Abstract
Tamoxifen, an estrogen receptor (ER) antagonist, is the mainstay treatment of breast cancer and the development of resistance represents a major obstacle for a cure. Although long non-coding RNAs such as HOTAIR have been implicated in breast tumorigenesis, their roles in chemotherapy resistance remain largely unknown. In this study, we report that HOTAIR (HOX antisense intergenic RNA) is upregulated in tamoxifen-resistant breast cancer tissues compared to their primary counterparts. Mechanistically, HOTAIR is a direct target of ER-mediated transcriptional repression and is thus restored upon the blockade of ER signaling, either by hormone deprivation or by tamoxifen treatment. Interestingly, this elevated HOTAIR increases ER protein level and thus enhances ER occupancy on the chromatin and potentiates its downstream gene regulation. HOTAIR overexpression is sufficient to activate the ER transcriptional program even under hormone-deprived conditions. Functionally, we found that HOTAIR overexpression increases breast cancer cell proliferation, whereas its depletion significantly impairs cell survival and abolishes tamoxifen-resistant cell growth. In conclusion, the long non-coding RNA HOTAIR is directly repressed by ER and its upregulation promotes ligand-independent ER activities and contributes to tamoxifen resistance.
Similar content being viewed by others
Introduction
Long non-coding RNAs (lncRNAs) are a major class of newly identified non-coding transcripts that are usually composed of more than 200 nucleotides. Accumulating evidence suggests that lncRNAs play critical roles in regulating a wide range of cellular processes by affecting various aspects of protein, DNA, and RNA expression and interactions.1, 2, 3, 4, 5 Large-scale RNA sequencing (RNA-seq) studies have revealed that lncRNAs are abundantly transcribed from the genome; a recent study comprehensively examined over 7000 RNA-seq libraries and uncovered nearly 60,000 lncRNAs from the human transcriptome.6 Out of numerous cancer-associated lncRNAs, HOTAIR (HOX antisense intergenic RNA) was among the most upregulated in breast cancer. Localized in chromosome 12, HOTAIR is 2.2 kb in length and transcribed from the antisense strand of the HOXC locus. It has been shown to interact with polycomb repressive complex 2 to reprogram the chromatin state and induce cancer metastasis.7, 8 In vivo experiments showed that HOTAIR is sufficient and required to promote invasion of breast carcinoma cells.7 Concordantly, HOTAIR and EZH2 expression levels were highly correlated in breast cancer tissues and high HOTAIR level is associated with worse prognosis.9, 10 In addition, these studies reported that strong HOTAIR expression correlated with estrogen receptor (ER) and PR positivity, and that HOTAIR expression is a strong predictor of poor clinical outcome especially in ER-positive breast cancer.9, 10
These results provide first lines of evidence that the lncRNA HOTAIR may play important roles in regulating breast cancer progression. Tamoxifen, an antagonist of the ER, is the most commonly used treatment for ER-positive breast cancer. Despite great success in improving the overall survival of breast cancer patients, development of tamoxifen resistance (TamR) is persistently seen in the clinic and is a major cause of breast cancer recurrence and mortality.11 Understanding the biological mechanisms underlying this acquired resistance to tamoxifen is thus of substantial clinical significance.12
ER is a hormonal transcription factor that is liganded and activated by estrogen. ER regulates target genes that control endocrine response and cell cycle progression.5, 13, 14 Tamoxifen competes with estrogen for binding to the ER protein, thereby inhibiting the conventional ER transcriptional program.5, 14, 15 Using chromatin immunoprecipitation sequencing (ChIP-seq), a recent study has mapped genome-wide ER binding profiles in primary breast cancers and found that ER is still recruited to the chromatin in TamR breast cancer, but to new regulatory regions associated with poor clinical outcome.16 This aberrant ER transcriptional activity is proposed to be regulated by various oncogenic mechanisms and have critical functions in mediating tamoxifen resistance and tumor progression. Here we report that HOTAIR is overexpressed in TamR breast cancer. It directly interacts with the ER protein to enhance ER transcriptional activity and thus ligand-independent breast cancer growth. Our study will not only inform about the mechanistic underpinnings of breast cancer progression but also provide evidence supporting therapeutic potentials of lncRNA targeting in breast cancer treatment.
Results
HOTAIR is upregulated in tamoxifen-resistant, ER-positive breast cancer
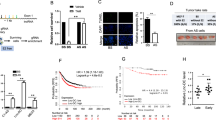
To determine lncRNAs that may contribute to breast cancer tamoxifen resistance, we re-analyzed publicly available data set profiling gene expression in wild-type MCF7 cells as well as its TamR derivatives treated with ethanol or 17β-estradiol (E2) for 4 h (GSE5840).17 Our analysis revealed 37 lncRNA genes that were repressed by estrogen and became upregulated in TamR cells (Figure 1a). Among the top deregulated lncRNAs are HOTAIR and TP53TG1. Although HOTAIR has been shown to be upregulated in metastatic breast cancer,7, 10 its role in TamR has not been investigated. To examine this, we performed in situ hybridization to probe the abundance of HOTAIR lncRNA in breast cancer tissues, comparing between matched primary and TamR breast carcinoma samples. Our results showed that HOTAIR localized primarily in the nuclei but was also present in the cytoplasm (Figure 1b). Most primary breast cancer tissues had weak HOTAIR staining, whereas TamR breast cancer generally exhibited moderate to strong HOTAIR staining. Overall, HOTAIR expression level was significantly higher in TamR breast cancer than primary, hormone-naive tumors (Figure 1c). Being consistent with this, quantitative reverse transcriptase−PCR analysis showed that tamoxifen treatment for 7 days significantly increased HOTAIR lncRNA levels in both MCF7 and T47D cells, while dramatically decreasing the expression of GREB1, a known ER-induced gene (Figures 1d and e). As tamoxifen is known to compete with estrogen to inhibit estrogen-induced ER activities, next we examined whether HOTAIR is a target of ER-mediated transcriptional regulation.
HOTAIR is upregulated in tamoxifen-resistant (TamR) breast cancer. (a) Heat map showing lncRNAs that are repressed by estradiol but upregulated in TamR MCF7 cells. Microarray data were downloaded from GEO with GSE5840 and re-analyzed for lncRNA expression. HOTAIR is shown in red. (b) HOTAIR in situ hybridization (ISH) staining in two representative pairs of primary and TamR breast cancers. (c) Boxplot showing HOTAIR ISH staining intensity in a set (n=13) of matched primary and TamR breast cancers. (d, e) HOTAIR expression is increased by tamoxifen treatment. Quantitative reverse transcriptase−PCR analysis of HOTAIR and GREB1 was done in MCF7 (d) and T47D (e) cells treated with increasing doses of tamoxifen for 7 days. Gene expression was normalized to GAPDH. Data shown are mean±s.e.m. and are representative of at least two independent experiments.
The lncRNA HOTAIR is directly repressed by estrogen receptor
To examine whether estrogen regulates HOTAIR expression, we carried out quantitative reverse transcriptase−PCR analysis of MCF7 cells treated with increasing doses of E2. HOTAIR expression was greatly inhibited for up to sevenfold, while GREB1 was increased as expected (Figure 2a and Supplementary Figure S1A). Estrogen inhibited HOTAIR expression in a dose- and time-dependent manner (Figures 2a and b). HOTAIR level was decreased about twofold after 4 h of E2 treatment and nearly 10-fold after 24 h of E2 treatment, whereas GREB1 was gradually induced by around 20-fold at 4 h and reached a plateau of >30-fold after 8 h (Supplementary Figure S1B). A similar trend of inhibition of HOTAIR expression by estrogen was observed in a different ER+ breast cancer cell line T47D, despite T47D being much less responsive to estrogen as indicated by much less GREB1 induction (Figures 2c and d). Furthermore, HOTAIR level is considerably restored in breast cancer cells following hormone deprivation, wherein GREB1 expression was lost (Figure 2e). Next, to determine whether estrogen inhibits HOTAIR expression through direct ER binding to HOTAIR regulatory elements, we re-analyzed a previously published study involving an ER ChIP-seq data set that was performed in MCF7 cells (GSE23893).18 We observed a very strong ER binding site at a genomic region about 14.5 kb upstream to the transcription start site of the HOTAIR gene (Figure 2f). In addition, this region is strongly occupied by H3K4me1 and H3K27ac (GSE40129), supporting its being an active enhancer (Supplementary Figure S2A). ER ChIP followed by quantitative reverse transcriptase−PCR analysis confirmed that estrogen stimulation significantly increased ER binding to this region as well as to positive control gene GREB1, but not to the negative control gene KIAA0066 (Figure 2g). Further, chromosome conformation capture (3C) experiment demonstrated estrogen-induced DNA looping between the transcription start site of the HOTAIR gene (anchor primer) and the ER-bound enhancer (P4; Supplementary Figure S2B). Taken together, our data showed that HOTAIR is directly repressed by estrogen and is therefore upregulated following hormone deprivation and in TamR breast cancer.
The lncRNA HOTAIR is directly repressed by estrogen through the estrogen receptor (ER). (a) Estrogen inhibits HOTAIR expression in a dose-dependent manner. MCF7 cells were hormone starved for 3 days and treated with increasing amounts of estradiol (E2) for 6 h. RNAs were then collected and subjected to quantitative reverse transcriptase−PCR (qRT−PCR) analysis of gene expression. (b) Estrogen inhibits HOTAIR expression in a time-dependent manner. MCF7 cells were treated with 1 nm E2 and collected at different time points for gene expression analysis by qRT−PCR. (c, d) Estrogen inhibits HOTAIR expression in T47D cells. T47D cells were hormone deprived for 3 days followed by E2 stimulation for up to 24 h. Cells were then collected at different time-points for qRT−PCR analysis of HOTAIR and GREB1 expression and normalized to GAPDH. (e) Estrogen depletion restores HOTAIR level. qRT−PCR analysis of HOTAIR and GREB1 in MCF7 cells subjected for hormone deprivation for up to 7 days. (f) Genome Browser view of ER binding events at the enhancer of the HOTAIR gene. ER chromatin immunoprecipitation sequencing (ChIP-seq) was performed in MCF7 cells stimulated with ethanol (Ethl) or E218 and re-analyzed using HOMER (GSE23893). (g) ChIP−qPCR showing ER binding to the HOTAIR distal enhancer. ER ChIP was performed in hormone-starved MCF7 cells stimulated with ethanol (vehicle) or E2 for 45 min. Enrichment of ER at specific genomic regions including GREB1 and HOTAIR enhancers was evaluated by qPCR. The KIAA0066 gene was utilized as a negative control as previously described.34 qPCR, quantitative PCR.
HOTAIR directly interacts with ER and enhances ER transcriptional activities
Next, we asked what is the role of elevated HOTAIR in breast cancer ER signaling and tamoxifen resistance. HOTAIR has been previously shown to directly interact with chromatin-modifying proteins such as EZH2 and LSD.15, 19, 20 As HOTAIR is upregulated in TamR breast cancer cells which often have altered ER program,16 we asked whether HOTAIR might regulate ER function. This may shed light on the mechanisms underlying recently reported correlation between HOTAIR expression and ER positivity in primary specimens.9, 10 To test this, we first examined whether HOTAIR lncRNA could physically interact with the ER protein using RNA pull-down assay. Briefly, we carried out in vitro transcription to synthesize biotinylated RNA probes from sense and antisense HOTAIR DNA templates, which were then incubated with MCF7 nuclear extracts to allow protein−RNA interactions and precipitated, along with its interacting proteins, with streptavidin beads. Western blot analysis demonstrated that the sense HOTAIR RNA probe, but not the antisense transcript, pulled down the ER protein (Figure 3a). On the other hand, we carried out RNA immunoprecipitation assay and found that the ER antibody significantly enriched for HOTAIR, as opposed to IgG control, whereas the negative control RNA U1 did not exhibit differential enrichment (Figure 3b).
HOTAIR interacts with the estrogen receptor (ER) protein and enhances ER genomic action. (a) HOTAIR lncRNA interacts with the ER protein. RNA pull-down assay was performed in MCF7 cells using biotin-labeled HOTAIR RNA probe transcribed in vitro. The antisense HOTAIR probe was used as negative control. (b) ER protein binds to HOTAIR lncRNA. MCF7 cells were subjected to RNA immunoprecipitation assay using an anti-ER antibody or IgG control. Immunoprecipitation-enriched RNA was then analyzed by quantitative reverse transcriptase−PCR. U1 RNA was utilized as a negative control. (c) HOTAIR overexpression increases ER protein level. MCF7 cell lysates were separated into the cytoplasm, nuclear, nucleoplasm, and chromatin-bound fractions and were detected by western blot analysis. GAPDH and H3 were utilized as loading controls for cytoplasmic and nuclear/chromatin fractions, respectively. Quantification was done by measuring band intensity with ImageJ (NIH, Bethesda, MD, USA) and normalizing to loading control. (d, e) Ectopic overexpression of HOTAIR increases nuclear ER level. ER immunostaining was performed in control and HOTAIR-overexpressing MCF7 cells grown in the presence (d) and absence (e) of estrogen. (f) Overlap of ER-binding sites detected by chromatin immunoprecipitation sequencing (ChIP-seq) in MCF7 cells with control or HOTAIR overexpression in the absence and presence of estrogen. (g) Heat map depicting ER ChIP-seq read intensity around (±5 kb) peak centers detected in control or HOTAIR-overexpressing MCF7 cells under hormone-starved condition. Average ER ChIP-seq read intensity around ER binding sites (±5 kb) is shown on the right.
Subsequently, we inquired into the consequences of HOTAIR−ER interaction, in order to speculate how ER activities may be affected as a result. By separating MCF7 cell lysates into cytoplasmic, nuclear, nucleoplasm and chromatin-bound fractions, we observed that ER, as expected, localized primarily within the nucleus as opposed to cytoplasm. HOTAIR overexpression substantially increased ER protein levels, suggesting potential roles of HOTAIR in enhancing ER transcriptional functions (Figure 3c and Supplementary Figure S3). Moreover, immunofluorescent staining confirmed noticeable increase of nuclear ER following HOTAIR overexpression (Figure 3d). Interestingly, this HOTAIR-mediated increase in nuclear ER level was also true under hormone-starved condition, in which there is only minimal estrogen present to activate ER translocation into the nucleus, suggesting the roles of HOTAIR in enhancing ligand-independent ER function (Figure 3e). To confirm the notion that HOTAIR may augment ER genomic targeting, we conducted ER ChIP-seq in MCF7 cells grown in the presence and absence of estrogen. As expected, the total number of ER binding sites was 4.6-fold higher in estrogen-stimulated vs hormone-deprived cells (Figure 3f). Importantly, upon overexpression of HOTAIR, global ER binding events were greatly increased under both conditions. Heat map and average intensity analysis of the various groups of ER peaks demonstrated a clear increase in ER ChIP-seq read intensity in both shared and HOTAIR-only groups, representing a majority of the ER binding events (Figure 3g and Supplementary Figure S4A). This HOTAIR-mediated increase of ER binding events was more prominent in the absence of estrogen, suggesting important functions of HOTAIR in regulating ligand-independent ER activities. Concordantly, quantitative PCR analysis of several previously reported ER target genes, such as GREB1, TFF1, PR, and CTSD, demonstrated that HOTAIR overexpression significantly increased ER occupancy at most of these genes (Supplementary Figure S4B). Similarly, the increase in ER binding at target genes was more prominent in hormone-deprived MCF7 cells (Supplementary Figure S4C). Next, we proceeded to investigate to what extent HOTAIR impacts ER-mediated transcriptional activities particularly in a hormone-deprived environment.
HOTAIR drives estrogen-independent ER transcriptional program
To identify HOTAIR- and estrogen-regulated genes, we conducted microarray profiling of hormone-deprived and estrogen-stimulated MCF7 cells with control or HOTAIR overexpression. Data analysis identified 132 and 112 genes that were induced and repressed by HOTAIR, respectively. Importantly, hierarchical clustering followed by heat map view revealed that a majority of HOTAIR-induced genes are also induced by E2 stimulation, whereas HOTAIR-repressed genes tend to become downregulated by estrogen (Figure 4a). Concordantly, gene set enrichment analysis demonstrated that E2-induced genes were significantly upregulated following HOTAIR overexpression, even in the absence of estrogen, whereas E2-repressed genes were strongly downregulated by HOTAIR (Figures 4b and c). Gene ontology analysis showed that HOTAIR-induced genes were significantly enriched for response to protein stimulus and regulation of cell death and apoptosis, being consistent with the functions of estrogen-mediated ER signaling (Figure 4d; Supplementary Tables S1 and S2). To confirm HOTAIR regulation of ER-mediated transcriptional program, we performed quantitative reverse transcriptase−PCR analysis of several known ER-target genes such as GREB1, TFF1, and c-MYC. Indeed, our data showed that HOTAIR overexpression induced ER-target gene expression in the absence of estrogen and further potentiated the effects of E2 (Figures 4e and g). Taken together, we provide evidence for a model by which the lncRNA HOTAIR increases ER protein level and enhances its chromatin binding and thus the ER transcriptional program, even in an estrogen-depleted environment. As HOTAIR is upregulated in TamR breast cancer, we next asked whether HOTAIR contributes to the development of tamoxifen resistance in breast cancer, wherein tamoxifen abolishes estrogen-mediated activation of ER, similar to hormone starvation.
HOTAIR overexpression enhances estrogen receptor (ER) transcriptional program. (a) HOTAIR-induced and repressed genes are, respectively, increased and decreased by estrogen. Expression microarray was utilized to profile gene expression in control and HOTAIR-overexpressing MCF7 cells that were hormone-deprived for 3 days followed by either ethanol or 1nM estradiol (E2) treatment for 6 h. Expression of genes induced or repressed by HOTAIR for at least twofold were clustered and visualized using heat map. (b, c) Estrogen-induced genes are significantly enriched for upregulation by HOTAIR (b), whereas estrogen-repressed genes are downregulated by HOTAIR (c). GSEA was carried out to determine the enrichment of E2-induced and -repressed gene sets in the expression data set comparing control and HOTAIR-overexpressing MCF7 cells. (d) HOTAIR-induced genes are enriched for cell growth and response to protein stimulus. Gene ontology (GO) analysis was performed using 132 genes that are increased by HOTAIR by at least twofold. Shown here are representative GO terms that are significantly enriched (P<0.05). (e−g) HOTAIR enhances ER-target gene expression. Quantitative reverse transcriptase−PCR analysis of representative ER-induced genes was performed in control and HOTAIR-overexpressing MCF7 cells stimulated with ethanol or estrogen. Data were normalized to GAPDH. Error bars: mean±s.e.m. *P<0.05 and **P<0.01.
LncRNA HOTAIR promotes tamoxifen-resistant breast cancer progression
To determine the role of HOTAIR in breast cancer, we first overexpressed HOTAIR in MCF7 cells (Figure 5a). Cell proliferation assay showed that HOTAIR overexpression increased MCF7 cell growth (Figure 5b). On the other hand, HOTAIR knockdown in T47D cells markedly reduced cell proliferation (Figures 5c and d). To provide direct evidence linking HOTAIR to tamoxifen resistance, we generated a TamR MCF7 cell line by continuously culturing the cells in the presence of 5 μm tamoxifen for several months. Consistent with previous HOTAIR staining results in TamR breast tumors, HOTAIR level showed a remarkable fourfold increase following long-term treatment of tamoxifen (Figure 5e). To determine whether this upregulated HOTAIR is critical for the TamR MCF7 cell growth, we performed HOTAIR knockdown using two independent short hairpin RNA constructs (Figure 5f). Subsequently, we performed cell proliferation assay to investigate to what extent HOTAIR contributes to tamoxifen resistance. As demonstrated in Figure 5g, knockdown of HOTAIR significantly decreased TamR MCF7 cell growth, suggesting that tamoxifen resistance may be reverted by targeting or depleting HOTAIR. Consistently, clonogenic assays showed that HOTAIR knockdown greatly inhibited the colony-formation abilities of the TamR cells, further supporting the role of HOTAIR in mediating TamR cell growth (Figure 5h).
HOTAIR promotes breast cancer cell growth and tamoxifen resistance. (a, b) HOTAIR overexpression increases breast cancer cell growth. HOTAIR was overexpressed in MCF7 cells through lentiviral transduction, with overexpression confirmed by quantitative reverse transcriptase−PCR (qRT−PCR) (a). Cell proliferation was determined by WST-1 cell growth assay (b). Data shown are mean±s.e.m. and are representative of at least two independent experiments, **P<0.01. (c, d) HOTAIR knockdown decreases breast cancer cell growth. HOTAIR was depleted in T47D cells through short hairpin RNA (shRNA) lentiviral transduction (c) and cell growth was evaluated by WST-1 assay (d). Data shown are mean±s.e.m. and are representative of at least two independent experiments, **P<0.01. (e) HOTAIR is upregulated in tamoxifen-resistant (TamR) breast cancer cells. MCF7 cells were continuously grown in medium containing 5 μm tamoxifen and periodically harvested for RNA isolation and qRT−PCR analysis. (f, g) HOTAIR knockdown decreases TamR breast cancer cell growth. HOTAIR was depleted in TamR MCF7 cells through lentiviral transduction of two shRNA constructs. Gene expression was determined by qRT−PCR (f) and cell growth by WST-1 assay (g). (h) HOTAIR knockdown inhibits colony formation abilities of breast cancer cells. Colony formation assays and quantifications were performed in TamR MCF7 cells stably expressing control or HOTAIR-targeting shRNAs.
Discussion
With the emergence of studies focusing on the functional attributes of nonprotein-coding RNA transcripts, such as lncRNAs, it has been revealed that these lncRNAs may contribute significantly to the biological processes involved in physiological as well as pathological conditions. Numerous lncRNAs have been identified as critical players during cancer development; some may be beneficial by acting as tumor or metastasis suppressors (for example, GAS5,21 MEG3,22 LIFR23), whereas others may be detrimental by promoting oncogenesis (for example, PCA3 or, as previously named, DD3,24 PCAT-1,25 SChLAP126). Previous studies have shown that lncRNAs exhibit great diversity in their functions and mechanisms of action, which include but are not limited to epigenetic transcriptional regulation, association with enhancer and chromosomal looping, and mRNA processing and translation.27 Several unique properties of lncRNAs make them highly useful in the clinic, with potential utilities including their use as diagnostic biomarkers due to their tissue specificity,25, 28 as well as in lncRNA-based therapies by means of RNA interference.29 Yet, lncRNAs have just begun to be identified and cataloged; a majority of them remain to be characterized.
Gupta et al.7 reported in 2010 that the lncRNA HOTAIR is notably increased in primary breast tumors as well as during metastases. Specifically, by interacting with EZH2 of the polycomb repressive complex 2 complex, which catalyzes trimethylation at histone H3 lysine 27 (H3K27me3) and is upregulated in a variety of aggressive cancers, HOTAIR was demonstrated to alter chromatin structure and regulate gene expression, thereby giving rise to an invasive cancer phenotype. In this study, we provide experimental evidence that HOTAIR is also critically involved in conferring tamoxifen resistance to MCF7 cells, which represents a major challenge in the clinic today. Tamoxifen, belonging to the class of selective ER modulators, is a competitive antagonist of ER that was developed in the 1970s and has been the mainstay treatment for ER-positive breast cancer, which accounts for at least 70% of all breast cancers.12 Despite its initial success in reducing disease mortality and improving survival, tamoxifen therapy frequently led to the onset of resistance, and recurrence was reported to occur within 15 years in one-third of patients treated with tamoxifen.12, 30 Thus, it has become imperative to understand the mechanisms for acquisition of tamoxifen resistance and to develop targeted therapies to improve treatment for breast cancer.
Our results showed that HOTAIR is highly upregulated in the tumors of TamR breast cancer patients compared to their primary tumors before treatment. Moreover, physical interaction between HOTAIR and the nuclear hormone receptor ER was detected, which in turn resulted in significant amount of nuclear ER even under estrogen-depleted conditions, thus allowing ER genomic targeting and consequently inducing the ER transcriptional program. Importantly, this phenomenon of HOTAIR-mediated activation of ER function in the absence of estrogen indicated a potential route to ligand independence that is manifested in TamR cells. Furthermore, by generating a TamR MCF7 cell model, we showed that HOTAIR was consistently upregulated over long periods of drug treatment. In addition, we demonstrated that HOTAIR significantly contributes to the growth of these TamR cells. Therefore, in our present study we provide evidence for a novel mechanism that is employed by the lncRNA HOTAIR to promote ER activation in the absence of estrogen and drive tamoxifen resistance. Because of this crucial role HOTAIR plays in the progression of breast cancer and development of drug resistance, it holds great promise as a useful biomarker and potential therapeutic target.
Materials and methods
Patient specimens and cell lines
All breast cancer tissue specimens (n=13) were collected via surgical resection or biopsy from patients diagnosed between January 2006 and February 2014 at the Cancer Center of Guangzhou Medical University. The study protocol was approved by the Ethics Committee of Cancer Center of Guangzhou Medical University. In general, with n=10, for a continuous outcome, there will be >89% power to reject the null hypothesis of no difference when the difference is 1.5 s.d. or more, using two-sided t-test and a type 1 error of 0.05. MCF7 and T47D cell lines were ordered from ATCC.
Plasmids, reagents, quantitative PCR and western blotting
HOTAIR sequence was amplified by PCR and subsequently cloned into the expression vector pCDH-MSCV-mcs-EF1-GFP-T2A-Pu (SBI) at EcoR1 and Not1 sites using Cold Fusion kit (SBI). The shHOTAIR was cloned into the pLKO lentivirus system. All PCR primers for cloning are listed in Supplementary Table S3 and high-fidelity enzyme Phusion was used for PCR amplification. All PCR products were verified by DNA sequencing. Specific antibodies used in this work include rabbit ER (06-935, Millipore, Billerica, MA, USA), mouse ER (sc-8002, Santa Cruz, Dallas, TX, USA), mouse GAPDH (ab9484, Abcam, Cambridge, MA, USA) and rabbit H3 (ab1791, Abcam). Other reagents include beta-estradiol (E8875, Sigma, St Louis, MO, USA) and 4-hydroxytamoxifen (H6278, Sigma-Aldrich, St Louis, MO, USA). All primers were designed using primer 3 and synthesized by Integrated DNA Technologies (Coralville, IA, USA; Supplementary Table 3). Quantitative PCR was performed using SYBR Green by StepOne Plus in three technical replicates and significance was determined by two-sided t-tests. Each experiment was repeated independently at least two times. Western blotting was carried out using standard protocol and repeated at least two times. Band intensity on western blot was quantified with ImageJ and normalized to each respective control to obtain the ratio of ER protein level.
3C assay
The digestion map of commonly used restriction enzymes around the enhancer/promoter region of HOTAIR locus (from –103 to +83 kb) and BglII was selected for digestion, as BglII sites show a distribution that will enable appropriate primers to be designed to generate 200−350 bp PCR products on re-ligation. All primers are designed based on the forward strand immediately upstream of a BglII restriction site (Supplementary Figure S2B and Supplementary Table S3). 3C experiments were conducted according to the standard 3C protocol as previously described.31 Briefly, fixed chromatin of hormone-starved or E2-treated MCF7 cells (1 × 107) was digested with BglII overnight and incubated with 50 units of T4 DNA ligase (10799009001, Sigma-Aldrich) overnight in a volume of 7 ml to keep the DNA concentration at 2−3 ng/ml to favor intramolecular ligation.
RNA pull-down assay
RNA pull-down was performed as previously described.20 Briefly, biotin-labeled RNAs were transcribed from DNA templates with biotin-UTP, NTP mix, and T7 RNA polymerase (Promega, Madison, WI, USA), treated with RNase-free DNase I (Promega) and purified with RNeasy Mini kit (QIAGEN). Nuclei were extracted from MCF7 cells and resuspended in 1 ml RIP buffer (150 mM KCl, 25 mM Tris pH 7.4, 0.5 mM dithiothreitol, 0.5% NP40, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor (Roche Complete Protease Inhibitor Cocktail Tablets)), and subsequently subjected to mechanical shearing using a dounce homogenizer. For precipitation assays, fragmented nuclear extract and the RNA probe were incubated at room temperature (RT) for 60 min, and 60 μl of Streptavidin agarose beads (Invitrogen, Grand Island, NY, USA) were added to each binding reaction and further incubated at RT for 1 h. After five times of washing with PBS, samples were boiled in SDS buffer and subjected to western blot analysis.
RNA immunoprecipitation
RIP protocol was derived from published reports.20 Briefly, cells were treated with 0.3% formaldehyde for 10 min at 37 ºC, then added with glycine to a final concentration of 0.125 M and then incubated at RT for 5 min. Cells were then washed twice in cold phosphate buffered saline and pellet was resuspended in 1 ml of RIPA buffer, which was incubated on ice with frequent vortexing for 30 min. Finally, the nuclear lysate was obtained by centrifugation at 13,000 r.p.m. for 10 min. To obtain bead and antibody complex, 20 μl protein beads were mixed with 1 μg antibody and rotated for 4 h at 4 ºC. The complex was added to nuclear lysates and incubated overnight at 4 °C and then incubated with RNase-free DNase I (Promega) at 37 °C for 15 min and proteinase K at 45 °C for 45 min. Lastly, RNA was extracted with 1 ml TRIzol (Invitrogen) and analyzed by quantitative PCR.
Immunofluorescent staining
Cells were fixed with 4% formaldehyde for 15 min at RT and then permeabilized in 0.1% Triton X-100 for 15 min at RT. Cells were then washed by PBS for three times, followed by incubation with 5% normal goat serum for 30 min at RT. Subsequently, cells were incubated with primary antibody, the anti-mouse ER antibody (Santa Cruz), for 1 h at RT. After washing three times with PBS, cells were incubated with secondary antibody, Alexa Fluor 594 goat anti-mouse IgG (Invitrogen), for 1 h at RT. Finally, cells were washed three times with PBS and mounted using Prolong Gold Antifade Reagent (Invitrogen).
LncRNA in situ hybridization
Biotin-labeled antisense HOTAIR RNAprobe /5Biosg/G+C+C+TTGCTCCCTT+G+CCTGCATTTCT+C+T+G was synthesized by EXIQON (Woburn, MA, USA). For paraffin-embedded tissue, after deparaffinization and rehydration, the samples were treated with peroxidase-quenching solution; proteinase K was added to digest tissues before prehybridization and hybridization, which were carried out at 56 °C for 30 min and 4 h, respectively. Then streptavidin–horseradish peroxidase was used to react with the bound biotin-labeled probe. The signal was further amplified using TSA amplification kit (PerkinElmer, Waltham, MA, USA). Finally, the signal was revealed with Ultra Vision One polymer and aminoethyl carbazole chromogen (Thermo Fisher Scientific, Waltham, MA, USA). The stainings were then scored by eye by two pathologists, on a three-tiered scoring system, using the following criteria for the three tiered system: 0=negative, 1=equivocal/uninterpretable, 2=weak positive and 3=strong positive.
Gene expression microarray and data analysis
Total RNAs were isolated using TRIzol reagent (Invitrogen). The integrity of the RNA was verified using Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). Microarray profiling was performed using HumanHT-12 v 4.0 Expression BeadChip (Illumina, San Diego, CA, USA). Bead-level data were processed using GenomeStudio (Illumina), and the expression values were quantile-normalized using the limma package in Bioconductor.32
Genes having at least twofold changes in HOTAIR-overexpressing ells compared with the control cells in the absence of estrogen were defined as HOTAIR-regulated gene set. Genes with at least twofold change between ethanol and estrogen-stimulated MCF7 cells were defined as estrogen-regulated genes. Gene ontology terms enrichment was analyzed using DAVID 6.7.33 Gene set enrichment analysis was performed as previously described.34
ChIP and ChIP-seq
ChIP experiments were carried out as previously described.35 Antibodies used are Rabbit ER (06-935, Millipore) and Rabbit IgG (sc-2027, Santa Cruz). ChIP-quantitative PCR enrichment of target loci was normalized to input DNA and reported as % input±s.e.m. ChIP DNA was prepared into libraries according to standard protocols using Bioo Scientific’s DNA Sample Kit (cat. no. 514101, Austin, TX, USA). Libraries were sequenced using Illumina Hi-Seq platforms. Sequence reads were aligned to the Human Reference Genome (assembly hg19) using Burrows−Wheeler alignment tool (bwa) version 0.6.1.36 Microarray and short-read sequencing data have been deposited in the GEO database with the accession number GSE61270.
Cell proliferation and clonogenic assay
Cell proliferation assay was carried out using the WST-1 kit according to the manufacturer’s instruction (Clontech, Mountain View, CA, USA). Briefly, 5000 cells were seeded in a 24-well plate. After adding 50 μl WST-1 reagents per well, cultures were incubated for 2 h and the absorbance at a wavelength of 440 nm was determined using a microplate reader. For clonogenic assay, 500 cells were plated in each well of a six-well plate. When there was visible colony by naked eye, cells were fixed with 4% formaldehyde and were stained with crystal violet (0.25%). Colonies were then counted.
References
Mercer TR, Dinger ME, Mattick JS . Long non-coding RNAs: insights into functions. Nat Rev Genet 2009; 10: 155–159.
Ponting CP, Oliver PL, Reik W . Evolution and functions of long noncoding RNAs. Cell 2009; 136: 629–641.
Wang KC, Chang HY . Molecular mechanisms of long noncoding RNAs. Mol Cell 2011; 43: 904–914.
Wilusz JE, Sunwoo H, Spector DL . Long noncoding RNAs: functional surprises from the RNA world. Genes Dev 2009; 23: 1494–1504.
Yoon JH, Abdelmohsen K, Kim J, Yang X, Martindale JL, Tominaga-Yamanaka K et al. Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat Commun 2013; 4: 2939.
Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 2015; 47: 199–208.
Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010; 464: 1071–1076.
Gupta S, Iljin K, Sara H, Mpindi JP, Mirtti T, Vainio P et al. FZD4 as a mediator of ERG oncogene-induced WNT signaling and epithelial-to-mesenchymal transition in human prostate cancer cells. Cancer Res 2010; 70: 6735–6745.
Chisholm KM, Wan Y, Li R, Montgomery KD, Chang HY, West RB . Detection of long non-coding RNA in archival tissue: correlation with polycomb protein expression in primary and metastatic breast carcinoma. PLoS One 2012; 7: e47998.
Sorensen KP, Thomassen M, Tan Q, Bak M, Cold S, Burton M et al. Long non-coding RNA HOTAIR is an independent prognostic marker of metastasis in estrogen receptor-positive primary breast cancer. Breast Cancer Res Treat 2013; 142: 529–536.
Ring A, Dowsett M . Mechanisms of tamoxifen resistance. Endocr Relat Cancer 2004; 11: 643–658.
Musgrove EA, Sutherland RL . Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer 2009; 9: 631–643.
Doisneau-Sixou SF, Sergio CM, Carroll JS, Hui R, Musgrove EA, Sutherland RL . Estrogen and antiestrogen regulation of cell cycle progression in breast cancer cells. Endocr Relat Cancer 2003; 10: 179–186.
Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M . Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 2000; 103: 843–852.
Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, Agard DA et al. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell 1998; 95: 927–937.
Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 2012; 481: 389–393.
Fan M, Yan PS, Hartman-Frey C, Chen L, Paik H, Oyer SL et al. Diverse gene expression and DNA methylation profiles correlate with differential adaptation of breast cancer cells to the antiestrogens tamoxifen and fulvestrant. Cancer Res 2006; 66: 11954–11966.
Joseph R, Orlov YL, Huss M, Sun W, Kong SL, Ukil L et al. Integrative model of genomic factors for determining binding site selection by estrogen receptor-alpha. Mol Syst Biol 2010; 6: 456.
Kaneko S, Li G, Son J, Xu CF, Margueron R, Neubert TA et al. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev 2010; 24: 2615–2620.
Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010; 329: 689–693.
Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT . GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene 2009; 28: 195–208.
Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R et al. Activation of p53 by MEG3 non-coding RNA. J Biol Chem 2007; 282: 24731–24742.
Chen D, Sun Y, Wei Y, Zhang P, Rezaeian AH, Teruya-Feldstein J et al. LIFR is a breast cancer metastasis suppressor upstream of the Hippo-YAP pathway and a prognostic marker. Nat Med 2012; 18: 1511–1517.
Bussemakers MJ, van Bokhoven A, Verhaegh GW, Smit FP, Karthaus HF, Schalken JA et al. DD3: a new prostate-specific gene, highly overexpressed in prostate cancer. Cancer Res 1999; 59: 5975–5979.
Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol 2011; 29: 742–749.
Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet 2013; 45: 1392–1398.
Prensner JR, Chinnaiyan AM . The emergence of lncRNAs in cancer biology. Cancer Discovery 2011; 1: 391–407.
Lee GL, Dobi A, Srivastava S . Prostate cancer: diagnostic performance of the PCA3 urine test. Nat Rev Urol 2011; 8: 123–124.
Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature 2010; 464: 1067–1070.
Clarke R, Leonessa F, Welch JN, Skaar TC . Cellular and molecular pharmacology of antiestrogen action and resistance. Pharmacol Rev 2001; 53: 25–71.
Wu L, Runkle C, Jin HJ, Yu J, Li J, Yang X et al. CCN3/NOV gene expression in human prostate cancer is directly suppressed by the androgen receptor. Oncogene 2014; 33: 504–513.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi WX et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015; 43 e47.
Huang, da W, Sherman BT, Lempicki RA . Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4: 44–57.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005; 102: 15545–15550.
Yu J, Yu J, Mani RS, Cao Q, Brenner CJ, Cao X et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell 2010; 17: 443–454.
Li H, Durbin R . Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics 2009; 25: 1754–1760.
Acknowledgements
Bioinformatic analysis was supported by the computational resources and staff contributions provided for the Quest high-performance computing facility at Northwestern University, which is jointly supported by the Office of the Provost, the Office for Research, and Northwestern University Information Technology. This work was supported by the the US Department of Defense W81XWH-13-1-0319 (to JY) and the Research Scholar Award RSG-12-085-01 (to JY) from the American Cancer Society. JK was supported in part by the National Institutes of Health Training Program in Oncogenesis and Developmental Biology (T32 CA080621), and YAY was supported in part by the National Institutes of Health/National Cancer Institute training grant T32 CA009560.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Xue, X., Yang, Y., Zhang, A. et al. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene 35, 2746–2755 (2016). https://doi.org/10.1038/onc.2015.340
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2015.340
- Springer Nature Limited
This article is cited by
-
LncRNA HOTAIR regulates autophagy and proliferation mechanisms in premature ovarian insufficiency through the miR-148b-3p/ATG14 axis
Cell Death Discovery (2024)
-
Circulating ESR1, long non-coding RNA HOTAIR and microRNA-130a gene expression as biomarkers for breast cancer stage and metastasis
Scientific Reports (2023)
-
circRNA-SFMBT2 orchestrates ERα activation to drive tamoxifen resistance in breast cancer cells
Cell Death & Disease (2023)
-
Long noncoding RNA LINC00173 induces radioresistance in nasopharyngeal carcinoma via inhibiting CHK2/P53 pathway
Cancer Gene Therapy (2023)
-
Landscape of NcRNAs involved in drug resistance of breast cancer
Clinical and Translational Oncology (2023)