Abstract
We have recently identified and characterized a novel oncogene, maelstrom (MAEL) from 1q24, in the pathogenesis of hepatocellular carcinoma. In this study, MAEL was investigated for its oncogenic role in urothelial carcinoma of the bladder (UCB) tumorigenesis/aggressiveness and underlying molecular mechanisms. Here, we report that overexpression of MAEL in UCB is important in the acquisition of an aggressive and/or poor prognostic phenotype. In UCB cell lines, knockdown of MAEL by short hairpin RNA is sufficient to inhibit cell growth, invasiveness/metastasis and suppressed epithelial–mesenchymal transition (EMT), whereas ectopic overexpression of MAEL promoted cell growth, invasive and/or metastatic capacity and enhanced EMT both in vitro and in vivo. We further demonstrate that MAEL could induce UCB cell EMT by downregulating a critical downstream target, the metastasis suppressor 1 (MTSS1) gene, ultimately leading to an increased invasiveness of cancer cells. Notably, overexpression of MAEL in UCB cells substantially enhanced the enrichment of DNA methyltrans-ferase (DNMT)3B and histone deacetylase (HDAC)1/2 on the promoter of the MTSS1, and thereby epigenetically suppressing the MTSS1 transcription. Downregulation of MTSS1 by MAEL in UCB cells is partially dependent on DNMT3B. Furthermore, we identify that beside the gene amplification of MAEL, miR-186 is a key negative regulator of MAEL and downregulation of miR-186 is another important mechanism for MAEL overexpression in UCBs. These data suggest that overexpression of MAEL, caused by gene amplification and/or decreased miR-186, has a critical oncogenic role in UCB pathogenesis by downregulation of MTSS1, and MAEL could be used as a novel prognostic marker and/or effective therapeutic target for human UCB.
Similar content being viewed by others
Introduction
Urothelial carcinoma of the bladder (UCB) is one among the most common and lethal urothelial malignancies in Western countries.1 Patients with muscle-invasive UCB have a poor prognosis with a 5-year overall survival of 48–67% even after radical cystectomy for localized disease.2 Despite advancement of the surgical technique and the development of novel drugs, approximately 50% of UCB patients develop metastatic disease within 2 years.3 Prevention of UCB metastasis is important for a favorable outcome of the disease. Thus, it is necessary to elucidate the molecular mechanism(s) underlying tumorigenesis and/or metastasis of UCB, so as to find novel therapeutic targets and develop new modalities of treatment.
Amplification of 1q is one of the most frequent chromosomal aberrations in human UCB4, 5, 6 and several types of other cancers including hepatocellular,7 lung,8 colorectal,9 breast10 and esophageal11 carcinomas. These suggest that human chromosome 1q contains oncogenes related to tumorigenesis and/or progression of human cancers. We have recently identified a novel oncogene, maelstrom (MAEL) from 1q24, one of the most frequent chromosomal alterations in hepatocellular carcinoma (HCC).12 The MAEL gene was originally identified in Drosophila, and functions in the establishment of oocyte polarity.13 In human cells and tissues, increased expression of MAEL protein has been examined in several types of cancer cell lines, including non-small-cell lung cancer and breast cancer cells. But the expression of MAEL in normal human tissues was only detected in the testis, and in breast cancer cells, the expression of MAEL is controlled by DNA methylation of a CpG island in its promoter.14 In addition, it was reported that MAEL is involved in the PIWI-interacting RNAs (piRNA) pathway.15 Composed of a high-mobility group domain and MAEL domain, MAEL has been identified to be essential for piRNA-mediated transcriptional transposon silencing.16 Recently, there is growing evidence that piRNA pathway has an important role in cancer occurrence and metastasis.17, 18 These data suggested that upregulation of MAEL may provide a selective advantage in the tumorigenesis and/or progression of human cancers. In human HCC of our previous study, we demonstrated that MAEL has an important oncogenic role in the development and/or progression of HCC by activating the AKT/GSK3b/Snail pathway.12 To date, however, the molecular status of MAEL and its potential oncogenic role and molecular mechanisms in UCB has not been elucidated. In this study, to investigate if abnormalities of MAEL are involved in the pathogenesis of UCB, the protein expression dynamics and amplification status of MAEL were first examined in a series of carcinomarous and non-neoplastic human bladder tissues and cells. The clinicopathologic/prognostic significance of MAEL expression in our UCB cohort was assessed. Moreover, the tumorigenicity of MAEL and the underlying molecular mechanisms involving the oncogenic role of MAEL and its regulation factors in UCB were investigated.
Herein, we provide evidence that overexpression of MAEL in UCB, caused by MAEL gene amplification and/or decreased microRNA (miR)-186, is important in the acquisition of an aggressive and/or poor prognostic phenotype. Silence of MAEL in UCB cells substantially inhibits cell growth, migration and invasion, whereas enforced overexpression of MAEL stimulates UCB cell growth, invasive and/or metastatic capacity both in vitro and in vivo. More importantly, we demonstrate that MAEL induces UCB cell epithelial–mesenchymal transition (EMT) by downregulating the metastasis suppressor 1 (MTSS1) gene, ultimately leading to an enhanced invasiveness of cancer cells. In addition, downregulation of MTSS1 by MAEL in UCB cells is partially dependent on DNA methyltrans-ferase (DNMT)3B. Our results, taken together, provide an explanation for the aggressive nature of UCB with overexpression of MAEL and the underling molecular mechanism that links the putative oncogene MAEL to the tumor-suppressor gene MTSS1.
Results
MAEL expression in bladder tissues and cells examined by western blotting
Of the eight bladder cell lines analyzed by western blot, endogenous MAEL was overexpressed in four UCB cell lines (that is, J82, EJ, T24 and UMUC3), whereas the human normal uroepithelial cell line SV-HUC-1 and the other three UCB lines (that is, 5637, Biu-87 and RT4) had low levels of MAEL (Figure 1a). In bladder tissues, 12/20 (60%) of primary UCB tissues showed upregulated MAEL expression (Figure 1a and Supplementary Figure 1).
Expression and amplification of MAEL in UCB cell lines and bladder tissues and its prognostic significance in UCB patients. (a) The levels of MAEL protein examined by western blotting in seven human UCB cell lines (that is, Biu-87, UMUC3, EJ, 5637, T24, RT4 and J82) and one normal uroepithelial cell line, SV-HUC-1 (left) and in 10 UCB tissues and paired normal bladder tissues. N, normal tissue; T, carcinoma tissue (right). (b) Representative IHC images show normal expression of MAEL in adjacent normal bladder tissue and overexpression of MAEL in UCB tissues (case 1 and case 2); scale bar, 100 μm. Amplification of MAEL gene was examined by FISH in UCB tissue case 2; scale bar, 20 μm. (c) Kaplan–Meier analysis indicating the correlation of MAEL overexpression with poorer overall survival and disease-free survival rates of 184 patients with UCB (log-rank test).
Expression and amplification of MAEL in bladder tissues and its correlation with UCB patient’s clinicopathological characteristics and survival
To investigate the potential clinical relevance of MAEL expression in UCB patients, we conducted immunohistochemistry (IHC) staining for MAEL in 184 formalin-fixed and paraffin-embedded (FFPE) UCB and 30 paired non-neoplastic bladder specimens. The expression of MAEL in all of the non-neoplastic bladder tissues was absent or at low levels, whereas 89/184 (48.4%) of primary UCBs were examined overexpression of MAEL. IHC staining of MAEL in representative samples of UCB and normal bladder tissues are shown in Figure 1b. In addition, overexpression of MAEL was positively associated with increased tumor size, T stage, lymph node metastasis and recurrence (P<0.05, Table 1). Survival analyses evaluated that overexpression of MAEL was a significant and independent prognostic factor for poor survival of UCB patients (P<0.01, log-rank test, Figure 1c and Supplementary Table 1; P=0.027, multivariate Cox regression analysis, Supplementary Table 2). The amplification of MAEL was examined in 11.1% (9/81) of the informative UCBs, and a significant correlation between overexpression and amplification of MAEL was evaluated (P=0.016, Supplementary Table 3).
MAEL shows oncogenicity function in UCB cells
To examine the oncogenic function of MAEL, two short hairpin RNAs (shRNAs) were used to suppress MAEL expression in two UCB cell lines (that is, T24 and UMUC3) that have high levels of MAEL. The efficiency of MAEL downregulation at the protein levels was evaluated by western blotting (Figure 2a). Cell growth assay showed that cell growth rates in the control UCB cells were significantly higher than those MAEL-silenced cells (P<0.01, Figure 2b). In the colony formation assay, T24-shMAEL and UMUC3-shMAEL cells also formed fewer and smaller colonies than that of control cells, respectively (Figure 2c), indicating that MAEL is involved in UCB cell proliferation. Furthermore, to determine whether or not ectopic overexpression of MAEL could enhance oncogenicity function of UCB cells, we constructed a 5637-MAEL cell line, which stably overexpressed MAEL (Figure 2d). Cell growth assay and colony formation assay revealed that overexpressing MAEL in 5637 line strongly provoked cell growth and number of foci formed (Figures 2d and e).
MAEL has strong oncogenic function in UCB cells. (a) Western blotting reveals that MAEL was efficiently knocked down by the treatment of MAEL-shRNA-1 or MAEL-shRNA-2 in T24 and UMUC3 UCB cells. (b) Rate of cell growth between shMAEL and shControl UCB cells by CCK-8 kit. *P<0.05, **P<0.01 by Student's t-test. (c) Representative images of decreased foci formation in monolayer culture induced by MAEL silenced in UCB cells. Data are the means±s.d. of three independent experiments. **P<0.01 by Student's t-test. (d) The levels of MAEL were substantially increased in 5637-MAEL cells compared with that in control 5637-vector cells by western blotting (left). Overexpression of MAEL promoted 5637 cells growth rate (right). **P<0.01 by Student's t-test. (e) Representative images of increased foci formation in monolayer culture induced by MAEL overexpression in 5637 cells. Data are the mean±s.d. of three independent experiments. **P<0.01 by Student's t-test. (f) Images of the xenograft tumors formed in nude mice injected with UMUC3-shControl, UMUC3-MAEL-shRNA-1 cells (upper) and 5637-vector, 5637-MAEL cells (down). Weights of xenograft tumors are the mean±s.d. **P<0.01 by Student's t-test.
To validate the in vivo effect of MAEL on tumor growth, a subcutaneous xenograft tumor mouse model was established. As shown in Figure 2f, the size and weight of tumors derived from UMUC3-shMAEL cells were significantly smaller and lighter than tumors from control cells (P<0.01). On the other side, tumors developed from 5637-MAEL cells were significantly larger and heavier than that of control 5637-vector cells (P<0.01).
The expression levels of MAEL influence the aggressive capacity of UCB cell lines in vitro
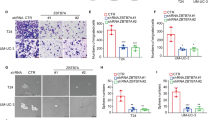
As overexpression of MAEL examined by IHC was positively associated with UCBs metastasis and/or ascending clinical stage, we furthered our study to determine the impact of MAEL on UCB cell migration and invasion. Wound-healing and Transwell invasion assays demonstrated that knockdown of MAEL markedly reduced the migratory speed and invasive ability of both T24 and UMUC3 cells (Figure 3a). On the other side, enforced overexpression of MAEL in 5637 cells substantially increased cell migration and invasive capacity (Figure 3b). These results clearly reveal that the elevated levels of MAEL are important for the aggressive nature of UCB cells.
The expression levels of MAEL influence the aggressive capacity and EMT of UCB cell lines. (a) Wound-healing and Transwell invasion assays show that MAEL-silenced T24 and UMUC3 cells had lower motility (upper) and invasive capacity (down) as compared with that in control T24 and UMUC3 cells. Data are the mean±s.d. of three independent experiments. *P<0.05, **P<0.01 by Student's t-test; scale bar, 100 μm. (b) Wound-healing and Transwell invasion assays demonstrate that 5637-MAEL cells had higher motility (upper) and invasive capacity (down) than that in 5637-vector cells. Columns: mean of triplicate experiments. **P<0.01 by Student's t-test; scale bar, 100 μm. (c) Representative lungs showing metastatic nodules (indicated by arrows) originated from 5637-vector and 5637-MAEL cells injected SCID-Beige mice, and hematoxylin and eosin (H&E) staining of lung metastatic tumors are shown, in which both the number and the volume of micrometastatic UCB lesions were markedly increased in the lungs of mice injected with 5637-MAEL cells (left). Number of metastatic nodules formed in the lung of mice 6 weeks after tail vein injection of 5637-vector and 5637-MAEL cells (eight mice per group; P<0.001; independent Student’s t-test) (right); scale bar, 100 μm. (d) Western blotting reveals that knockdown of MAEL by shRNAs resulted in an increased expression of E-cadherin and β-catenin and a decreased expression of fibronectin and vimentin in both T24 and UMUC3 cells, compared with that in control shC treated cells. Decreased levels of E-cadherin and β-catenin and increased levels of fibronectin and vimentin were examined in 5637-MAEL cells compared with that in control 5637-vector cells. (e) IF staining shows a downregulated expression of E-cadherin and an upregulated expression of vimentin in 5637-MAEL cells compared with that in control 5637-vector cells with a corresponding change in the number of positive staining cells.
Overexpression of MAEL enhances metastatic potential of UCB cell line in vivo
To investigate if MAEL overexpression is causative in an in vivo experimental metastasis model, 5637-MAEL cells were injected into tail vain of SCID mice, whereas 5637-vector cells were used as control (eight mice per group). Six weeks after cell injection, mice were killed and metastatic tumor nodules formed in the lung and in the liver were examined. We did not detect tumor nodule formed in the liver of all mice examined. However, metastatic tumor nodules were examined in the lung of mice, and overexpression of MAEL significantly increased 5637 cells lung metastasis (P<0.001, Figure 3c).
MAEL induces EMT in UCB cells
Our recent study demonstrated that MAEL could promote HCC cell metastasis by inducing EMT,12 we thus assessed if MAEL also induces EMT in UCB cells. Western blot analysis showed that after knockdown of MAEL in T24 and UMUC3 cells, the expression of the epithelial markers E-cadherin and β-catenin increased, whereas the expression of the mesenchymal markers fibronectin and vimentin decreased (Figure 3d). On the other hand, after enforced overexpression of MAEL in 5637 cells, the levels of E-cadherin and β-catenin were downregulated, whereas the levels of fibronectin and vimentin were upregulated (Figure 3d). Immunofluorescent (IF) staining further confirmed the downregulated expression of E-cadherin and upregulated expression of vimentin in the 5637-MAEL cells with a corresponding change in the number of positively staining cells (Figure 3e). These findings indicate that the expression levels of MAEL influence UCB cells EMT.
The expression levels of MAEL do not affect Akt/GSK-3β/Snail signaling in UCB cells
We previously provided evidence that overexpression of MAEL in HCC cells could promote cancer cell aggressiveness by activating the AKT/GSK-3β/Snail signaling pathway.12 In the present study of our UCB cells, however, we did not observe apparent differences in the levels of phosphorylated Akt, phosphorylated GSK-3β (active form) and Snail between 5637-MAEL cells and control 5637-vector cells (Supplementary Figure 2). Moreover, the expression levels of phosphorylated Akt, phosphorylated GSK-3β and Snail were not affected after knockdown of MAEL in T24 cells (Supplementary Figure 2). These results imply that MAEL do not affect Akt/GSK-3β/Snail signaling in UCB cells.
MAEL regulates the expression of MTSS1 in UCB cells
In an effort to determine the potential downstream targets regulated by MAEL in promoting UCB cells invasion and/or metastasis, mRNA expression profiles of T24-shMAEL cells were compared with that of control T24-shControl cells using a Human Tumor Metastasis RT2 Profiler PCR Array containing 84 cell metastasis-related genes. As shown in Figure 4a, three upregulated genes (that is, EWSR1, MTSS1 and MDM2) and four downregulated genes (that is, MMP2, Flt4, CCL7 and FGFR4), classified as such by a more than twofold change in mRNA levels, were identified in T24-shMAEL cells compared with that in T24-shControl cells (Supplementary Table 4). The two targets MTSS1 and Flt4 were validated by a western blot assay (Figure 4b).
MTSS1 is responsible for MAEL-induced UCB cell invasiveness and EMT. (a) The seven genes, EWSR1, MTSS1, MDM2, MMP2, Flt4, CCL7 and FGFR4, were examined >2-fold mRNA differential expression in T24-MAEL-shRNA-1 cells compared with that in T24-shControl cells by using a Human Tumor Metastasis RT2 Profiler PCR Array. (b) Silence of MAEL by shRNA-1 in T24 cells substantially upregulated MTSS1 expression and downregulated Flt4 expression as examined by western blotting. (c) Overexpression of MAEL and low-level expression of MTSS1 was examined by IHC in a UCB case; scale bar, 100 μm. (d) Western blot shows that MTSS1 was efficiently knocked down by the treatment of siMTSS1 in T24-MAEL-shRNA-1 cells. (e) Wound-healing and Transwell assays show that knockdown of MTSS1 substantially increased the migration (upper) and invasion (down) abilities of T24-MAEL-shRNA-1 cells. Data are the means±s.d. of three independent experiments; scale bar, 100 μm. *P<0.05, **P<0.01 by Student's t-test. (f) Western blotting shows that after silence of MTSS1 by specific siRNA in T24-MAEL-shRNA-1 cells, the levels of E-cadherin and β-catenin decreased, whereas the levels of fibronectin and vimentin increased (left). IF staining demonstrates a downregulated expression of E-cadherin and an upregulated expression of vimentin in T24-MAEL-shRNA-1 cells, after knockdown of MTSS1 (right).
In addition, a significant negative correlation between the expression of MAEL and MTSS1 was evaluated in our cohort of UCB tissues (Figure 4c, P=0.022, Supplementary Table 5). There were no significant difference of Flt4 expression between the MAEL overexpression and normal expression groups (P=0.141, Supplementary Table 5). Furthermore, similar to that in T24 cells, knockdown of MAEL in other three UCB cell lines (UMUC3, J82 and EJ) also substantially increased the levels of MTSS1 (Supplementary Figure 3).
Knock down of MTSS1 re-enhances MAEL-silenced UCB cells invasiveness and EMT
To address whether or not MTSS1 is involved in MAEL-induced UCB cell invasiveness and EMT, a rescue experiment was performed. We used RNA interference to knockdown MTSS1 expression in MAEL-silenced T24 cells in which MTSS1 level was upregulated (Figure 4d). Wound-healing and Transwell assays showed that after siMTSS1 treatment, the suppressed migrative and invasive capacities of MAEL-silenced T24 cells were all rescued (Figure 4e), and meanwhile, the inhibited EMT of MAEL-silenced T24 cells was also markedly re-enhanced as evidenced by decreased expression of epithelial markers (E-cadherin and β-catenin) and increased expression of mesenchymal markers (fibronectin and vimentin). IF staining of E-cadherin and vimentin also confirmed this result (Figure 4f). These data provide evidence that decreased expression of MTSS1 is responsible for the MAEL-induced UCB cell invasiveness and EMT.
Downregulation of MTSS1 by MAEL in UCB cells is partially dependent on DNMT3B
As that DNMT3B has previously been reported to directly regulate the levels of MTSS1 by binding to the promoter of MTSS1,19 and DNMT3B can cooperate with histone deacetylase (HDAC)1/2 to silence certain tumor-suppressor genes in colorectal cancer,20 these prompted us to investigate if MAEL downregulated MTSS1 expression is mediated by DNMT3B in UCB cells. As anticipated, the chromatin immunoprecipitation results showed that the enrichment of DNMT3B, as well as HDAC1 and HDAC2, on the promoter of MTSS1 was substantially enhanced in 5637-MAEL cell line, concurrent with decreased levels of histone H3 acetylation (AC-H3), as compared with that in control 5637-vector cells (Figure 5a). But we did not observe the altered protein levels of DNMT3B, HDAC1 and HDAC2 after ectopic overexpression of MAEL (Figure 5b). On the other side, when MAEL was knocked down in T24 cells, the enrichment of DNMT3B, HDAC1 and HDAC2 on the promoter of MTSS1 was largely reduced, concurrent with increased levels of histone H3 acetylation. But the levels of DNMT3B, HDAC1 and HDAC2 proteins were not altered after MAEL depletion (Supplementary Figures 4a and b). In addition, when endogenous DNMT3B was silenced by siRNA in 5637-MAEL cells, the enrichment of DNMT3B, HDAC1 and HDAC2 on the MTSS1 promoter was substantially reduced and meanwhile, the levels of AC-H3 on the MTSS1 promoter were markedly increased (Figure 5c). Furthermore, the dual luciferase reporter assay showed that knocking down DNMT3B by specific siRNA partially blocked the inhibitory effect of MAEL on the MTSS1 transcription (downregulated ration 3.66 vs 1.79, Figure 5d). These results indicate that MAEL transcriptionally downregulates MTSS1 expression via DNMT3B in UCB cells. Further functional studies showed that MAEL-mediated downregulation of MTSS1, enhanced migrative/invasive capacity and EMT of 5637-MAEL cells were all prevented, when DNMT3B was knocked down (Figures 5e–h).
Downregulation of MTSS1 by MAEL in UCB cells is partially dependent on DNMT3B, and MAEL-mediated cell aggressiveness is inhibited after silence of DNMT3B. (a) The chromatin immunoprecipitation (ChIP) assay was performed to analyze the enrichment of DNMT3B, HDAC1, HDAC2 and the levels of acetylated histone H3 on the promoter of MTSS1 between 5637-vector and 5637-MAEL cells. (b) The expression levels of MAEL, DNMT3B, HDAC1 and HDAC2 proteins in 5637-vector and 5637-MAEL cells examined by western blotting. (c) 5637-MAEL cells were transfected with siDNMT3B or negative control. Forty-eight hours later, the ChIP experiment was carried out to assess the enrichment levels of DNMT3B, HDAC1, HDAC2 and the levels of acetylated histone H3 on the MTSS1 promoter. The knocking down efficiency of DNMT3B with specific siRNA was assessed by western blotting (insert). (d) 5637 cells were firstly transfected with control siRNA or siDNMT3B. After 36 h, the cells were co-transfected with the pGL3-MTSS1 promoter luciferase, pRL-TK Renilla luciferase construct and control plasmid pEZ-Lv201 vector or pEZ-Lv201-MAEL plasmid. Thirty-six hours later, luciferase activity of the MTSS1 promoter was measured and normalized relative to luciferase activity (RLA). The bar shows the average±s.d. of three independent experiments. (e) Western blotting shows that after silence of DNMT3B in 5637-MAEL cells, the levels of MTSS1 increased. (f) Wound-healing assay shows that the enhanced migrative ability in 5637-MAEL cells was inhibited by silence of DNMT3B. (g) Transwell assay demonstrates that the increased invasive capacity of 5637-MAEL cells was suppressed by silence of DNMT3B; scale bar, 100 μm. Data are the means±s.d. of three independent experiments. *P<0.05 by Student's t-test. (h) Western blotting reveals that after silence of DNMT3B in 5637-MAEL cells, the levels of E-cadherin and β-catenin increased, whereas the levels of fibronectin and vimentin decreased (left). IF staining shows an upregulated expression of E-cadherin and a downregulated expression of vimentin in 5637-MAEL cells, after silence of DNMT3B in 5637-MAEL cells.
To test if HDAC affects MTSS1 expression in UCB cells, we further treated 5637-MAEL cells with three different gradient concentrations of trichostatin A (an effect HDAC inhibitor). We observed that inhibition of HDAC by trichostatin A caused an increased expression levels of MTSS1 in a dose-dependent manner (Supplementary Figure 4c). This result reveals the downregulation of MTSS1 mediated by MAEL in UCB cells could be blocked by the silence of HDAC.
MiR-186 is a negative regulator of MAEL and downregulated miR-186 induces MAEL overexpression in UCB cells
It is known that overexpression of an oncogene in human cancers is often caused by gene amplification.21, 22 In our 81 informative UCB cases examined simultaneously by both IHC analysis and fluorescence in situ hybridization (FISH), overexpression of MAEL was detected in 88.9% (8/9) of UCBs that had MAEL amplification. However, amplification of MAEL was not observed in 29 other UCBs with overexpression of MAEL (Supplementary Table 3). These results indicate that overexpression of MAEL was associated with MAEL gene amplification, but it did not always coincide, suggesting that other mechanisms, including transcriptional or post-translational regulation (for example, microRNA (miRNA)), may have critical roles in MAEL regulation. Thus, to investigate the potential miRNA regulators of MAEL, we first performed bioinformatics analyses and overlapped the predicted miRNA regulators with the downregulated miRNAs from miRNA expression profiles of UCBs.23 The result showed that miR-186 was singled out as a potential regulator of MAEL (Supplementary Figure 5).
Next, our quantitative reverse transcriptase–PCR analysis showed that the levels of miR-186 were frequently reduced in fresh UCB tissues and cell lines compared with that in adjacent normal bladder tissues and normal uroepithelial cell line (SV-HUC-1), respectively (P<0.001, Figure 6a). These results was consistent with a previous report, in which miR-186 in was markedly downregulated in UCB tissues and cells, and miR-186 could suppress UCB cells proliferation and invasion.24 Moreover, a significant inverse correlation between the levels of miR-186 and MAEL mRNA was evaluated in these UCB tissues and cells (P<0.001, Figure 6b). Furthermore, in a cohort of our 184 FFPE UCB tissues, we revealed that the IHC score of MAEL protein expression in UCBs with low expression of miR-186 was significantly larger than that in UCBs with high expression of miR-186 (P<0.01, Student's t-testFigure 6c).
The expression levels of miR-186 in bladder tissues and cell lines, and miR-186 is a negative regulator of MAEL in UCB. (a) Quantitative reverse transcriptase–PCR analysis of miR-186 expression in 20 fresh UCB samples compared with adjacent normal bladder mucosal tissues (left). The levels of miR-186 in one human normal uroepithelial cell line SV-HUC-1 and 7 UCB cell lines (right). (b) Spearman's correlation analysis demonstrates that the levels of miR-186 expression was inversely correlated with the mRNA levels of MAEL in 20 fresh UCB tissues(left) and 8 bladder cell lines (right). (c) In 184 FFPE UCB tissues, the IHC score of MAEL protein expression in UCBs with low-expression of miR-186 was significantly larger than that in UCBs with high expression of miR-186. **P<0.01 by Student's t-test. (d) Predicted miR-186 target sequence in 3′UTR of MAEL (MAEL-3′UTR) and mutant containing three altered nucleotides in the seed sequence of miR-186 (miR-186-mut). (e) Luciferase assay of pGL3-MAEL-3′UTR reporters co-transfected with increasing amounts (10, 20 and 50 nM) of miR-186 mimic and mutant oligonucleotides in T24 and UMUC3 cell lines, or with increasing amounts (20, 50 and 100 nM) of miR-186 inhibitor oligonucleotides in 5637 cell line. (f) Quantitative reverse transcriptase–PCR analysis shows that overexpression of miR-186 significantly suppressed MAEL mRNA expression levels in both T24 and UMUC3 cells, whereas inhibition of miR-186 led to a significantly increase in MAEL mRNA expression level of 5637 cell. *P<0.05 by Student's t-test. (g) Western blotting analysis demonstrates that miR-186 tranfection markedly decreased MAEL protein levels of T24 and UMUC3 cells, whereas anti-miR-186 increased MAEL protein expression in 5637 cells. (h) Schematic diagram depicting a proposed model for a major mechanism of MAEL and its upregulation in the promotion of UCB cell aggressiveness.
In UCB cells, we further observed that overexpression of miR-186 decreased, and inhibition of miR-186 increased, the luciferase activity linked with the 3′untranslated region (3′UTR) of MAEL (Figures 6d and e). Meanwhile, the mRNA and protein levels of MAEL were all substantially reduced after overexpression of miR-186 in UCB cells, whereas inhibition of miR-186 markedly increased the expression of MAEL (Figures 6f and g). These data, collectively, provided evidence that miR-186 directly suppress MAEL expression and decreased miR-186 may induce MAEL overexpression in UCBs.
Discussion
Recently, we have identified and characterized a novel candidate oncogene, MAEL from chromosome 1q24, in the pathogenesis of human HCC and it was suggested as a potential therapeutic target of the disease.12 Amplification of 1q, in which the MAEL gene located in, is also one of the most frequent chromosomal aberrations in human UCB.4, 5, 6 To date, however, the molecular status of MAEL and its potential oncogenic role and underlying mechanisms in UCB remain unclear. In this study, by using western blot and IHC, we found that MAEL was frequently overexpressed in UCB cells and tissues. Statistical analyses evaluated that high expression of MAEL in UCBs was positively associated with an advanced clinical stage, tumors metastasis and/or recurrence and was a strong and an independent predictor of poor patient survival. Thus, the examination of MAEL expression by IHC could be used as an additional tool in identifying those UCBs at increased risk of tumor invasion and/or metastasis, and have a predictive potential in UCB patient clinical outcome. These findings underscore a potentially important role of MAEL in the development and/or progression of UCB.
Subsequently, a series of in vitro and in vivo assays were used in our study to investigate the biological function of MAEL in regulating UCBs malignant phenotype. Our results show that enforced knockdown or ectopic overexpression of MAEL in UCB cells substantially repressed or promoted the capacities of cell growth, migration, invasion and EMT, respectively. In a tail vein injection mouse model of cancer metastasis, we further observed that ectopic overexpression of MAEL in UCB cells led to a significant increase in the number of lung metastatic lesions. Following the emerging view that MAEL is an important oncogenic factor in UCB, the next step was to characterize the underling biological mechanisms of MAEL to promote UCB cell aggressiveness. In a recent study of ours using HCC cells, we found that MAEL could enhance EMT through the activation of the AKT-GSK-3β-Snail signaling pathway, and thus inducing cancer cell invasiveness.12 In the current study of UCB cells, however, we did not examine the altered levels of active AKT, active GSK-3β and Snail before and after MAEL overexpression or knockdown. It does appear, therefore, that in our UCB cells, MAEL promotes aggressiveness through regulation of targets and/or pathways other than the activation of AKT-GSK-3β-Snail signaling, suggesting that the mechanism(s) by which MAEL mediates cancer progression may be tumor-type specific.
Thus, to better understand the downstream molecular events involving MAEL and UCB invasiveness and/or metastasis, a Human Tumor Metastasis RT2 Profiler PCR Array containing 84 cell metastasis-related genes was used to compare mRNA expression profiles between T24-shControl cells and T24-shMAEL cells. We found and validated that MTSS1 was the downstream target of MAEL in UCB cells. In our large cohort of UCB tissues, we further evaluated a significant negative correlation between expression of MAEL and MTSS1. These results suggest that in UCB cells, MAEL may regulate cell aggressiveness via the regulation of MTSS1.
To date, the gene functions of MTSS1 in human cancers are complicated and it appears to be dependent to different tumor types. The MTSS1 acts as a tumor suppressor in certain human cancers, such as bladder, lung, prostate and gastric cancers.25, 26, 27, 28, 29 In melanoma and breast cancer, however, it was reported to exert oncogenic functions.30, 31 Similar to previous reports of MTSS1 in UCBs,25, 26 we observed that low-level expression of MTSS1 was closely associated with UCBs aggressiveness and/or poor survival (Supplementary Table 6 and Supplementary Figure 6), suggesting that MTSS1 exerts tumor-suppressive functions in human UCB. Next, to determine if MTSS1 is a downstream target involving in MAEL-induced UCB cell aggressiveness, a rescue experiment where UCB T24 cells that stably repressed MAEL with upregulated expression of MTSS1 and dually transfected with siMTSS1, successfully enhanced the cells migrative/invasive ability and EMT. These data suggest that MTSS1 is a critical downstream target of MAEL and decreased expression of MTSS1 is one of the main reasons responsible for the MAEL-induced invasiveness and EMT in UCB cells.
Up to date, however, the molecular mechanisms by which MAEL regulates MTSS1 expression have not been elucidated. Recently, Fan et al.19 have found DNMT3B can bind directly to the promoter of the MTSS1 gene to inhibit its transcription in tumor cells through a DNA methylation-independent mechanism. More recently, it was identified that DNMT3B can cooperate with HDAC1/2 to silence certain tumor-suppressor genes in colorectal cancer.20 In our current study, we further found that after ectopic overexpression of MAEL in 5637 UCB cells, the enrichment of DNMT3B on the promoter of MTSS1 was substantially enhanced, concurrent with decreased levels of acetylated H3. Although we did not observe the altered levels of DNMT3B, as well as HDAC1 and HDAC2 after MAEL overexpression, we did find that the recruitments of DNMT3B, HDAC1 and HDAC2 to the promoter of MTSS1 were all increased. Furthermore, when DNMT3B was knocked down by siRNA in 5637-MAEL cells, the enrichment of DNMT3B, HDAC1 and HDAC2 on the MTSS1 promoter was clearly reduced and the levels of acetylated H3 were largely increased, and meanwhile, the MAEL-mediated downregulation of MTSS1, enhanced cell migration/invasion and EMT in 5637-MAEL cells were all substantially prevented. Several studies have demonstrated that DNMTs including DNMT3B can transcriptionally represses gene expression by recruiting transcriptional co-repressors and chromatin remodeling enzymes including HDACs.32, 33, 34 In our study, we further identified that silence of HDAC could block the MAEL-mediated MTSS1 downregulation through DNMT3B. Our data, in combination with others results,19, 20, 32, 33, 34 provided evidence that MAEL enhances the binding of DNMT3B to the promoter of MTSS1; such could recruit the two histone deacetyltransferases HDAC1 and HDAC2 to deacetylate H3, thereby resulting in downregulation of MTSS1 to promote the aggressive phenotypes of UCB cells.
In our study, one critical question was then raised: how is MAEL upregulated in UCBs? It is well established that gene amplification is a common pathological mechanism of oncogene overexpression in human cancers.21, 22 In our informative UCB cases examined by FISH, most UCBs with amplification of MAEL overexpressed MAEL protein. However, the majority of UCBs that had overexpression of MAEL was not examined MAEL amplification, suggesting that other mechanisms than MAEL amplification may have critical roles in MAEL regulation. We know that a class of miRNAs has emerged as important regulators of gene expression at the transcriptional or post-translational level.35, 36, 37 In this study, we further revealed that miR-186 is a key negative regulator of MAEL and downregulated miR-186 is another important mechanism for MAEL overexpression in UCBs. This finding is based on several lines of evidence: (1) there is a conserved miR-186 binding site in the 3′UTR of MAEL; (2) MAEL 3′UTR-mediated luciferase activity is specifically responsive to overexpressed or suppressed miR-186; (3) the overexpression or suppression of miR-186 reduced or enhanced the expression of MAEL, respectively, at both mRNA and protein levels; and (4) a significant inverse correlation between the levels of miR-186 and MAEL expression was evident in our UCB cells and in a large cohort of UCB tissues. Although it was reported that the methylation status of MAEL promoter may regulate of MAEL expression in certain breast cancer cell lines,14 as a result of our collective present data, herein we propose a model for the major molecular mechanism of MAEL and its upregulation in promoting UCB cell aggressiveness and it is illustrated in Figure 6h.
To sum up, our report describes, for the first time, the protein expression and amplification dynamics of MAEL in human UCBs. Overexpression of MAEL, caused by gene amplification and/or decreased miR-186, may be important in the tumorigenesis and acquisition of an aggressive/poor prognostic phenotype of UCB. In addition, functional and mechanistic studies of MAEL as provided in this study, suggest a critical oncogenic role of MAEL in the support of UCB cell EMT and invasion/metastasis by downregulating MTSS1 through DNMT3B, an activity that may be responsible, at least partially, for the development and/or progression of human UCBs.
Materials and methods
Patients and specimen characteristics
In this study, FFPE tissues of 184 UCB patients, who underwent radical cystectomy between February 2000 and October 2009, were obtained from the archives of the Department of Pathology of Sun Yat-sen University Cancer Center (Guangzhou, China). Written informed consent was obtained from all patients before the study. The median follow-up time was 69 months (range 6–176 months) and the clinicopathological characteristics are summarized in Table 1. In addition, 20 pairs of fresh UCB and adjacent normal bladder mucosa specimens were collected in 2013. Tumor stage was defined according to the criteria of the sixth edition of the TNM classification of the International Union Against Cancer (UICC, 2002). For mRNA and protein extraction, UCB tissue samples were processed by dissection according to each hematoxylin and eosin slide and comprised at least 70% of tumor cells. Studies using human tissues were reviewed and approved by Ethical Committee of Sun Yat-sen University Cancer Center (Guangzhou, China).
Cell lines and cell cultures
Eight bladder cell lines (that is, Biu-87, UMUC3, EJ, 5637, T24, RT4, J82 and SV-HUC-1) were selected and cultured in this study. Human UCB cell line T24, 5637, UMUC3, J82 and RT4, and human normal uroepithelial cell line SV-HUC-1 was purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). UCB cell line Biu-87 and EJ were a gift from the First Affiliated Hospital of Sun Yat-sen University. Cell lines EJ, T24, Biu-87, RT4, SV-HUC-1 and 5637 were cultured in RPMI-1640 (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (GE Healthcare - HyClone Laboratories Inc., South Logan, UT, USA). UMUC3 and J82 were cultured in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (HyClone). All cells were grown in a humidified incubator at 37 °C with 5% CO2.
Fluorescence in situ hybridization
Two-color FISH was applied to the sections of FFPE UCB tissues using spectrum red-labeled BAC clone (RP4-782G3) containing the MAEL gene and a chromosome 1 centromere probe was labeled by spectrum green (Vysis, Downers Grove, IL, USA) and used as internal control. The FISH reaction was performed as described previously.38, 39 The criteria for MAEL gene amplification was defined as the presence of >3 times as many gene signals than centromere signals of chromosome 1 (Figure 1b).
Construction of the recombinant lentiviral vector
The construction of the MAEL shRNA lentiviral expression vector were made in psi-LVRH1MP (GeneCopoeia Company, Rockville, MD, USA) vector using standard protocols. shRNA control vector (HSH021057-LVRH1MP) were purchased from the GeneCopoeia Company. The T24 and UMUC3 cells had high expression of MAEL and they were infected with retroviruses carrying psi-LVRH1MP-MAEL-shRNA. The target sequences of MAEL for constructing lentiviral shRNA were as follows: shRNA1: 5′-GGAACTGGCCACCTATCTACT-3′; shRNA2: 5′-GAGTCAACTGGTGTTTGAAGC-3′. Puromycin was used to select stable cells.
Plasmid constructs and transfection
The construction of a plasmid expressing human MAEL (pcDNA-MAEL) was conducted as described in our previous study.12 In brief, full-length human MAEL complementary DNA was amplified by PCR and cloned into pcDNA3.1 (+) expression vector (Invitrogen), and then transfected into 5637 cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Cells transfected with empty vector were used as controls. Stable MAEL-expressing clones were selected by Geneticin (Roche Diagnostics, Indianapolis, IN, USA).
Experimental in vivo metastasis model
Eight 4-week-old male severe combined immunodeficient (SCID-Beige) mice in each experimental group were injected with 5637-Vec and 5637-MAEL cells, respectively. Briefly, 2 × 105 cells were injected intravenously through tail vein into each mouse in a laminar flow cabinet. Six weeks after cell injection, mice were killed and examined.
MiR-186 mimic, antagomir-186, plasmids and transient transfection
The 3′UTRs of MAEL were amplified and cloned downstream to the luciferase gene in a modified pGL3 control vector. MiR-186 mimic and antagomir-186 were purchased from RIBOBIO Company (GuangZhou, China). MiR-186 mimic, antagomir-186 and their corresponding control oligonucleotides were transfected into UCB cells cultured in six-well plates using Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions.
Statistical analysis
Data were analyzed using SPSS17.0 software (SPSS, Chicago, IL, USA). Differences between variables were assessed by the Chi-square test or Fisher's exact test. Survival curves were plotted by Kaplan–Meier analysis and compared by the log-rank test. Cox regression analysis was carried out to assess the significance of variables for survival. Bivariate correlations between study variables were calculated by Spearman’s rank correlation coefficients. Data derived from cell line experiments are presented as mean ±s.d. (X±s.d.) and assessed by the two-tailed Student’s t-test. P-values of <0.05 were considered statistically significant.
References
Burger M, Catto JW, Dalbagni G, Grossman HB, Herr H, Karakiewicz P et al. Epidemiology and risk factors of urothelial bladder cancer. Eur Urol 2013; 63: 234–241.
Malkowicz SB, van Poppel H, Mickisch G, Pansadoro V, Thuroff J, Soloway MS et al. Muscle-invasive urothelial carcinoma of the bladder. Urology 2007; 69: 3–16.
Sternberg CN, Bellmunt J, Sonpavde G, Siefker-Radtke AO, Stadler WM, Bajorin DF et al. ICUD-EAU International Consultation on Bladder Cancer 2012: chemotherapy for urothelial carcinoma-neoadjuvant and adjuvant settings. Eur Urol 2013; 63: 58–66.
Hopman AH, Moesker O, Smeets AW, Pauwels RP, Vooijs GP, Ramaekers FC . Numerical chromosome 1, 7, 9, and 11 aberrations in bladder cancer detected by in situ hybridization. Cancer Res 1991; 51: 644–651.
Poddighe PJ, Ramaekers FC, Smeets AW, Vooijs GP, Hopman AH . Structural chromosome 1 aberrations in transitional cell carcinoma of the bladder: interphase cytogenetics combining a centromeric, telomeric, and library DNA probe. Cancer Res 1992; 52: 4929–4934.
Lopez V, Gonzalez-Peramato P, Suela J, Serrano A, Algaba F, Cigudosa JC et al. Identification of prefoldin amplification (1q23.3-q24.1) in bladder cancer using comparative genomic hybridization (CGH) arrays of urinary DNA. J Transl Med 2013; 11: 182.
Ma NF, Hu L, Fung JM, Xie D, Zheng BJ, Chen L et al. Isolation and characterization of a novel oncogene, amplified in liver cancer 1, within a commonly amplified region at 1q21 in hepatocellular carcinoma. Hepatology (Baltimore, MD) 2008; 47: 503–510.
Ma J, Gao M, Lu Y, Feng X, Zhang J, Lin D et al. Gain of 1q25-32, 12q23-24.3, and 17q12-22 facilitates tumorigenesis and progression of human squamous cell lung cancer. J Pathol 2006; 210: 205–213.
Zhou CZ, Qiu GQ, Fan JW, Wang XL, Tang HM, Huang L et al. Refined mapping of loss of heterozygosity on 1q31.1-32.1 in sporadic colorectal carcinoma. World J Gastroenterol 2008; 14: 1582–1587.
Mesquita B, Lopes P, Rodrigues A, Pereira D, Afonso M, Leal C et al. Frequent copy number gains at 1q21 and 1q32 are associated with overexpression of the ETS transcription factors ETV3 and ELF3 in breast cancer irrespective of molecular subtypes. Breast Cancer Res Treat 2013; 138: 37–45.
Maru DM, Luthra R, Correa AM, White-Cross J, Anandasabapathy S, Krishnan S et al. Frequent loss of heterozygosity of chromosome 1q in esophageal adenocarcinoma: loss of chromosome 1q21.3 is associated with shorter overall survival. Cancer 2009; 115: 1576–1585.
Liu L, Dai Y, Chen J, Zeng T, Li Y, Chen L et al. Maelstrom promotes hepatocellular carcinoma metastasis by inducing epithelial-mesenchymal transition by way of Akt/GSK-3beta/Snail signaling. Hepatology 2014; 59: 531–543.
Clegg NJ, Frost DM, Larkin MK, Subrahmanyan L, Bryant Z, Ruohola-Baker H . Maelstrom is required for an early step in the establishment of Drosophila oocyte polarity: posterior localization of grk mRNA. Development (Cambridge, England) 1997; 124: 4661–4671.
Xiao L, Wang Y, Zhou Y, Sun Y, Sun W, Wang L et al. Identification of a novel human cancer/testis gene MAEL that is regulated by DNA methylation. Mol Biol Rep 2010; 37: 2355–2360.
Aravin AA, van der Heijden GW, Castaneda J, Vagin VV, Hannon GJ, Bortvin A . Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet 2009; 5: e1000764.
Matsumoto N, Sato K, Nishimasu H, Namba Y, Miyakubi K, Dohmae N et al. Crystal structure and activity of the endoribonuclease domain of the piRNA pathway factor maelstrom. Cell Rep 2015; 11: 366–375.
Cheng J, Deng H, Xiao B, Zhou H, Zhou F, Shen Z et al. piR-823, a novel non-coding small RNA, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells. Cancer Lett 2012; 315: 12–17.
Lim SL, Ricciardelli C, Oehler MK, Tan IM, Russell D, Grutzner F . Overexpression of piRNA pathway genes in epithelial ovarian cancer. PloS One 2014; 9: e99687.
Fan H, Chen L, Zhang F, Quan Y, Su X, Qiu X et al. MTSS1, a novel target of DNA methyltransferase 3B, functions as a tumor suppressor in hepatocellular carcinoma. Oncogene 2012; 31: 2298–2308.
Cai Y, Geutjes EJ, de Lint K, Roepman P, Bruurs L, Yu LR et al. The NuRD complex cooperates with DNMTs to maintain silencing of key colorectal tumor suppressor genes. Oncogene 2014; 33: 2157–2168.
Stark GR, Debatisse M, Giulotto E, Wahl GM . Recent progress in understanding mechanisms of mammalian DNA amplification. Cell 1989; 57: 901–908.
He LR, Liu MZ, Li BK, Jia WH, Zhang Y, Liao YJ et al. High expression of EZH2 is associated with tumor aggressiveness and poor prognosis in patients with esophageal squamous cell carcinoma treated with definitive chemoradiotherapy. Int J Cancer 2010; 127: 138–147.
Yoshino H, Chiyomaru T, Enokida H, Kawakami K, Tatarano S, Nishiyama K et al. The tumour-suppressive function of miR-1 and miR-133a targeting TAGLN2 in bladder cancer. Br J Cancer 2011; 104: 808–818.
Yao K, He L, Gan Y, Zeng Q, Dai Y, Tan J . MiR-186 suppresses the growth and metastasis of bladder cancer by targeting NSBP1. Diagn Pathol 2015; 10: 146.
Du P, Ye L, Ruge F, Yang Y, Jiang WG . Metastasis suppressor-1, MTSS1, acts as a putative tumour suppressor in human bladder cancer. Anticancer Res 2011; 31: 3205–3212.
Nixdorf S, Grimm MO, Loberg R, Marreiros A, Russell PJ, Pienta KJ et al. Expression and regulation of MIM (missing in metastasis), a novel putative metastasis suppressor gene, and MIM-B, in bladder cancer cell lines. Cancer Lett 2004; 215: 209–220.
Kayser G, Csanadi A, Kakanou S, Prasse A, Kassem A, Stickeler E et al. Downregulation of MTSS1 expression is an independent prognosticator in squamous cell carcinoma of the lung. Br J Cancer 2015; 112: 866–873.
Loberg RD, Neeley CK, Adam-Day LL, Fridman Y St, John LN, Nixdorf S et al. Differential expression analysis of MIM (MTSS1) splice variants and a functional role of MIM in prostate cancer cell biology. Int J Oncol 2005; 26: 1699–1705.
Liu K, Wang G, Ding H, Chen Y, Yu G, Wang J . Downregulation of metastasis suppressor 1(MTSS1) is associated with nodal metastasis and poor outcome in Chinese patients with gastric cancer. BMC Cancer 2010; 10: 428.
Mertz KD, Pathria G, Wagner C, Saarikangas J, Sboner A, Romanov J et al. MTSS1 is a metastasis driver in a subset of human melanomas. Nat Commun 2014; 5: 3465.
Giacobbe A, Compagnone M, Bongiorno-Borbone L, Antonov A, Markert EK, Zhou JH et al. p63 controls cell migration and invasion by transcriptional regulation of MTSS1. Oncogene 2015; 35: 1602–1608.
Bachman KE, Rountree MR, Baylin SB . Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J Biol Chem 2001; 276: 32282–32287.
Rountree MR, Bachman KE, Baylin SB . DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet 2000; 25: 269–277.
Fuks F, Burgers WA, Godin N, Kasai M, Kouzarides T . Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. EMBO J 2001; 20: 2536–2544.
Bartel DP . MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116: 281–297.
Kottakis F, Polytarchou C, Foltopoulou P, Sanidas I, Kampranis SC, Tsichlis PN . FGF-2 regulates cell proliferation, migration, and angiogenesis through an NDY1/KDM2B-miR-101-EZH2 pathway. Mol Cell 2011; 43: 285–298.
Zheng F, Liao YJ, Cai MY, Liu TH, Chen SP, Wu PH et al. Systemic delivery of microRNA-101 potently inhibits hepatocellular carcinoma in vivo by repressing multiple targets. PLoS Genet 2015; 11: e1004873.
Xie D, Sham JS, Zeng WF, Lin HL, Bi J, Che LH et al. Correlation of AIB1 overexpression with advanced clinical stage of human colorectal carcinoma. Hum Pathol 2005; 36: 777–783.
Tong ZT, Cai MY, Wang XG, Kong LL, Mai SJ, Liu YH et al. EZH2 supports nasopharyngeal carcinoma cell aggressiveness by forming a co-repressor complex with HDAC1/HDAC2 and Snail to inhibit E-cadherin. Oncogene 2012; 31: 583–594.
Acknowledgements
This work was supported by grants from the Nature Science Foundation of China (no. 81225018 and no.81472385), the Fundamental Research Funds for the Central Universities (Sun Yat-Sen University Young Teachers Plan) (no. 14ykpy39), Natural Science Foundation of Guangdong Province (no. 2015A030313009), China and Science and Technology Foundation of the Guangdong Province, China (grant no. 2013B021800173).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Li, XD., Zhang, JX., Jiang, LJ. et al. Overexpression of maelstrom promotes bladder urothelial carcinoma cell aggressiveness by epigenetically downregulating MTSS1 through DNMT3B. Oncogene 35, 6281–6292 (2016). https://doi.org/10.1038/onc.2016.165
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2016.165
- Springer Nature Limited
This article is cited by
-
Maelstrom promotes tumor metastasis through regulation of FGFR4 and epithelial-mesenchymal transition in epithelial ovarian cancer
Journal of Ovarian Research (2022)
-
MAEL as a diagnostic marker for the early detection of esophageal squamous cell carcinoma
Diagnostic Pathology (2021)
-
MAEL Cancer-Testis Antigen as a Diagnostic Marker in Primary Stages of Gastric Cancer with Helicobacter pylori Infection
Journal of Gastrointestinal Cancer (2020)
-
Computational analysis of the evolutionarily conserved Missing In Metastasis/Metastasis Suppressor 1 gene predicts novel interactions, regulatory regions and transcriptional control
Scientific Reports (2019)
-
N6-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis
Nature Communications (2019)