenesis, autoimmunity, infection and tumorigenesis.29, 30 Notably, increased SDF-1 and CXCR4 expression have been reported in several types of cancer and is frequently associated with increased distant metastasis and poor prognosis.31 Collectively, our data demonstrate that inhibition of the SDF-1/CXCR4 pathway represents a novel therapeutic approach for the treatment of this deadly disease.
Results
KLF10 is not required for normal pancreas development in mice
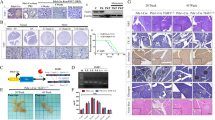
In the pancreas of wild-type mice, KLF10 was observed in islets and periacinar fibroblast-like cells. Moderate or strong nuclear KLF10 expression levels were found during PanINs progression and PDAC formation; however, reduced KLF10 staining intensity was observed in the metastatic PDAC of Pdx-1CreLSL-KrasG12Dp53L/L (PKP) mice, consistent with observations in human specimens (Figure 1a and Supplementary Figure 1a).32 To gain insight into the functional roles of KLF10 deficiency on the development of the pancreas, we obtained a KLF10 flox/flox mouse developed by Drs. Hawse and Subramaniam at Mayo Clinic, USA, in which the conditional KLF10 loxp/loxp allele was engineered to sustain Cre-mediated excision of exons 1 and 2 (Figure 1b). We first crossed KLF10 flox/flox mice with Pdx-1Cre transgenic mice to direct Cre recombinase expression to the endodermal progenitor lineages of the pancreas (Pdx-1-positive subpopulation). The Cre recombinase induced DNA rearrangement of the KLF10 gene locus in the pancreas from Pdx-1Cre KLF10L/L mice was determined by allele-specific PCR genotyping and by detection of KLF10 protein expression on western blot and immunohistochemistry (IHC) analysis (Figures 1a, c and d) to confirm the deletion of KLF10 expression in the pancreas. Pdx-1Cre KLF10L/Lmice (N=23) were born at the expected Mendelian ratio, indicating that Pdx-1Cre-mediated inactivation of KLF10 in the pancreas did not cause embryonic lethality. Appearance of the pancreas, body weight and pancreas weight did not reveal any differences between Pdx-1Cre KLF10L/Land wild-type mice (Figure 1e and data not shown). Histological analysis of pancreatic tissues of Pdx-1Cre KLF10L/L mice at early and late time points showed normal exocrine granular and ductal structural components and islet formation after verification that KLF10 expression was lost in the pancreas from Pdx-1Cre KLF10L/L (Figures 1a and f). Amylase, insulin, glucagon and pancreatic ductal markers, CK19 and DBA showed normal expression between the two genotypes of mice and functional analysis for glucose tolerance revealed no significant differences in the Pdx-1Cre KLF10L/L compared to WT controls (Figures 1g and h). These data revealed that KLF10 inactivation does not prominently affect pancreas development or play any role in the induction of pancreatic carcinogenesis.
Depletion of KLF10 in the mouse pancreas does not perturb pancreatic development and function. (a) Immunohistochemical detection of KLF10 protein in normal mouse pancreas (Pdx-1Cre), neoplastic lesions of PK, PKP+, PKP and Pdx-1Cre KLF10L/L mice. (b) Pdx-1Cre-mediated deletion of KLF10 in the pancreas of Pdx1-Cre KLF10L/L mice. Structure of the KLF10 floxed allele: Loxp sites were inserted into the introns surrounding exons 1 and 2. (c) Specific genotyping PCR analysis to detect KLF10 wild types and loxp alleles from the tail DNA of the Pdx-1Cre KLF10wt, Pdx-1Cre KLF10L/+ and Pdx-1Cre KLF10L/Loffspring. Recombined allele; loxp-flanked allele (L); wild-type allele (+) of KLF10 gene; M, 100 bp DNA marker. (d) Western blot analyses for detection of KLF10 protein expression in pancreatic lysates from Pdx-1Cre control, Pdx1-Cre KLF10L/L mice. β-actin is shown as a loading control. (e) The gross appearance of pancreas isolated from Pdx1-Cre KLF10L/L mice is shown. (f) H&E-stained histological sections of pancreatic tissue from Pdx1-Cre KLF10L/L and wild-type mice. Scale bar is 50 μm. (g) IHC and IF staining demonstrating that conditional knockout of KLF10 in the mouse pancreas does not alter insulin, amylase, glucagon, cytokeratin 19 (CK19) and dolichos biflorus agglutinin (DBA) expression in the pancreas compared to wild-type controls. (h) No differences in blood glucose levels were found when blood samples from overnight-fasted Pdx-1Cre KLF10L/L mice were compared to wild-type mice. Data are means±SE obtained from six mice/group. H&E, hematoxylin and eosin; IF, immunofluorescence; IHC, immunohistochemical.
KLF10 loss in the pancreas rapidly provokes mutant Kras-induced PanIN to PDAC
An active Kras mutation (KrasG12D) was frequently found in the majority of human PDAC during early carcinogenesis.33 The conditional expression of KrasG12D in the pancreas results in the development of focal PanIN lesions but no PDAC formation up to 25 weeks in Pdx-1Cre LSL-KrasG12D (PK) model.7, 8 To study whether KLF10 inactivation affects the progression and development of mutant Kras-induced PanIN lesions in mice, we generated Pdx-1Cre LSL-KrasG12DKLF10L/L(PKK) compound mice by crossing Pdx-1Cre KLF10L/L mice with LSL-KrasG12D KLF10L/L mice. In the aging mouse cohorts, PKK mice were killed when they developed fatigue or wasting syndrome, mostly at 20–24 weeks of age. We observed that half of the compound mice had progressed to invasive PDAC formation compared to PK mice which only displayed early to mid-stages of PanIN lesions in the pancreas (n=27 mice/genotype) (Figures 2a and bi–ii).
Concomitant KLF10 loss and activated KrasG12D to drive rapid malignant PDAC development in mice. (a) Gross anatomy images of PDAC in a 20-week-old PKK mouse. Gross images of distant metastases sites; lung, thymus and stomach developed from widely metastatic PDAC of PKK mice. (b) Histological analysis of pancreas from PKK and PK mice at different ages by H&E staining (i). Quantification of PanINs grading and PDAC lesions in 8-week-old (ii) and 20-week-old (iii) PK and PKK mice. A minimum of six nonconsecutive H&E sections from each mouse (three mice per genotype) were examined. Error bars are the means±s.e.m. *P< 0.05, **P<0.01, ***P<0.001. (c) Kaplan–Meier curve showing significantly reduced survival time of PKK mice compared to PK mice. (d) H&E histological examination revealing a 20-week-old PKK mouse with distant lung (i) and thymus (ii) metastasis. Scale bar is 50 μm. (e) Relative intensity of the expression levels of E-cadherin (E-cad) and CK19 in pancreatic lesions of indicated genotypes as detected by IHC analysis (i). Alcian blue staining indicating mucin content and IHC analysis for α-smooth muscle actin (SMA) in PDAC from the indicated genotypes of mice (ii), and immunostaining of pancreatic lesions from mice of indicated genotypes with anti-Ki67, TGFβ1, shh and EGFR antibodies (iii). Scale bar is 100 μm. H&E, hematoxylin and eosin; IHC, immunohostochemical; shh, sonic hedgehog.
As indicated by Kaplan–Meier analysis, PKK mice exhibited a shortened lifespan compared with PK mice (P<0.01) (Figure 2c). All observed animals with malignant PDAC had metastasis to distant sites, including mesenteric lymph nodes, stomach, liver, lung and thymus, which were confirmed by examining hematoxylin and eosin-stained sections, and representative metastases were shown in Figures 2a and d. Histological examination of tumors revealed well or moderately differentiated PDAC lesions that are reminiscent of human PDAC and were positive for CK 19 and E-cadherin, but negative for amylase and insulin by IHC staining (Figures 2e and i and data not shown). Metaplastic ductal lesions of the pancreas in PKK mice exhibited positive staining for Alcian blue, and further characterization of the neoplastic stromal compartment revealed marked induction of α-smooth muscle actin (SMA) expression in PKK PDAC models compared with PK models (Figures 2e–ii). In this setting, we also observed increased expression of Ki67 in the neoplastic ductal lesions of PKK pancreas compared with PK and wild-type controls (Figures 2e–iii and Supplementary Figure 1b). Furthermore, increased expression of pathway components known to be involved in PDAC development and progression, such as TGFβ1, sonic hedgehog and epidermal growth factor receptor (EGFR) were observed in the pancreatic ductal lesions of PKK mice as compared with that of PK mice (Figures 2e–iii). In particular, sonic hedgehog signaling has been shown to play important roles in pancreatic development and malignancy.34 Collectively, these data demonstrated that KLF10 loss in the setting of activated KrasG12D results in rapid PDAC formation and metastatic disease.
Homozygous deletion of KLF10 accelerates development of metastatic PDAC in PKP model
Mutations of tumor-suppressor p53 have been found in more than 50% of late stage human PDAC, and have been associated with the progression of metastatic PDAC.33 We and Hingorani and colleagues previously reported a well-characterized PDAC model that faithfully mimics the human disease in which there is rapid development of PDAC and metastatic disease in p53 homozygous mutant animals on a mutant KrasG12D background.6, 9 Thus, in the next set of experiments, we aimed to elucidate the role of KLF10 in promoting metastasis by crossing Pdx-1Cre KLF10L/Lp53L/L mice with LSL-KrasG12DKLF10L/Lp53L/L mice to generate compound triple mutant Pdx-1-Cre LSL-KrasG12DKLF10L/Lp53L/L (PKKP) mice. As expected, all PKKP mice developed extensive invasive and metastatic PDAC with high incidence of biliary obstruction and hemorrhagic ascites as early as 7 weeks (n=34 mice/genotyped) with 100% penetrance (Figure 3a). The Kaplan–Meier curves of overall survival of the four compound mouse cohorts, PKP; PKKP; Pdx-1Cre LSL-KrasG12Dp53L/+(PKP+); Pdx-1-Cre LSL-KrasG12D KLF10L/L p53L/+(PKKP+) are shown for comparison in Figure 3b. A pronounced cooperative effect of KLF10 loss was noted with PKKP+ mice showing significantly reduced survival compared to PKP+ controls (P<0.01).
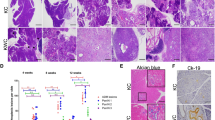
KLF10 deletion promotes metastatic PDAC in cooperation with KrasG12D activation and P53 deficiency. (a) Gross anatomy indicating the effects of metastatic PDAC from PKKP with obstructive jaundice and distant metastasis to the lung. (b) Kaplan–Meier survival curves representing the life span of PK,PKP+, PKKP+, PKKP, PKP mice. (c) High magnification of tumor sections from PKKP mice indicating abnormal anaphase cells (scale bar, 20 μm). (d) Macroscopic examination showing representative primary and metastatic PDAC tumor masses in pancreas, stomach, lung, lymph node, thoracic diaphragm, liver, spleen and thymus peritoneal metastases. (e) H&E staining of pancreas, stomach, lung, liver, lymph node and diaphragm indicating metastatic lesions from PKKP PDAC mice. Scale bar, 50 μm. H&E, hematoxylin and eosin.
At autopsy, most PKKP tumors displayed a diagnostic feature of well-differentiated PDAC with prominent nuclear atypia and abundant mitoses, and these were further verified by a biliary pancreatic pathologist (Figure 3c). Histologic examination of invasion of adjacent organs and distant metastases to the stomach, liver, lymph nodes, spleen, diaphragm, lung and thymus is shown in Figures 3d and e. The distribution of metastatic involvement in comparison between PKP and PKKP tumors is summarized in Supplementary Table 1. For instance, murine PDAC metastasized to the liver in 6 of 34 PKKP and 4 of 17 PKP mice (P>0.05). In 23 of 34 PKKP and 3 of 17 PKP mice, the tumor metastasized to the lung (P<0.01), and in 20 of 34 PKKP and 4 of 17 PKP mice developed metastasis to the lymph nodes (P<0.05). Of note, these findings are consistent with the pattern of metastatic PDAC observed in humans. Furthermore, primary tumors and metastatic PDAC lesions in distant organs were collected and primary murine PDAC cell lines were isolated and utilized for cellular and molecular studies.
Pathological and immunohistological analysis of PDAC lesions derived from PKKP model
To further characterize signaling pathway changes occurring due to loss of KLF10 expression in PKKP pancreata, we performed a set of immunostaining experiments. First, an immunohistochemical study verified that both primary PDAC tumors and distant PDAC metastatic lesions in the PKKP mice stained strongly positive with an epithelial marker (CK19) (Figures 4a and b). In addition, conditional KLF10 deletion induced significant increases in the levels of several proteins important for developmental pathways such as Notch1 and Wnt1 expression in the PDAC from PKKP mice as compared to that of PKP mice (Figure 4a). High TGFβ1 staining levels were observed in both PKKP and PKP groups (Figure 4a). Western blots provided confirmation of results in Figure 4a (see Supplementary Figure 1c). Furthermore, the tumors from PKKP mice showed focally strong immunostaining for EGFR, a key upstream effector of p-Akt and pERK (p-p44/42) pathways (Figure 4a). High power views of IHC images revealed significant increase in nuclear p-Akt and p-p44/42 staining in PDAC from PKKP mice as compared to that of PKP (Supplementary Figure 2a). We also evaluated the vascularization of PDAC tissues with CD31, LYVE-1 and VEGF-A staining which revealed high vascularization in PDAC from PKKP mice (Supplementary Figure 2b). Additionally, at distant metastatic sites, IHC analysis demonstrated that lung and peritoneal metastases displayed higher intensity for CD44, Twist2 and c-Myc immunostaining (Figure 4b and data not shown). Of note, β-catenin staining was mainly localized to the cytosol of PDAC lesions of PKP mice, whereas the PDAC lesions of PKKP mice exhibited stronger nuclear β-catenin staining, suggesting that activation of the Wnt-β-catenin pathway might in part be responsible for PDAC progression upon KLF10 loss (Supplementary Figure 2c). Moreover, histological analysis revealed a decrease in T lymphocytes and macrophages in PDAC lesions of KLF10 mutant mice (PKKP) compared with PKP PDAC (Supplementary Figure 3).
IHC characterization of metastatic PDAC in PKKP mice. (a) Malignant PDAC lesions from PKKP and PKP mice were stained with anti-CK19, Notch1, WNT1, TGFβ1, EGFR, p-Akt and anti-p44/42 antibodies. Note the increasingly intensive immunoreactivity of Notch1, WNT1, TGFβ1 and EGFR, and strong nuclear expression of p-p44/42 and p-Akt in ductal epithelium of PDAC. Scale bar is 50 μm. (b) IHC analysis indicating intensive staining for CK19, CD44 and Twist2 staining in PDAC from PKKP mice with associated hepatic, lymph node, diaphragm, thymus and lung metastasis when compared with PDAC from PKP mice. Scale bar is 50 μm. IHC, immunohistochemical.
Resistance to TGFβ-mediated growth inhibition and induction of EMT to enhance cell migration and invasion in KLF10-null PDAC cells
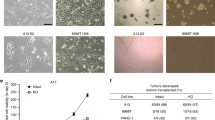
To characterize the effect of KLF10 loss on the growth, migration and invasion of murine PDAC cells in vitro, we utilized established primary murine PDAC cells derived from PKP and PKKP mice. Examination with a phase-contrast microscope revealed morphological differences between cells derived from mice with different genotypes. Specifically, loss of KLF10 expression in PDAC cells resulted in a more spindle/fibroblast-like morphology compared to cells derived from PKP mice which exhibited a more cobblestone/epithelial like morphology (Figure 5a). Western blot analysis confirmed deletion of KLF10 in PKKP PDAC cells as compared to PKP PDAC cells expressing wild-type KLF10 (Figure 5b). In subsequent experiments, we next determined whether KLF10 loss affects proliferation of PDAC cells in vitro and found no significant difference between the two cell lines (Figure 5c). However, our results indicated that KLF10 may be involved in suppressing colony sphere formation (modulated cancer stem cell activity) of PDAC cells as indicated by tumor sphere formation assays (P<0.01, Figure 5d). Next, to evaluate the effect of KLF10 loss on the migration of PDAC cells in vitro, scratch-wound healing assays were performed in PKKP and PKP cells. Our results revealed that KLF10-null PDAC cells exhibit significantly increased rates of cell migration compared with KLF10-proficient cells (Figure 5e). Similar effects were observed in transwell invasion assays (P<0.01, Figure 5f). Taken together, our in vitro results demonstrate that KLF10 loss enhances cell migration and invasion of PDAC cells. Thus, our in vitro cell culture models corroborate with our in vivo KLF10-null PDAC model in which increased metastatic potential was observed.
Loss of KLF10 modulates TGFβ1 and WNT pathways to promote EMT and cell motility in PDAC. (a) Representative morphology of primary murine PDAC cell lines from PKKP and PKP mice by phase-contrast light microscopy. Scale bar is 100 μm. (b) Immunoblot analysis confirmed that the loss of KLF10 protein expression in primary PKKP PDAC cell lines. (c) Cell viability assay indicating that KLF10 loss does not significantly affect in vitro growth rate of PDAC cells. (d) Loss of KLF10 in PDAC cells facilitates anchorage independent growth and tumor sphere formation in ‘sphere’-forming assays (i). Quantification of the number and size of tumor spheres in PKKP and PKP groups. Data are expressed as Mean±s.e.m., N=3, ** P<0.01 (ii). (e) Wound healing assays showed that the in vitro migration ability of PKP and PKKP PDAC cells. The results showed that KLF10 loss enhances wound healing in PDAC cells. Scale bar, 100 μm. (f) Transwell invasion assays depicting the invasive ability of PKP and PKKP PDAC cells. The bar graph showed that the ability of invasion was significantly increased in PKKP PDAC cells as compared to PKP cells. (g) RT–qPCR analysis of EMT marker genes following the loss of KLF10 expression in PDAC. (h) Western blot analysis showing EMT marker protein expression following the loss of KLF10 in PDAC cells. (i) IHC analysis showing the staining intensities of E-cadherin, SMA and vimentin protein in PDAC lesions from mice of the indicated genotypes. (j) SBE4 luciferase activity assay revealed the inactivation of KLF10 increases the activation of TGFβ1-SMAD signaling pathways in murine PDAC cells (Mean±s.e.m.; n=3). (k) Reporter assays were performed using TOP/FOP reporters in PKKP and PKP PDAC cells. (l) Analysis of the activation of Wnt signaling pathway by western blot that revealed KLF10 loss increases the levels of active−β-catenin and c-myc proteins in PKKP as compared to PKP PDAC cells. (m) RT–qPCR analysis of Wnt target gene expression in PKP and PKKP PDAC cells. (n) PKKP and Pdx-1Cre LSL-KrasG12D SMAD4L/L p53L/L (PKSP) PDAC cells are resistant to TGF-β1-mediated growth inhibition as compared to PKP cells. *P<0.05. IHC, immunohistochemical.
Epithelial-to-mesenchymal transition is an important process by which malignant tumor cells activate invasion and metastasis that is required for cancer cell migration and invasion, and TGFβ1 and WNT pathways are important in the regulation of EMT program. Therefore, we next investigated whether KLF10 loss modulates EMT in PDAC cells. We performed RT–qPCR and western blot analyses to examine the expression pattern of EMT markers in KLF10-deficient and -proficient PDAC cells. These included mesenchymal markers such as vimentin, snail and SMA, and the epithelial marker, E-cadherin. As shown in Figures 5g and h, the results of our study indicated that KLF10 loss in PDAC cells suppressed the mRNA and protein expression levels of E-cadherin, and increased the levels of SMA and vimentin protein expression. These results were also further verified by IHC staining to confirm the related EMT marker expression levels in the PDAC of PKP and PKKP mice (Figure 5i).
Additionally, we also determined whether loss of KLF10 resulted in modulation of the TGFβ1 and Wnt signaling pathways by using Wnt-responsive TOP-Flash and TGFβ1-responsive SBE4-luc reporter assays. Our results indicated that inactivation of KLF10 induces activation of TGFβ1 and Wnt-β-catenin signaling pathways (Figures 5j and k). The increased expression levels of Wnt target genes such as active-β-catenin (ABC), c-Myc, Survivin, Cyclin D1 and Twist2 were demonstrated by using RT–qPCR and western blot analysis (Figures 5l and m). Meanwhile, we also examined whether inactivation of KLF10 modulates the TGFβ1-mediated growth inhibition in murine PDAC cells. Interestingly, we observed that PKP PDAC cells were efficiently growth arrested by TGFβ1, whereas the PKKP cells exhibited no growth inhibitory effects in response to TGFβ1 (Figure 5n). Therefore, the inactivation of KLF10 in PDAC significantly blocked the TGFβ1-induced growth inhibition of PDAC cells in vitro. Taken together, these results revealed that KLF10 can suppress the Wnt/β-catenin signaling pathway to reduce in vitro cell migration effects that were further verified by the use of human Panc-1 PDAC cells (Supplementary Figure 4). Furthermore, our western blot and immunostaining experiments also demonstrated that loss of KLF10 results in increased levels of the pluripotent markers, Nanog and Sox2 protein expression in PDAC, but not that of Oct4. Cancer stem cell markers, CD44, Nestin and side population marker, ABCG2 have been reported to be associated with metastatic phenotype of malignant PDAC. In the present study, we also observed that KLF10 loss activates Wnt-associated cancer stem cell markers, CD133, CD44, Nestin and ABCG2 in PDAC. Thus these observations implicate that KLF10 loss in our model system promotes cancer stem cell properties in PDAC (Supplementary Figure 5).
Characterization of the gene expression profiles of PKKP PDAC tumor cells
To find potential target genes affected by KLF10 loss in PDAC, we compared the profiles of differentially expressed genes using cDNA microarray analysis of our primary murine PDAC cell lines. The cDNA microarray analysis consisted of three different pairs of PDAC cell lines derived from PKP vs PKKP mice. Using the Significance Analysis of Microarrays algorithm, we identified a total of 362 transcripts (Fold Change filtering:⩾2FC) that were significantly differentially expressed between the PKKP and PKP cell lines (Figure 6a). The signatures of genes displaying a⩾2-fold change in expression upon KLF10 loss was further deposited in the Gene Expression Omnibus database and are listed in Supplementary Table S2. The top 10 upregulated and downregulated genes in the PKKP cell compared to the PKP cells are highlighted in Figure 6a. Specifically, the most induced genes following loss of KLF10 expression include CXCL12 (35.4-fold increase; P=0.017), IRX2 (26.5-fold increase; P=0.017), SORL1 (22.3-fold increase), HUNK (15.6-fold increase; P=0.028), Eva1a (13.7-fold increase; P=0.01), Arhgap8 (12.8-fold increase; P=0.012), BMP7 (11.9-fold increase; P=0.004), Serpina1d (10.8-fold increase; P=0.038), Hey1 (10.7-fold increase; P=0.042) and Pcsk9 (10-fold increase; P=0.02).
cDNA microarray analysis of primary PDAC cells from PKKP and PKP mice. (a) Heat-map showing the gene profiling of primary PDAC cells established from three independent of PKKP and PKP mice indicating genes were significantly increased (red), intermediate (black) or decreased (green) expression levels. (b) RT–qPCR analysis confirmed the most significant genes regulated by KLF10 that showed changes in the microarray analysis. Data represent the means±s.d. of triplicate samples. *P<0.05; **P<0.01, t-test. (c) Western blot analysis to verify alterations in protein expression levels of CXCL12(SDF-1), SORL1, HUNK and IRX2 from tumor lysates of PKKP and PKP mice. β-actin served as a loading control.
As expected, the KLF10 transcript was the most downregulated (−72-fold; P=0.0001) in PKKP PDAC cells as compared to PKP cells. To validate the microarray analysis results, we reanalyzed the expression of randomly selected and various known tumorigenic- and metastatic-related genes (CXCL12, IRX2, HUNK, BMP7, SOX17 and KLF10, and so on) by RT–qPCR analysis. All analyzed genes confirmed the microarray data as they were significantly increased or decreased in the PKKP cells relative to the PKP PDAC cells (8-, 2.5-, 7-, 3-, 7- and 3.5-fold increase for CXCL12, IRX2, SORL1, HUNK, EVA1a and BMP7, respectively; 8-, 4-, 3-, 4-, 7-, 8- and 3-fold reduce for KLF10, AVIL, THBS4, HSD3B2, SOX17, GSPT2 and KLHL30 respectively, P<0.01) (Figure 6b). Subsequently, the protein levels of CXCL12/SDF-1, IRX2, HUNK and SORL1 in KLF10-deficient PDAC cells were further confirmed by western blot analysis (Figure 6c).
To understand the potential biological relevance of the upregulated set of genes, we employed KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis to identify biologic pathways that were altered in KLF10-depleted PDAC cells. Based on KEGG pathway enrichment analysis, the most significant pathway associated with differentially expressed genes affected by KLF10 loss in PDAC cells was the TGFβ1 signaling pathway (CXCL12, INHBA, PPP2CA, BMP7, THBS4). Moreover, we also observed three highly significant networks of potentially interacting upregulated genes affected by KLF10 deficiency in PDAC cells, and they are as follows: Pancreatic Cancer Pathway (PLD1, CDKN2A, RALA, BRCA2, AKT2, RAD51), WNT signaling pathway (CHD8, SIAH1B, PPP2CA, FRAT2, CAMK2B, SOX17, TCF7L1, FZD6, NFATC1) and cysteine and methionine metabolism pathway (LDHA, AHCY, ENOPH1, DNMT3B, CBS). Based on these bioinformatics results, we hypothesized that loss of KLF10 increases CXCL12 (SDF-1) expression in PDAC, thereby enhancing the activation of TGFβ1/Wnt signaling pathways, and further amplifying cysteine and methionine metabolism pathways resulting in a pro-metastatic phenotype.
KLF10 loss induces SDF-1 expression to promote in vitro cell migration of PDAC
Currently, there are a number of reports in the literature demonstrating involvement of SDF-1 in cancer migration and invasion, including PDAC. To confirm the elevated levels of SDF-1 protein in response to KLF10 loss in PDAC, we further employed the cytokine protein array to validate different chemokine receptor or cytokines protein expression differences between the tumor lysates from PKP and PKKP PDAC mice. As shown in Figure 7a, the expression level of SDF-1 was appreciably increased in the KLF10-deficient murine PDAC tumors (P<0.01). In addition, the upregulation of the SDF-1/CXCR4 axis in KLF10-null PDAC was further confirmed by using IHC validation of anti-mouse SDF-1 and CXCR4 antibodies in murine PDAC (Figure 7b). Subsequently, western blot analysis also confirmed the direct upregulation of SDF-1 and CXCR4 protein expression and increasing activation of their downstream transcription factors including c-Jun, c-Fos and JunB in primary PDAC cell lines from PKKP PDAC model as compared to PKP groups (Figure 7c). Furthermore, all metastatic primary PDAC cells, isolated from a variety of distant metastatic sites, such as liver, lymphoid node, peritoneal ascites and lung in KLF10-null PDAC mice, displayed high levels of SDF-1 and CXCR4 protein expression, indicating that the SDF-1/CXCR4 axis may have a pivotal role in mediating PDAC metastasis in mice (Supplementary Figure 6a).
KLF10 loss induces SDF-1 upregulation resulting in increased c-Jun phosphorylation and enhanced cell migration of PDAC in vitro. (a) Detection of mouse cytokine expression profile from the PDAC lysate of PKKP mice and one matched PKP control (i). Template alignment of the mouse cytokines in the array represent (ii): POS, positive; NEG, negative; IL, interleukin; SDF-1, stromal cell-derived factor 1; BLC, B-lymphocyte chemo-attractant; TAC, protachykinin; TCA-3, small inducible cytokine A1; TIMP, tissue inhibitors of metalloproteinase; LIX, LPS-induced CXC chemokine; MCSF, macrophage colony stimulating factor; MCP-1, monocyte chemotactic protein 1; MIG, mitogen-inducible gene; MIP-1, macrophage inflammatory protein 1. Quantitation of the intensity of SDF-1 protein expression in mouse cytokine arrays revealed a sixfold increase in PDAC from PKKP mice compared to the PKP group. **P<0.01 (iii). (b) IHC staining verified that high expression of SDF-1 and CXCR4 proteins in PDAC from PKKP mice compared to the PKP group. (c) Immunoblotting analysis showing increased of SDF-1, CXCR4, and activated c-Jun, c-Fos and JunB expression in PKKP PDAC cells as compared to PKP cells. (d) In vitro scratch assays demonstrating strong promigratory effects for PKKP conditional medium (CM) compared to control PKP culture medium. Scale bar is 200 μm. (e) In vitro detection of secreted SDF-1 protein measured by enzyme-linked immunosorbent assay, increased SDF-1 was observed in the conditional medium of PKKP PDAC cells as compared to PKP group *P<0.05. (f) Proliferation assays indicating the effects of overexpression of SDF-1 on the growth rates of PDAC cells in vitro. (g) In vitro scratch assay indicating the effects of overexpression of SDF-1 on the migratory ability of PKP PDAC cells. Scale bar, 200 μm. (h) Western blot analysis confirming that overexpression of SDF-1 in PKP PDAC cells activates p-Akt, p-44/42 and c-jun proteins. IHC, immunohistochemical.
Since SDF-1 is a secreted factor, we sought to determine the effects of conditioned media isolated from PKKP PDAC cells on PKP PDAC cells in wound healing. As shown in Figure 7d, the conditioned media of PKKP enhanced cell migration in the wound healing assays compared to that of PKP control media. In compliance with the above data, enzyme-linked immunosorbent assay analysis confirmed that there was a threefold increase in SDF-1 protein in the conditioned medium from PKKP PDAC cells compared with the PKP condition medium (*P<0.01; Figure 7e). Furthermore, overexpression of SDF-1 augmented the migratory ability of PKP PDAC cells but had no effect on their proliferation rates (Figures 7f and g). Similarly, use of SDF-1 shRNA for stable suppression of SDF-1 did not alter cell growth, but significantly inhibited the migration of PKKP PDAC cells from our in vitro scratch wound healing analysis (Supplementary Figure 6b). Subsequently, immunoblot analysis demonstrated that overexpression of SDF-1 resulted in an increase in the phosphorylation levels of p44/42, Akt and c-Jun in PKP PDAC cells (Figure 7h). Taken together, these data suggest that upregulation of SDF-1 expression promotes cell migration without affecting proliferation in PDAC cells.
The SDF-1/CXCR4 antagonist, Plerixafor, delays PDAC development and metastasis in mice
Based on the above data, we hypothesized that the SDF-1/CXCR4 pathway is likely to be a metastasis-promoting pathway in PDAC. Having observed an increase in SDF-1 expression in our primary murine PDAC cells which influenced PDAC cell migratory potential, we next assayed the efficacy of plerixafor (AMD3100),35 a bicyclam molecule that antagonizes the binding of SDF-1 to its cognate receptor CXCR4, on modulation of proliferation and migration in PDAC cells (KLF10 proficient and deficient). Following 5 days of treatment, we observed that PDAC cell proliferation was suppressed by plerixafor in a dose-dependent manner (0.05–20 uM) (Figure 8a). Flow cytometry analysis revealed that plerixafor treatment for 48 h significantly increased G1 phase cell cycle arrest and reduced the percentage of cells in S phase as compared with control (Figure 8b). In addition, non-adherent sphere assays showed that plerixafor treatment significantly reduced the number and size of tumor spheroids formed in murine PDAC cells indicating that the SDF-1/CXCR4 axis may be involved in the maintenance of cancer stemness properties of PDAC cells (Figure 8c). Next, we determined the effects of plerixafor treatment on the migration of PDAC cells, and we observed that plerixafor significantly reduced the migration of murine PDAC cells in vitro (Figure 8d). Furthermore, western blot analysis confirmed significant reductions in all of the SDF-1/CXCR4 axis effector proteins in response to plerixafor treatment. Our data indicated decreased levels of p-Akt and p-Jun in both genotypes of PDAC cell lines treated with plerixafor as compared with those of phosphate-buffered saline controls (Figure 8e). These results indicate that plerixafor treatment has minor effects on PDAC cell proliferation but elicits marked effects on cell migration.
Inhibition of SDF-1/CXCR4 signaling blocks PDAC formation in PKKP and PKP mice. (a) Cell proliferation rates were suppressed in PKKP and PKP PDAC cells treated for 5 days with plerixafor inhibitor, as compared to untreated groups *P<0.05. (b) Representative fluorescence-activated cell sorting profiles showing PKKP and PKP PDAC cells following plerixafor (5 μM) treatment compared with PBS control groups. The percentages of cells in G1, S or G2/M phases of cell cycle are indicated next to each histogram. Mean±s.e.m.; n=3. (c) Sphere formation assays revealed plerixafor treatment effectively reduced the number and the size of tumor sphere formed in both PKKP and PKP PDAC cells (i). Quantitation of tumor spheres on PKP and PKKP PDAC cells after plerixafor treatment as compared to PBS treatment. Mean±s.e.m.; n=3. **P<0.01. ***P<0.001 (ii). (d) In vitro wound healing assays showed the reduced wound closure abilities in PKKP followed by plerixafor (50 nM) treatment, as compared with PKP groups (i). Scale bar is 200 μm. Quantitation of wound healing rates at indicated conditions (ii). (e) Immunoblotting analysis indicating decreased p-c-Jun and p-Akt, protein levels in PKKP PDAC cells following 5 μM of plerixafor treatment. (f) Macroscopic appearance and H&E histological analysis of pancreatic, lung and spleen lesions in PKP+ and PKKP+ mice treated intraperitoneally with plerixafor or vehicle (PBS) for 15 weeks (i). Scale bar, 50 μm. Quantification of PanINs grading and PDAC lesions in PK+ and PKK+ mice with or without plerixafor treatment (ii). (g) IHC analysis indicating the reduction of staining intensity for Ki67 and p-c-Jun proteins in the pancreatic lesions after plerixafor treatment as compared with PBS-treated control groups. (h) Proposed models depicting the molecular mechanisms of action of KLF10 in TGFβ1-induced PDAC progression. SB431542, TGFβ1 pathway inhibitor; IWP-2 and FH535, Wnt pathway inhibitor; Plerixafor, SDF-1/CXCR4 antagonist. H&E, hematoxylin and eosin; IHC, immunohistochemical; PBS, phosphate-bufferef saline.
Given these in vitro findings, we next evaluated the anti-tumor effects of plerixafor on PDAC progression and metastasis in vivo. Towards this goal, we employed our PKP+ and PKKP+ PDAC models to evaluate the in vivo efficacy of plerixafor treatment with regard to inhibition of tumor initiation and progression. In our study design, groups of mice were treated with plerixafor or phosphate-buffered saline through intraperitoneal injection starting at 6 weeks of age, a time point at which only PanINs are known to be present in these compound mutant mice. Mice were randomized and dose twice a week (lasting for 4 weeks effects with control solvent or 1 mg/kg plerixafor treatment). At the end of treatment, mice from each experimental group were killed and the tumors were collected for macro and pathohistological examination. We observed a significant extension of tumor-free survival in all mice treated with plerixafor and there was no sign of any PDAC lesions in plerixafor-treated animals (Figure 8f). Of note, body weight was not significantly different between vehicle and plerixafor-treated groups. Necropsy analysis also did not indicate significant side effects or toxicity associated with plerixafor treatment based on histopathological and biochemical evaluation of liver and kidney specimens (data not shown). Additionally, plerixafor treatment of wild-type control animals had no effect on pancreatic mass or histology (data not shown), indicating a specific effect of plerixafor on PDAC development. IHC analysis demonstrated that plerixafor treatment resulted in reduced levels of Ki67 and p-c-Jun expression in pancreata of PDAC models (Figure 8g and Supplementary Figure 7). These results indicate that plerixafor exerts a significant anti-tumor effect against PDAC development in vivo suggesting that sustained SDF-1 activity is required for the development of premalignant lesions and their progression to advanced PDAC in mice.
Discussion
PDAC is one of the most deadly of all types of human malignancies and the average life expectancy after diagnosis with malignant PDAC disease is 3–6 months.36 Dissemination of malignant cells from a primary tumor to the distal sites in the body is the major causes of death in PDAC patients. However, the molecular mechanisms governing the metastatic spread of tumor cells remain largely unresolved. For instance, increased levels of TGF-β1 have been detected during PDAC progression in vivo. During early stages of tumor development, TGFβ1 functions to inhibit cell cycle progression and suppress tumor growth. However, in later stages, human tumor cells generally develop resistance to TGFβ1-mediated growth inhibition. TGFβ1 ceases to function in tumor suppression and adopts the converse role of enhancing metastatic spread.37, 38 The overall role of TGFβ1 in stimulating distant metastasis appears highly complex and remains incompletely resolved due to limited focus on this topic. Although there has been significant progress in elucidating the association between genetic alterations in the SMAD and TGFβ receptor genes with PDAC, the nature of defects involving other TGFβ1-induced downstream effectors have been elusive.38, 39
KLF10 was originally discovered from human osteoblasts40, 41 and has been shown to play important roles in mediating TGFβ signaling.20 KLF10 is also known to play distinct roles in TGFβ1 signaling in CD4+ and CD8+ T-cell development and has been implicated in macrophage differentiation and activation as well.42 KLF10 has also been demonstrated to elicit apoptotic responses when overexpressed in pancreatic adenocarcinoma cell lines.43 Importantly, a recent report has stated that inactivation of KLF10 in advanced PDAC through epigenetic regulation is associated with the presence of distant metastases in clinical pancreatic cancer tissue specimens.32 In the current study, we demonstrated that in the mouse pancreas, KLF10 was strongly expressed in islet and periacinar fibroblast cells, but was not or was weakly expressed in acinar and ductal cells. In our mutant KrasG12D PDAC model, we observed that KLF10 protein was strongly expressed in all neoplastic ductal cells (PanINs lesions), even in the high-grade PanINs and in foci of microinvasive carcinoma. However, weak or complete loss of KLF10 expression was observed in metastatic PDAC. Similarly, Chang et al. observed that the protein level of KLF10 decreased locally in advanced PDAC particularly at the leading edges of PDAC and in metastasis.32 These data indicate that inactivation of KLF10 may be a critical event in acquisition of pro-metastatic characteristics in pancreatic cancer; however, the molecular basis of a tumor suppressor role of KLF10 in inhibiting advanced metastatic PDAC remains unclear.
In the present study, using a pancreas-specific knockout mouse model, we selectively deleted KLF10 expression in the pancreatic progenitor linage population. Our results indicated that KLF10 loss alone does not affect mouse pancreas development or result in initiation of neoplastic lesion formation in aged Pdx-1Cre KLF10L/L animals. To further examine the functional consequence of KLF10 loss in PanINs formation, we crossed the Pdx-1Cre KLF10L/L mice with mice harboring a conditionally expressed allele of mutant KrasG12D. Our results unequivocally demonstrated that inactivation of KLF10 cooperates with activated mutant KrasG12D to induce rapid onset of advanced malignant PDAC. Hence, our study here provides the first line of evidence linking a strong genetic interaction between KrasG12D and KLF10 in the development of PDAC. Since mice with combined conditional mutant KrasG12D expression and KLF10 loss exhibit 50% penetrance of malignant PDAC at 25weeks, we also examined the effect of KLF10 loss in the setting of p53 deletion. Our PKKP mice display 100% penetrance of metastatic PDAC implicating important roles of KLF10, in combination with other genetic mutations, in facilitating distant metastases.
Converging lines of evidence suggest that the canonical Wnt signaling pathway, through nuclear translocation of β−catenin, plays an important role in regulating EMT in different cancer types.44, 45 The EMT-like transition in cancer cells enhances tumor invasion and distant metastases, and is associated with chemoresistance.46 In the present study, we also identified KLF10 as a novel player in modulating EMT through the TGFβ1/ Wnt pathways in PDAC. Additionally, in vivo inactivation of KLF10 was shown to result in activation of the Wnt/β-catenin pathway in PDAC. In this context, our subsequent molecular studies confirmed that loss of KLF10 increased the expression of the mesenchymal markers, vimentin and SMA, and inhibited E-cadherin expression in PDAC. Surprisingly, we did not observe any significant changes in cell proliferation rates upon loss of KLF10 in PDAC, but tumor sphere assays revealed that KLF10 loss led to a robust increase in self-renewal capacity and stemness. Thus, we hypothesize that inactivation of KLF10 in later stages of PDAC further promotes malignant progression of PDAC possibly by enriching cancer stemness. To fully understand the effects of loss of KLF10 expression on the gene expression profiles in PDAC, cDNA microarray analyses were performed in primary murine PDAC cells. Our results further confirm an important role for KLF10 in modulating TGFβ1 and Wnt signaling pathways as well as EMT and cancer stemness. The top differentially expressed genes, which have been shown to influence cancer progression and the metastatic processes, with coordinal KLF10 loss were CXCL12, Irx2, Sorl1, BMP7, CCL9 and Sox17.
CXCL12, also named SDF-1, is an inflammatory chemokine and neutrophil migration molecule.47 Accumulating evidence has unveiled that SDF-1 selectively binds to the G-protein-coupled receptor CXCR4 and activates a number of cellular responses such as calcium mobilization, actin polymerization and chemotaxis in migrating cells.48 Several prior reports simultaneously confirmed that the presence of a positive relationship between SDF-1/CXCR4 axis and distant metastasis in several types of human malignancy.48, 49, 50 For instance, primary tumor IHC analysis from PDAC patients has revealed that high expression of SDF-1 is associated with microvessel density which may play crucial roles in the metastasis and progression of pancreatic cancer.51 In breast cancer, CXCR4-positive tumor cells exhibit enhanced metastatic properties in the setting of increased SDF-1 levels.52 Of relevance, Tan et al have shown that SDF-1 induces LHSCC cell invasion through paracrine-activated CXCR4, to stimulate ERK/c-Jun-dependent upregulation of MMP-13.53 Additionally, the use of AMD3100, a compound that targets the SDF-1/CXCR4 signaling system, has been examined in clinical trials of patients with lymphomas or multiple myelomas.54 Similarly, Nervi et al demonstrated that treatment of leukemic mice with chemotherapy plus AMD3100 eradicated drug-resistant leukemic blasts, markedly decreased tumor burden and improved overall survival compared with mice treated with chemotherapy alone.55 In line with its potential role in various cancers, SDF-1 was identified as the most upregulated gene in this study, suggesting that activation of this gene could be highly involved in the invasion and metastasis of KLF10-null PDAC cells.
An additional interesting finding in our study is that some of our compound PKP and PKKP mice spontaneously develop gastric carcinoma. Analysis of the stomach in moribund PKK mice showed that 10 out of 25 animals developed squamous cell carcinoma and adenocarcinoma mixed type gastric tumors that arose from the antrum and corpus regions of the stomach (unpublished data). The increase in the number of gastric carcinomas in the PKK mice suggests that suppression of KLF10, in the context of KrasG12D mutation in the Pdx-1-positive endodermal progenitor subpopulation increases the risk of gastric tumorigenesis. Since TGFβ1 signaling is a critical pathway for gastrointestinal development in mice, further research is necessary to determine the role of TGFβ1/SMAD4/KLF10 in mediating the development of gastric neoplasia in mice.
In conclusion, this study provides in vivo evidence that KLF10 functions as a tumor suppressor gene in PDAC. Our results using novel PDAC animal models provide compelling evidence that KLF10 acts to halt the progression of mouse PanINs and that inactivation of KLF10 in the context of KrasG12D mutation and/or in combination with a p53-null background strongly drive an invasive and metastatic phenotype of PDAC with distant metastases. Furthermore, our data identified that the upregulation of SDF-1/CXCR4 signaling following KLF10 deletion is responsible for promoting distant metastasis of PDAC through the activation of TGFβ1/Wnt/beta-catenin and c-Jun/activator protein-1 (AP-1) pathways (Figure 8h and Supplementary Figure 8). The c-Jun proto-oncogene encodes the founding member of the AP-1 family, and c-Jun overexpression is common in human tumors that contribute to increase tumor cell proliferation and invasiveness.56 Thus, the current studies demonstrate for the first time that inactivation of KLF10 enhances activation of c-Jun to enhance PDAC cell growth, invasion and cancer stem cell expansion. Notably, in the present study, we also demonstrate that blockade of the SDF-1/CXCR4 pathway with plerixafor, a specific CXCR4 antagonist, attenuates PDAC development and metastasis in vivo. These results demonstrate that inhibition of the SDF-1/CXCR4-mediated signaling pathway represents a novel therapeutic strategy for the treatment of PDAC.
Materials and methods
Genetically modified mice and mouse genotyping
Pdx-1Cre, LSL-KrasG12D and p53Loxp/Loxp mice were obtained from the Mouse Models of Human Cancers Consortium (MMHCC) under material transfer agreements and were made available by Drs. Andrew M Lowy, Tyler Jacks and Anton Berns respectively.9 KLF10Loxp/Loxp (KLF10L/L) mice were generously provided by Drs. John Hawse and Malayannan Subramaniam at the Mayo Clinic. All compound mutant mice were on a mixed genetic 129Sv x C57BL/6 background, and detailed genotyping protocols are available on request. All studies were approved by the Animal Care Committee of the University of Kaohsiung Medical University (animal permit number 100186). Mice were anesthetized by intraperitoneal administration of 2.5% Avertin (0.38 ng/kg body wt. 2,2,2-tribromoethyl alcohol), and followed by full necropsy, and tumor tissues were collected for further analysis. Pancreatic tissue samples were fixed in 10% buffered formalin overnight, washed with 1 × phosphate-buffered saline buffer, and transferred to 70% ethanol before paraffin embedding, sectioning, and hematoxylin and eosin staining.
The methods that describe glucose tolerance tests, primary murine PDAC cell culture, human PDAC cell culture, RNA extraction, cDNA microarray, bioinformatics, immunofluorescence, IHC, mouse cytokine array, quantitative RT–qPCR, SDF-1 enzyme-linked immunosorbent assay, shRNA knockdown, luciferase reporter, western blotting, cell proliferation, wound healing, soft agar colony formation, plerixafor treatment, flow cytometry and statistical analysis are included in the online supplementary methods.
Accession codes
Microarray data are available in the Gene Expression Omnibus with accession numbers GSE85521.
References
Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, Depinho RA . Genetics and biology of pancreatic ductal adenocarcinoma. Genes dev 2006; 20: 1218–1249.
Hidalgo M . Pancreatic cancer. N Engl j med 2010; 362: 1605–1617.
Chen LT, Wang-Gillam A, Von Hoff DD, Bayever E, Belanger B . Nanoliposomal irinotecan in metastatic pancreatic cancer—authors' reply. Lancet 2016; 387: 1997.
Jemal A, Siegel R, Xu J, Ward E . Cancer statistics, 2010. CA: Cancer j clin 2010; 60: 277–300.
Bardeesy N, DePinho RA . Pancreatic cancer biology and genetics. Nat Rev Cancer 2002; 2: 897–909.
Bardeesy N, Aguirre AJ, Chu GC, Cheng KH, Lopez LV, Hezel AF et al. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc Natl Acad Sci USA 2006; 103: 5947–5952.
Aguirre AJ, Bardeesy N, Sinha M, Lopez L, Tuveson DA, Horner J et al. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes dev 2003; 17: 3112–3126.
Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer cell 2003; 4: 437–450.
Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer cell 2005; 7: 469–483.
Massaous J, Hata A . TGF-beta signalling through the Smad pathway. Trends cell biol 1997; 7: 187–192.
Romero-Gallo J, Sozmen EG, Chytil A, Russell WE, Whitehead R, Parks WT et al. Inactivation of TGF-beta signaling in hepatocytes results in an increased proliferative response after partial hepatectomy. Oncogene 2005; 24: 3028–3041.
Siegel PM, Massague J . Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat Rev Cancer 2003; 3: 807–821.
Iacopetta BJ, Welch J, Soong R, House AK, Zhou XP, Hamelin R . Mutation of the transforming growth factor-beta type II receptor gene in right-sided colorectal cancer: relationship to clinicopathological features and genetic alterations. J pathol 1998; 184: 390–395.
Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science 1995; 268: 1336–1338.
Tian F, Byfield SD, Parks WT, Stuelten CH, Nemani D, Zhang YE et al. Smad-binding defective mutant of transforming growth factor beta type I receptor enhances tumorigenesis but suppresses metastasis of breast cancer cell lines. Cancer res 2004; 64: 4523–4530.
David CJ, Huang YH, Chen M, Su J, Zou Y, Bardeesy N et al. TGF-beta tumor suppression through a lethal EMT. Cell 2016; 164: 1015–1030.
Lamouille S, Xu J, Derynck R . Molecular mechanisms of epithelial–mesenchymal transition. Nat Rev Mol Cell Biol 2014; 15: 178–196.
Subramaniam M, Harris SA, Oursler MJ, Rasmussen K, Riggs BL, Spelsberg TC . Identification of a novel TGF-beta-regulated gene encoding a putative zinc finger protein in human osteoblasts. Nucleic acids res 1995; 23: 4907–4912.
Subramaniam M, Hawse JR, Rajamannan NM, Ingle JN, Spelsberg TC . Functional role of KLF10 in multiple disease processes. BioFactors 2010; 36: 8–18.
Hawse JR, Iwaniec UT, Bensamoun SF, Monroe DG, Peters KD, Ilharreborde B et al. TIEG-null mice display an osteopenic gender-specific phenotype. Bone 2008; 42: 1025–1031.
Tau KR, Hefferan TE, Waters KM, Robinson JA, Subramaniam M, Riggs BL et al. Estrogen regulation of a transforming growth factor-beta inducible early gene that inhibits deoxyribonucleic acid synthesis in human osteoblasts. Endocrinology 1998; 139: 1346–1353.
Jin W, Chen BB, Li JY, Zhu H, Huang M, Gu SM et al. TIEG1 inhibits breast cancer invasion and metastasis by inhibition of epidermal growth factor receptor (EGFR) transcription and the EGFR signaling pathway. Mol cell biol 2012; 32: 50–63.
Spittau G, Happel N, Behrendt M, Chao TI, Krieglstein K, Spittau B . Tieg1/Klf10 is upregulated by NGF and attenuates cell cycle progression in the pheochromocytoma cell line PC12. J Neurosci Res 2010; 88: 2017–2025.
Hawse JR, Subramaniam M, Monroe DG, Hemmingsen AH, Ingle JN, Khosla S et al. Estrogen receptor beta isoform-specific induction of transforming growth factor beta-inducible early gene-1 in human osteoblast cells: an essential role for the activation function 1 domain. Mol endocrinol 2008; 22: 1579–1595.
Spittau B, Krieglstein K . Klf10 and Klf11 as mediators of TGF-beta superfamily signaling. Cell Tissue Res 2012; 347: 65–72.
Papadakis KA, Krempski J, Reiter J, Svingen P, Xiong Y, Sarmento OF et al. Kruppel-like factor KLF10 regulates transforming growth factor receptor II expression and TGF-beta signaling in CD8+ T lymphocytes. Am J Physiol Cell Physiol 2015; 308: C362–C371.
Tachibana I, Imoto M, Adjei PN, Gores GJ, Subramaniam M, Spelsberg TC et al. Overexpression of the TGFbeta-regulated zinc finger encoding gene, TIEG, induces apoptosis in pancreatic epithelial cells. J clin invest 1997; 99: 2365–2374.
Nagasawa T, Hirota S, Tachibana K, Takakura N, Nishikawa S, Kitamura Y et al. Defects of B-cell lymphopoiesis and bone-marrow myelopoiesis in mice lacking the CXC chemokine PBSF/SDF-1. Nature 1996; 382: 635–638.
Ma Q, Jones D, Borghesani PR, Segal RA, Nagasawa T, Kishimoto T et al. Impaired B-lymphopoiesis, myelopoiesis, and derailed cerebellar neuron migration in CXCR4- and SDF-1-deficient mice. Proc Natl Acad Sci USA 1998; 95: 9448–9453.
Feng Y, Broder CC, Kennedy PE, Berger EA . HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 1996; 272: 872–877.
Duda DG, Kozin SV, Kirkpatrick ND, Xu L, Fukumura D, Jain RK . CXCL12 (SDF1alpha)-CXCR4/CXCR7 pathway inhibition: an emerging sensitizer for anticancer therapies? Clin cancer res 2011; 17: 2074–2080.
Chang VH, Chu PY, Peng SL, Mao TL, Shan YS, Hsu CF et al. Kruppel-like factor 10 expression as a prognostic indicator for pancreatic adenocarcinoma. Am J Pathol 2012; 181: 423–430.
Hruban RH, Goggins M, Parsons J, Kern SE . Progression model for pancreatic cancer. Clin cancer res 2000; 6: 2969–2972.
Thayer SP, di Magliano MP, Heiser PW, Nielsen CM, Roberts DJ, Lauwers GY et al. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature 2003; 425: 851–856.
Dubrovsky L, Pankov D, Brea EJ, Dao T, Scott A, Yan S et al. A TCR-mimic antibody to WT1 bypasses tyrosine kinase inhibitor resistance in human BCR-ABL+ leukemias. Blood 2014; 123: 3296–3304.
Conroy T, Desseigne F, Ychou M, Bouche O, Guimbaud R, Becouarn Y et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl j med 2011; 364: 1817–1825.
Derynck R, Akhurst RJ, Balmain A . TGF-beta signaling in tumor suppression and cancer progression. Nat Genet 2001; 29: 117–129.
Massague J . TGFbeta in cancer. Cell 2008; 134: 215–230.
Padua D, Massague J . Roles of TGFbeta in metastasis. Cell res 2009; 19: 89–102.
Johnsen SA, Subramaniam M, Monroe DG, Janknecht R, Spelsberg TC . Modulation of transforming growth factor beta (TGFbeta)/Smad transcriptional responses through targeted degradation of TGFbeta-inducible early gene-1 by human seven in absentia homologue. J biol chem 2002; 277: 30754–30759.
Johnsen SA, Subramaniam M, Janknecht R, Spelsberg TC . TGFbeta inducible early gene enhances TGFbeta/Smad-dependent transcriptional responses. Oncogene 2002; 21: 5783–5790.
Cao Z, Wara AK, Icli B, Sun X, Packard RR, Esen F et al. Kruppel-like factor KLF10 targets transforming growth factor-beta1 to regulate CD4(+)CD25(-) T cells and T regulatory cells. J biol chem 2009; 284: 24914–24924.
Jiang L, Chen Y, Chan CY, Wang X, Lin L, He ML et al. Down-regulation of stathmin is required for TGF-beta inducible early gene 1 induced growth inhibition of pancreatic cancer cells. Cancer lett 2009; 274: 101–108.
Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T . Opinion: migrating cancer stem cells - an integrated concept of malignant tumour progression. Nat Rev Cancer 2005; 5: 744–749.
Kalluri R, Neilson EG . Epithelial–mesenchymal transition and its implications for fibrosis. J clin invest 2003; 112: 1776–1784.
Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 2015; 527: 525–530.
Bleul CC, Fuhlbrigge RC, Casasnovas JM, Aiuti A, Springer TA . A highly efficacious lymphocyte chemoattractant, stromal cell-derived factor 1 (SDF-1). J exp med 1996; 184: 1101–1109.
Singh S, Singh UP, Grizzle WE, Lillard JW Jr. . CXCL12-CXCR4 interactions modulate prostate cancer cell migration, metalloproteinase expression and invasion. Lab invest 2004; 84: 1666–1676.
Im KS, Graef AJ, Breen M, Lindblad-Toh K, Modiano JF, Kim JH . Interactions between CXCR4 and CXCL12 promote cell migration and invasion of canine hemangiosarcoma. Vet Comp Oncol 2017; 15: 315–327.
Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell 2005; 121: 335–348.
Cui K, Zhao W, Wang C, Wang A, Zhang B, Zhou W et al. The CXCR4-CXCL12 pathway facilitates the progression of pancreatic cancer via induction of angiogenesis and lymphangiogenesis. J surg res 2011; 171: 143–150.
Mukherjee D, Zhao J . The Role of chemokine receptor CXCR4 in breast cancer metastasis. Am j cancer res 2013; 3: 46–57.
Gu H, Yang L, Sun Q, Zhou B, Tang N, Cong R et al. Gly82Ser polymorphism of the receptor for advanced glycation end products is associated with an increased risk of gastric cancer in a Chinese population. Clin cancer res 2008; 14: 3627–3632.
Devine SM, Flomenberg N, Vesole DH, Liesveld J, Weisdorf D, Badel K et al. Rapid mobilization of CD34+ cells following administration of the CXCR4 antagonist AMD3100 to patients with multiple myeloma and non-Hodgkin's lymphoma. J clin oncol 2004; 22: 1095–1102.
Nervi B, Ramirez P, Rettig MP, Uy GL, Holt MS, Ritchey JK et al. Chemosensitization of acute myeloid leukemia (AML) following mobilization by the CXCR4 antagonist AMD3100. Blood 2009; 113: 6206–6214.
Sioletic S, Czaplinski J, Hu L, Fletcher JA, Fletcher CD, Wagner AJ et al. c-Jun promotes cell migration and drives expression of the motility factor ENPP2 in soft tissue sarcomas. J Pathol 2014; 234: 190–202.
Acknowledgements
This work was supported by grants MOST 104-2320-B-110-003 and 105-2320 -B-110-004 (to KH Cheng), MOST 101-2628-B-400-003-MY2 (to LT Chen) and MOST 105-2321-B-400-010 (to LT Chen and KH Cheng) from the Ministry of Science and Technology, Taiwan ROC, and grants KMU-TP104G00, KMU-TP105G00 (to KH Cheng) from Kaohsiung Medical University, Kaohsiung, Taiwan, and grant R01 DE14036 (to JRH and MS) from the National Institutes of Health, USA.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Weng, CC., Hawse, J., Subramaniam, M. et al. KLF10 loss in the pancreas provokes activation of SDF-1 and induces distant metastases of pancreatic ductal adenocarcinoma in the KrasG12D p53flox/flox model. Oncogene 36, 5532–5543 (2017). https://doi.org/10.1038/onc.2017.155
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2017.155
- Springer Nature Limited
This article is cited by
-
Krüppel-like factor 10 modulates stem cell phenotypes of pancreatic adenocarcinoma by transcriptionally regulating notch receptors
Journal of Biomedical Science (2023)
-
Genome-wide aberrant methylation in primary metastatic UM and their matched metastases
Scientific Reports (2022)
-
Upregulating sirtuin 6 ameliorates glycolysis, EMT and distant metastasis of pancreatic adenocarcinoma with krüppel-like factor 10 deficiency
Experimental & Molecular Medicine (2021)
-
ERas regulates cell proliferation and epithelial–mesenchymal transition by affecting Erk/Akt signaling pathway in pancreatic cancer
Human Cell (2020)
-
Loss of the transcriptional repressor TGIF1 results in enhanced Kras-driven development of pancreatic cancer
Molecular Cancer (2019)