Abstract
The resistance of melanoma to current treatment modalities represents a major obstacle for durable therapeutic response, and thus the elucidation of mechanisms of resistance is urgently needed. The crucial functions of activating transcription factor-2 (ATF2) in the development and therapeutic resistance of melanoma have been previously reported, although the precise underlying mechanisms remain unclear. Here, we report a protein kinase C-ɛ (PKCɛ)- and ATF2-mediated mechanism that facilitates resistance by transcriptionally repressing the expression of interferon-β1 (IFNβ1) and downstream type-I IFN signaling that is otherwise induced upon exposure to chemotherapy. Treatment of early-stage melanomas expressing low levels of PKCɛ with chemotherapies relieves ATF2-mediated transcriptional repression of IFNβ1, resulting in impaired S-phase progression, a senescence-like phenotype and increased cell death. This response is lost in late-stage metastatic melanomas expressing high levels of PKCɛ. Notably, nuclear ATF2 and low expression of IFNβ1 in melanoma tumor samples correlates with poor patient responsiveness to biochemotherapy or neoadjuvant IFN-α2a. Conversely, cytosolic ATF2 and induction of IFNβ1 coincides with therapeutic responsiveness. Collectively, we identify an IFNβ1-dependent, cell-autonomous mechanism that contributes to the therapeutic resistance of melanoma via the PKCɛ–ATF2 regulatory axis.
Similar content being viewed by others
Introduction
Human melanoma, characterized by aggressive metastatic behavior and the ability to rapidly develop therapeutic resistance, represents one of the most lethal forms of skin cancer. Despite the advent of effective targeted monotherapies, such as the mutant B-RAF kinase inhibitors vemurafenib (PLX4720) and dabrafenib, most melanomas eventually develop therapeutic resistance that drives relapse and progression.1 A number of studies have identified genetic and epigenetic mechanisms through which melanomas can acquire resistance to mutant B-RAF inhibitors, including mutation of RAS, MAP2K1 and ERK, and upregulation of PDGFR and MAP3K82, 3, 4—all of which contribute to reactivation of the mitogen-activated protein kinase/extracellular-signal-regulated kinase signaling pathway. Other therapeutic modalities for melanoma include agents that inhibit immune response checkpoints, such as CTLA-4 (cytotoxic T-lymphocyte-associated protein 4)5, 6, 7 and PD1 (programmed cell death protein 1),8, 9 and immunomodulatory cytokines such as interleukin-2 (IL-2) and interferon (IFN)-α2a10 have exhibited variable efficacy. In addition, chemo- and biochemotherapeutic regimens (for example, chemotherapeutic agents cisplatin, vinblastine or dacarbazine, alone or in combination with IFN-α2a or IL-2), have been limited in efficacy and are considered as palliative modalities for late-stage metastatic melanoma patients.11, 12, 13 In general, the overall therapeutic success for melanomas has been limited by our insufficient understanding of mechanisms—beyond the mitogen-activated protein kinase signaling pathway—that facilitate resistance and by our inability to identify patients who might be most responsive to specific therapies.
Activating transcription factor-2 (ATF2), a member of the activator protein-1 (AP1) helix-loop-helix transcription factor family, elicits both oncogenic and tumor suppressor functions, depending on its subcellular localization. We previously reported that in melanoma cells subjected to genotoxic stress (a common outcome of most anticancer therapies), ATF2 localizes to the cytoplasm where it acts as a tumor suppressor by perturbing the VDAC1/HK1 (voltage-dependent anion channel 1/hexokinase 1) complex at the mitochondrial outer membrane and promoting apoptosis.14 In contrast, phosphorylation of ATF2 on threonine 52 (T52) by protein kinase C-ɛ (PKCɛ) promotes the nuclear localization and transcriptional activity of ATF2, rendering the cells resistant to chemotherapeutic stress. In successive stages of melanoma progression, levels of both PKCɛ and nuclear ATF2 are increased and correlate with poorer clinical outcome,14 suggesting that the PKCɛ–ATF2 signaling axis contributes to tumorigenesis and chemoresistance. Notably, PKCɛ was previously identified among the top 10 kinases that can confer resistance to BRAF inhibition in melanoma2 and, importantly, a recent study identified ATF2 as a crucial mediator of resistance to Sorafenib in liver cancer, demonstrating that loss of ATF2 is sufficient to revert resistance.15 Consistent with this notion, synthetic peptides or small-molecule inhibitors that attenuate the phosphorylation of ATF2 by PKCɛ, promote its cytoplasmic localization, and thus inhibit its transcriptional activity, can sensitize melanoma cells to death.16, 17 However, the precise transcriptional program coordinated by PKCɛ and ATF2 to drive chemoresistance is not yet known. Here, we report that the PKCɛ–ATF2 signaling axis facilitates resistance in melanoma by repressing the tumor-suppressive, therapeutic stress-induced expression of IFNβ1.
Results
PKCɛ–ATF2 signaling represses chemotherapy-induced IFNβ1 expression
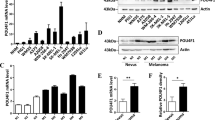
We previously showed that phosphorylation of ATF2 by PKCɛ on threonine 52 (pATF2) promotes its nuclear retention and transcriptional activation in melanoma cells, conferring resistance to chemotherapeutic stress.14 Indeed, the expression of either the phosphomimic ATF2T52E or a constitutively active form of PKCɛ (caPKCɛ) renders WM793 melanoma cells resistant to the chemotherapeutic (genotoxic) stress induced by etoposide (ETO) or cisplatin (CIS), reducing cell death by ~50% (Figure 1a and Supplementary Figure 1a, lower; 14). To determine the mechanism by which activation of the PKCɛ–ATF2 signaling axis imparts chemotherapeutic resistance, we performed gene expression profiling of WM793 cells that were first depleted of endogenous ATF2 and then reconstituted with either ATF2T52E or the nonphosphorylatable mutant ATF2T52A that is predominantly cytoplasmic/mitochondrial and is transcriptionally inactive14 (Supplementary Figure 1b). We identified the top 100 genes that were significantly upregulated or downregulated upon ETO treatment of ATF2T52E-expressing compared with ATF2T52A-expressing cells (Supplementary Table 1). Of these genes, IFN signaling was identified as the most significantly altered canonical signaling pathway (Supplementary Figure 1b, left), in which IFNβ1 and the IFN-related genes SP110, IRF9 and IFI144L were significantly downregulated in ETO-treated ATF2T52E-expressing cells (Supplementary Figure 1b, right)—suggesting that pATF2 represses IFN pathway components. Quantitative real-time reverse transcription–PCR analysis of IFNβ1 transcripts confirmed that ETO and CIS treatments increased IFNβ1 mRNA levels by ~4- to 5-fold and ~3-fold, respectively, in WM793 cells (Figure 1b, left). This induction of IFNβ1 by ETO or by CIS was reduced by ~60–70% or ~40–60% by the expression of ATF2T52E or wild-type ATF2, respectively (Figure 1b, right). Consistent with these observations, ETO treatment increased the expression of both IFNβ1 and SP110 by ~4- to 5-fold in melanoma cells with low levels of PKCɛ/pATF2 (WM793) and by ~2-fold in cells with intermediate levels (LU1205), but had no effect in cells with high levels of PKCɛ/pATF2 (501Mel; Supplementary Figures 1a and c–e). This relationship between PKCɛ/pATF2 levels and IFN-related gene expression was also observed for the downstream IFNβ1 effectors IFIT2, ISG56, OAS1 and PKR, thereby establishing that PKCɛ/pATF2 levels repress chemotherapeutic stress-induced IFNβ1 expression and signaling (Supplementary Figure 1f). In addition, treatment of WM793 cells with SBI-410, a small-molecule inhibitor of PKCɛ-mediated phosphorylation of ATF2,17 dose-dependently induced IFNβ1 transcription (~2- to 9-fold; Supplementary Figure 1g). Moreover, the small interfering RNA-mediated knockdown of PKCɛ resulted in increased expression of IFNβ1 transcripts, both at baseline and during ETO treatment (Supplementary Figure 1h) Finally, we confirmed that ETO treatment increased IFNβ1 protein production by ~2.5-fold in WM793 cells (Figure 1c), and to a lower degree in LU1205 (~1.8-fold) and 501Mel (~1.2-fold) cells. Similar effects were observed following CIS treatment (Figure 1c). This induction was also blocked by the expression of ATF2T52E or caPKCɛ (Figure 1d). Together, these data indicate that the PKCɛ–ATF2 pathway represses the stress-induced expression of IFNβ1 and its downstream signaling effectors.
PKCɛ-phosphorylated ATF2 confers chemotherapeutic stress resistance and reduces IFNβ1 expression. (a, left) Representative Annexin-V (AV)/propidium iodide (PI) fluorescence-activated cell sorting (FACS) plots of WM793 melanoma cells transfected with empty vector (EV), ATF2T52E or constitutively active PKCɛ (caPKCɛ) for 48 h and treated with dimethyl sulfoxide (DMSO), 10 μM ETO or 10 μM CIS for 24 h. (a, right) Quantitation of Annexin-V- and/or PI-positive cells. *P≤0.01. (b, left) Quantitative real-time reverse transcription–PCR (qRT–PCR) analysis of IFNβ1 expression in WM793 cells treated with ETO or CIS normalized to levels in DMSO-treated cells. (b, right) qRT–PCR analysis of IFNβ1 transcripts in ATF2-depleted WM793 cells reconstituted with ATF2WT, ATF2T52A or ATF2T52E for 48 h and then treated with ETO or CIS for 24 h. The IFNβ1 transcript levels shown are relative to cells reconstituted with ATF2T52A. (c, left) Representative FACS histogram showing intracellular IFNβ1 protein expression in DMSO-, ETO- and CIS-treated WM793 cells. (c, right) FACS quantification of induction of intracellular IFNβ1 for WM793, LU1205 and 501Mel cells treated with DMSO or ETO. (d) Intracellular IFNβ1 protein levels in WM793 cells expressing EV, caPKCɛ or ATF2T52E and treated with DMSO or ETO. *P<0.05; #P=0.0032. The results shown represent the mean values±s.d. of experiments performed in biological triplicate.
PKCɛ-phosphorylated ATF2 binds the IFNβ1 promoter to repress transcription
To determine whether pATF2 directly represses IFNβ1 transcription in melanoma cells subjected to chemotherapeutic stress, we examined ATF2 binding to the IFNβ1 promoter in 501Mel cells that express high levels of PKCɛ and pATF2 and that failed to upregulate IFNβ1 in response to stress (Supplementary Figure 1a and e). Chromatin immunoprecipitation–PCR analysis revealed high levels of the IFNβ1 5′ promoter sequence in ATF2 immunoprecipitates from cells incubated with or without ETO, suggesting that ATF2 binds constitutively to the IFNβ1 promoter to repress transcription in cells exhibiting high levels of PKCɛ (Figure 2a, left). In LU1205 cells that moderately upregulated IFNβ1 following chemotherapeutic stress, chromatin immunoprecipitation analysis revealed a ~20% reduction of ATF2 binding to the IFNβ1 promoter (Figure 2a, right), supporting the notion that the presence of ATF2 on the IFNβ1 promoter suppresses its transcription, whereas its absence following genotoxic stress promotes it.
PKCɛ-phosphorylated ATF2 binds to and represses transcription of the IFNβ1 promoter. (a) Chromatin immunoprecipitation (ChIP)–PCR analysis of ATF2-associated 5′-IFNβ1 promoter in 501Mel (left) and LU1205 (right) cells treated with dimethyl sulfoxide (DMSO) or 10 μM etoposide (ETO) for 24 h. (b, upper) Schematic of the 5′-untranslated region (UTR) of the human IFNβ1 gene showing E2 and E3 AP1 sites on the forward strand and E1 and E4 AP1 sites on the reverse strand. (b, middle) IFNβ1 promoter luciferase constructs are shown below. ΔE1 contains a mutagenized E1 site. (b, lower) Quantitation of full-length (FL) IFNβ1 promoter luciferase construct activity upon treatment with DMSO, ETO or 10 μM CIS for 24 h at the indicated concentrations. *P<0.001. (c) Luciferase assays of WM793 cells expressing the indicated IFNβ1 promoter luciferase constructs and treated with DMSO or ETO overnight. Results are expressed relative to the luciferase activity in cells expressing the DMSO-treated FL construct. *P<0.0005. (d) Luciferase assays performed as in (c) on WM793 cells co-transfected with the IFNβ1 promoter luciferase constructs and either empty vector (EV) or ATF2T52E. *P=0.0005. (e) Luciferase assays performed as in (c) with WM793 cells co-transfected with the indicated IFNβ1 promoter luciferase constructs and either EV or ATF2T52E and treated with DMSO or ETO. *P=0.0018; #P=0.03. The results shown represent the mean values±s.d. of experiments performed in biological triplicate.
The IFNβ1 5′-promoter contains four putative AP1 sites that could serve as ATF2 binding sites. To test this, we generated luciferase constructs containing varying combinations of the IFNβ1 promoter AP1 sites (E1, E2, E3 and E4; Figure 2b), and evaluated their activity in WM793 cells coexpressing empty vector or ATF2T52E. ETO and CIS treatment induced luciferase expression ~2-fold in cells expressing the full-length construct (containing E1–E4) (Figure 2b, lower, and Figure 2c), consistent with the effects of chemotherapeutic stress on endogenous IFNβ1. Constructs consisting of the E3 domain alone (#3) or E3+E4 domains (#4) exhibited an ~2- to 4-fold higher luciferase activity compared with cells expressing the full-length construct, in the presence or absence of ETO (Figure 2c), suggesting the presence of a transcriptional activating element. However, only the E1-containing full-length and E1–3 (#123) constructs were transcriptionally repressed by ATF2T52E, suggesting that the E1 element is required for pATF2-mediated transcriptional suppression (Figure 2d). Indeed, mutation of the AP1 consensus-binding motif in E1 abrogated the ATF2T52E transcriptional repression (▵E1; Figure 2e). These data indicate that pATF2 represses IFNβ1 transcription by binding to the E1 element within the 5′-untranslated region of the IFNβ1 gene.
IFNβ1 induction results in delayed S-phase and a senescence-like phenotype
To determine the biological action of IFNβ1 in melanoma cells, we examined two- and three-dimensional growth in WM793 cells overexpressing IFNβ1 or 501Mel cells treated with exogenous recombinant human IFNβ1, respectively, and found that both treatments significantly impaired melanoma cell proliferation when grown in standard tissue culture conditions as well as in three-dimensional spheroid culture (Figure 3a, left and right). Furthermore, IFNβ1 treatment effectively doubled the proportion of LU1205 cells in the S and G2 phases of the cell cycle compared with untreated cells (Figure 3b). In addition, the number of cells in S phase was ~10% higher in cultures treated with both IFNβ1 and ETO compared with ETO alone (Figure 3b). These data indicate that IFNβ1 treatment promotes the accumulation of cells in S phase and is sufficient, when applied as a single agent, to double the S-phase population. We posited that this effect of IFNβ1 might be because of activation of the intra-S-phase checkpoint. Indeed, immunostaining for the DNA repair protein Mre11 revealed that IFNβ1 treatment increased the frequency of bulky Mre11-positive DNA repair foci by ~40% (Figure 3c), consistent with the accumulation of cells in late S phase.18 These data therefore demonstrate that IFNβ1 impairs S/G2-phase progression.
IFNβ1 signaling impairs melanoma growth and proliferation and slows S/G2 transit. (a, left) Growth of WM793 cells was measured at indicated times after transfection with empty vector (EV) or vector encoding IFNβ1. (a, right) The three-dimensional growth of 501Mel spheroids was measured at the indicated times after transfer to soft agar and addition of dimethyl sulfoxide (DMSO) or 300 IU/ml human IFNβ1. Media and fresh IFNβ1 was replenished after 72 h. *P<0.0001. (b) Cell-cycle fluorescence-activated cell sorting (FACS) profiles of LU1205 cells treated as indicated for 24 h. (c, left) Immunofluorescent staining for Mre11 in WM793 cells treated as indicated for 24 h. DNA was stained with 4',6-diamidino-2-phenylindole (DAPI). Scale bar=10 μm. (c, right) Quantification of nuclei containing Mre11+ foci. Results are the mean values±s.d. of 50 nuclei per sample. (d) Cell cycle-phase distribution of LU1205 cells infected with EV (shEV) or short hairpin RNAs (shRNAs) targeting IFNAR1 (shA4 and shH3) or IFNAR2 (shF3). Cells were incubated with DMSO or 10 μM etoposide (ETO) for 24 h. Cell-cycle analysis was performed as in (b). *P<0.0001; #P<0.001 compared with shEV+ETO. (e) Quantitation of Annexin-V and propidium iodide staining of LU1205 cells treated as indicated for 32 h. For all FACS, N=10 000 cells per sample, and the results shown represent the mean values±s.d. of experiments performed in biological triplicate.
We next asked whether inhibition of IFNβ1 signaling might reverse the cell-cycle effects of ETO. To this end, we performed short hairpin RNA-mediated knockdown of the IFNβ1 receptor subunit IFNAR2 in WM793 cells (Supplementary Figure 2a) and assessed cell-cycle progression and viability under chemotherapeutic stress. Control cells expressing scrambled short hairpin RNA exhibited an ~30% increase in S/G2-phase cells and increased dead/dying cells following exposure to ETO (Figures 3d and e). In contrast, knockdown of IFNAR2 increased the percentage of ETO-treated cells in G1 (~15–20%), and reduced S-phase accumulation and cell death by ~10% and ~13%, respectively, compared with control ETO-treated cells (Figures 3d and e). A similar reduction of chemotherapeutic stress-induced cell death and increase in G1-phase cells was observed in cells overexpressing caPKCɛ or ATF2T52E compared with empty vector-expressing cells (Supplementary Figure 2b), consistent with the cell-cycle changes being driven by PKCɛ–ATF2-mediated signaling.
Interestingly, the effects of IFNβ1 and ETO on the cell cycle were accompanied by a senescence-like phenotype characterized by the increased activity of senescence-associated β-galactosidase and the expression of senescence markers including DEC1, DCR2 and p21, but not p16, p53 or p27 (Figure 4a). Despite this profile, the cells treated with ETO and/or IFNβ1 continued to proliferate, although at a greatly reduced rate (Figure 3a and data not shown). The mutant B-RAF inhibitor PLX4720 has also been shown to induce senescence-associated β-galactosidase activity.19 In agreement, PLX4720 treatment of WM793 cells increased senescence-associated β-galactosidase activity (Figure 4a) and also induced the transcription of IFNβ1 (Figure 4b), similar to the effects of ETO and CIS. These results indicate that the therapeutic stress-induced expression of IFNβ1 in melanoma cells results in impaired S/G2 transit that is characterized by increased DNA repair foci and a senescence-like phenotype.
IFNβ1 induces a senescence-like phenotype and enhances the toxicity of etoposide and PLX4720. (a, left panel) Senescence-associated β-galactosidase (SA-β-gal) activity in WM793 cells treated as indicated overnight. (a, right panel) Quantitative real-time reverse transcription–PCR (qRT–PCR) analysis of DCR2, DEC1 and p16 transcripts in WM793 cells treated with dimethyl sulfoxide (DMSO) or 10 μM ETO overnight. *P<0.05. (b) qRT–PCR analysis of IFNβ1 transcripts in WM793 cells treated with DMSO or 5 μM PLX4720 overnight. For (a, b), results represent the mean values±s.d. of triplicates and are expressed relative to cells treated with DMSO. (c) Quantification of dead cell (sub-G1) population of WM793 cells treated with 600 IU/ml human IFNβ1 (6 h of pretreatment)±10 μM ETO or 5 μM PLX4720 (PLX) and analyzed by flow cytometry at the indicated times. #P<0.005. (d) Quantitation of Annexin-V and propidium iodide staining of empty vector (EV), caPKCɛ and WM793 cells co-overexpressing EV or IFNβ1 that were incubated in the presence of DMSO or ETO for 32 h.
IFNβ1 cotreatment enhances chemotherapeutic efficacy
We hypothesized that the IFNβ1-induced S-phase accumulation might enhance the sensitivity of melanoma cells to chemotherapeutic agents or PLX4720. Indeed, we found that whereas treatment with IFNβ1 alone was not significantly cytotoxic, the toxicity of ETO (by ~30%) and PLX4720 (by ~40%) was significantly enhanced by cotreatment with IFNβ1 (Figure 4c and Supplementary Figures 2c and d). These results suggest that the cell cycle-altering effects of IFNβ1 can sensitize melanoma cells to chemotherapeutic agents as well as to PLX4720. In support of this notion, whereas the overexpression of caPKCɛ or ATF2T52E suppressed ETO-induced cell death to ~13% or 10%, respectively, compared with ~27% cell death in EV-expressing cells, the co-overexpression of IFNβ1 was sufficient to significantly overcome caPKCɛ- or ATF2T52E-mediated resistance to ETO, restoring cell death induction to ~35% (Figure 4d).
Taken together, these results indicate that IFNβ1 signaling status contributes to the responsiveness of melanoma cells to stress via intact IFNβ1 signaling, which sensitizes cells to stress, whereas loss of IFNβ1 signaling under stress conditions is protective. These findings support a tumor suppressor role for cell-autonomous IFNβ1 signaling in melanoma cells—consistent with the previously observed function of PKCɛ-phosphorylated ATF2.
Effects of chemotherapeutic stress-induced IFNβ1 expression in melanoma cells are cell autonomous
We next asked whether chemotherapeutic stress-induced IFNβ1 expression in melanoma cells might also elicit cell nonautonomous effects, such as immune cell-mediated tumor clearance. To test this, we performed co-culture experiments using the B6-derived murine Pten:Braf:Cdkn2a mutant melanoma cell line YUMM1.320 and splenic lymphocytes from syngeneic wild-type B6 mice carrying YUMM1.3 tumors. The YUMM1.3 cells were modified to express green fluorescent protein (GFP) and ATF2T34E (murine ATF2T52E equivalent) or empty vector, allowing direct assessment of the contribution of pATF2 (Figure 5a).
Chemotherapeutic stress-induced expression of IFNβ1 in melanoma cells does not activate lymphocytes to mediate tumor clearance. (a) Immunofluorescence images of green fluorescent protein (GFP)-expressing YUMM1.3 (B6 syngeneic) murine melanoma cells, either untreated or exposed to 5 Gy IR and cultured for 24 h with or without lymphocytes (melanoma to lymphocyte ratio: 1:30) from the spleens of B6 mice that were previously inoculated subcutaneously with YUMM1.3 tumors (for 1 month). Anti-IFNβ1 antibody (aIFNβ1) was added at 10 μg/ml as indicated. Images were acquired after 24 h of culture. Scale bar=100 μm. (b) Viability of GFP-expressing YUMM1.3 cells co-transfected with empty vector (EV) or vector encoding ATF2T34E and treated as in (a). Viability was measured as described in the Materials and methods section after 24 h of culture. The results represent the mean values±s.d. of triplicates. #P<0.005.
Treatment of YUMM1.3 cells with ionizing irradiation (IR; 5 or 10 Gy for 24 h) alone, a therapeutic stress that can be targeted specifically to the melanoma cells but not to the lymphocytes, effectively reduced cell viability by ~40%. The addition of a neutralizing anti-IFNβ1 antibody attenuated cell death (~25% increased viability) in cells exposed to 5 Gy IR, and to a lesser extent, after 10 Gy IR, indicating that IR-induced death was partially dependent on IFNβ1 (Figures 5a and b and Supplementary Figure 3). Although the addition of lymphocytes decreased (~20%) the viability of nonirradiated YUMM1.3 cells, the lymphocytes did not significantly reduce the viability of irradiated YUMM1.3 cells compared with irradiated YUMM1.3 cells alone. Furthermore, the addition of an anti-IFNβ1 antibody to these co-cultures did not affect cell viability compared with YUMM1.3 cells cultured in the absence of lymphocytes. Intriguingly, the addition of lymphocytes to cultures of ATF2T34E-expressing YUMM1.3 cells, which were refractory to irradiation, did not decrease cell viability (Figures 5a and b and Supplementary Figure 3). These results demonstrate that the IR-induced expression of IFNβ1 in the melanoma cells elicited minimal additional effects in triggering lymphocyte-mediated tumor cell death in our co-culture system. Furthermore, flow cytometric analysis of the co-cultures revealed that the irradiated YUMM1.3 cells did not further activate CD4+ or CD8+ T cells, NKp46+ NK cells or GR-1+ lymphocytes (Supplementary Figures 4a–d) compared with nonirradiated cells, whereas the neutralizing IFNβ1 antibody abolished their basal activation state (Supplementary Figure 4).
Together, these data indicate that the changes observed in melanoma cells following altered IFNβ1 expression are predominantly because of cell-autonomous effects.
Subcellular localization of ATF2 and expression of IFNβ1 in human melanomas correlates with therapeutic responsiveness
We next investigated whether ATF2 and IFNβ1 status in melanoma tumors might associate with clinical outcome. We hypothesized that after treatment, tumors exhibiting cytoplasmic accumulation of ATF2 and induction of IFNβ1 would correlate with therapeutic responsiveness, whereas those exhibiting nuclear enrichment of ATF2 and no induction or reduction of IFNβ1 would correlate with nonresponsiveness to therapy. To this end, we examined pre- and post-treatment melanoma tumor samples obtained from three medical centers.
The first cohort of patients specimens assessed were paired pre- and post-treatment melanoma tumor sections obtained from elective surgeries on 9 AJCC (American Joint Committee on Cancer) stage IIIc/IV cutaneous melanoma patients who were administered combinatorial biochemotherapy (cisplatin, dacarbazine, vinblastine, IL-2 and IFN-α2a),21, 22 dacarbazine, temodar, and/or had radiation (gamma knife) with several years of follow-up (John Wayne Cancer Institute, Santa Monica, CA, USA). ATF2 localization (nuclear vs cytoplasmic) and IFNβ1 levels were examined and compared, by blinded analysis, between the patient-matched pre- and post-treatment tissue sections by immunofluorescence microscopy and were found to correlate with clinical responsiveness (time to progression) for 7 of the 9 patients assessed as follows: 3 of 3 patients exhibiting poor responses and 4 of 6 patients exhibiting favorable responses (Supplementary Figures 5a and b and 6a). Furthermore, a separate cohort of 8 nontreated melanoma patient specimens and 15 drug-treated nonresponder specimens (from patients who had progressed/recurred rapidly on treatment) demonstrated a ninefold reduction in IFNβ1 expression, as determined by qRT-PCR analysis, consistent with the transcriptional repression of IFNβ1 in nonresponsive patients (Supplementary Figure 6c). Drug therapies for the treated patients included ipilimumab, MEK inhibitor, vemurafenib, IFN-α2a, abraxane and/or avastin.
As pATF2 was associated with downregulated IFNβ1 and its downstream effectors, we assessed the possibility that the status of ATF2 and IFNβ1 might correlate with responsiveness to IFN-based therapy. Thus, we examined pre- and post-treatment tumor sections from an independent cohort of 12 melanoma patients who were administered single-agent neoadjuvant IFN-α2a (University of Pittsburgh Cancer Institute Melanoma Center). Blinded scoring and comparison of the patient-matched sections for changes in ATF2 localization and IFNβ1 expression before and after treatment revealed a strong association with clinical responsiveness (time to progression) in 8 of 12 patients: 3 of 4 patients exhibited poor responses and 5 of 8 patients exhibited favorable responses (Figures 6a and b and Supplementary Figure 6b). Consistent with these findings, analyses of pre- and post-IFN-α2a-treated tumor samples from five nonresponder patients (from the University of Zurich) revealed unchanged or reduced IFNβ1 and nuclear ATF2 staining (Supplementary Figure 8).
IFNβ1 expression, ATF2 subcellular localization and the presence of CD8+ T cells in melanoma patient tumor samples correlates with therapeutic responsiveness. Immunofluorescence staining for ATF2 (red) and IFNβ1 (green) in representative melanoma tumor sections from a responder (a) and nonresponder (b) patient from the University of Pittsburgh Cancer Center (UPCC) cohort, before (Pre-Tx; upper) and after (Post-Tx; lower) IFN-α2a treatment as indicated in the text, is shown. Immunofluorescence staining for CD8 in representative sections from a nonresponder (c) or responder (d) patient from the John Wayne Cancer Institute (JWCI) cohort, before (Pre-Tx; upper) and after (Post-Tx; lower) biochemotherapy treatment as indicated in the text, is shown. Nuclei were counterstained with 4',6-diamidino-2-phenylindole (DAPI). Scale bars represent 100 μm.
CD8+ T-cell infiltration and intratumoral proliferation represents an important component of antitumor immunity,23 and thus we examined whether the samples from the biochemotherapy and IFN-α2a patient cohorts exhibited a correlation between the presence of CD8+ T cells, patient responsiveness and ATF2/IFNβ1 status. In this analysis, we hypothesized that the presence of CD8+ T cells after treatment would correlate with responsiveness, whereas lack of CD8+ T cells would correlate with nonresponsiveness. By immunofluorescence assessment of the biochemotherapy cohort samples, we found that 5 of 9 patients exhibited increased CD8+ T-cell populations in responders but not in nonresponders (Figures 6c and d and Supplementary Figures 6a and b). In the IFN-α2a cohort, 6 of 12 patients exhibited increased CD8+ T-cell populations in responders but not in nonresponders (Figures 6c and d and Supplementary Figures 6a and b). Notably, in both cohorts, the patients who exhibited a correlation of CD8+ T-cell populations with responsiveness or nonresponsiveness were also patients whose responses also correlated with ATF2 and IFNβ1 status. These observations suggest that although the therapeutic stress-induced expression of IFNβ1 that we observe in melanoma cells is insufficient to trigger lymphocyte activation/infiltration, it does not preclude the presence/proliferation of intratumoral CD8+ T-cell populations that are likely affected by mechanisms that are independent of the therapy-induced, ATF2-regulated expression of IFNβ1.
Taken together, the analyses of patient samples support our mechanistic data, indicating that the transcriptional repression of IFNβ1 expression by PKCɛ–ATF2 drives melanoma resistance. In addition, we also assessed the status of ATF2 and IFNβ1 in a cohort of 17 melanoma patients who were treated with IL-2 at the Anschutz Medical Campus of the University of Colorado, and we did not observe a significant correlation between these proteins and patient responsiveness (data not shown), suggesting that this mechanism of resistance might apply to specific therapeutic modalities. Nonetheless, in the other treatment cohorts, we find that the profile of ATF2 localization and IFNβ1 expression appears to correlate with clinical responsiveness (time to progression) in ~70% of the patients examined; additional independent larger-cohort studies are required to further substantiate these observations.
Discussion
The development of resistance to advanced and specific therapies represents a continuing challenge for effective and durable treatment responses of melanoma. Improving our understanding of the crucial mechanisms that propel resistance in melanoma will aid in the improvement and development of current and future therapeutic modalities as well as potentially patient stratification that would represent a major advance for the treatment of melanoma. Here, we report a previously undisclosed molecular mechanism that facilitates the development of the resistance of melanoma to therapeutic stress.
Our data demonstrate that PKCɛ-phosphorylated ATF2 downregulates IFNβ1 expression (and signaling) that promotes the resistance of melanoma cells to chemotherapeutic agents, as well as the BRAF inhibitor PLX4720 (the model compound for PLX4032/vemurafenib). Phosphorylated ATF2 directly represses IFNβ1 transcription, and attenuating ATF2-mediated suppression enables the induction of IFNβ1 and its tumor-suppressive effects. We found that the stress-induced expression of IFNβ1 elicits predominantly cell-autonomous effects on melanoma cells—which might be explained by the fact that chemotherapy induces ~2.5-fold increases in IFNβ1 protein levels, whereas infectious stimuli (that is, viral infection or lipopolysaccharide exposure) that elicit immune responses have been reported to induce IFNβ1 to levels on the order of thousands of fold increase.24, 25, 26 The chemotherapy-induced IFNβ1 impairs growth and proliferation by altering cell-cycle dynamics and inducing the accumulation of cells in S/G2 phases and sensitizing melanoma cells to death.
Melanomas are notorious for their propensity for developing resistance to a range of therapies, including DNA-damaging agents.10, 27, 28, 29 We found that the upregulation of PKCɛ–ATF2 signaling and subsequent repression of IFNβ1 in cells exposed to chemotherapeutic stress is sufficient to alter cell-cycle dynamics and to reduce the population of cells in S phase, identifying one mechanism by which melanomas develop resistance to chemotherapeutic agents. Indeed, the expression of constitutively active PKCɛ or pATF2 renders melanoma cells with low endogenous levels of PKCɛ/pATF2 less responsive to the cytotoxic effects of exogenous IFNβ1 and/or ETO (Supplementary Figure 7). It is possible that, by driving the nuclear localization and activity of ATF2, PKCɛ might also promote the DNA repair function of ATF2,30 further contributing to enhanced melanoma survival during therapeutic stress—a point that deserves further investigation. That PKCɛ–ATF2 reduced IFNβ1/ETO responsiveness (measured by cell death) by only ~50% suggests that other downstream ATF2-dependent and ATF2-independent mechanisms also contribute to the development of therapeutic resistance. Of note, the correlation of intratumoral CD8+ T-cell populations, ATF2/IFNβ1 status and patient responsiveness that we observed in the biochemotherapy cohort samples, but not in our co-culture studies, implies an effect of PKCɛ–ATF2 on the proliferation of intratumoral CD8+ T cells rather than the recruitment of CD8+ T cells. Further studies on how ATF2 modulates intratumoral immune responses are warranted.
Our findings are corroborated by the analyses of samples from melanoma patients before and after biochemotherapy/chemotherapy/radiation treatment, as well as neoadjuvant IFN-α2a therapy. Accordingly, tumors from patients classified as therapeutic responders exhibited increased levels of cytoplasmic (transcriptionally inert) ATF2 and correspondingly increased IFNβ1 levels. Conversely, nonresponders exhibited nuclear ATF2 and unaltered or reduced levels of IFNβ1 after treatment. These findings therefore offer mechanistic insight into the development of therapeutic resistance through downregulation of a mitogen-activated protein kinase pathway-independent molecular mechanism that would otherwise sensitize tumor cells to chemotherapeutic/therapeutic stress. Such alterations correlated with the clinical responsiveness of the majority of patients we evaluated, suggesting that PKCɛ–ATF2–IFNβ1 signaling plays an important role in melanoma resistance and progression in patients. Further, our data suggest that ATF2 and IFNβ1 might represent novel markers of clinical responsiveness, a possibility that requires further assessment in large cohort studies. Findings from recent clinical studies support the possibility that the mechanism (and ATF2/IFNβ1 profile) that we have identified extends beyond melanoma to other types of cancers. For example, the expression of type-1 IFNs (including IFNβ1) was recently found to correlate with biochemical recurrence and metastasis in prostate and breast cancer patients, respectively, although the underlying mechanism was unclear in those studies.31, 32 Furthermore, recent in vitro studies suggest that tumor stroma-derived IFNβ1, such as that from adipose tissue, also elicits tumor-suppressive effects,33 and the treatment of other cancer cell types, including hepatocellular carcinoma cells, with IFNβ1, was recently found to elicit similar cell-cycle alterations and cell death as those identified in our study.34, 35
Importantly, the results of this study also suggest the potential therapeutic value of agents that can promote ATF2 cytoplasmic localization, thereby derepressing IFNβ1 transcription. We previously showed that the inhibition of PKCɛ-mediated phosphorylation of ATF2 promotes its accumulation at the mitochondrial outer membrane, where it contributes to stress-induced mitochondrial leakage.14 Our subsequent high-content microscopy-based screen identified two compounds that trigger the cytoplasmic/mitochondrial localization of ATF2, simultaneously blocking its transcriptional activity.17 Of those, SBI-0089410 effectively induced the expression of IFNβ1 (Supplementary Figure 1g), supporting the possibility that such compounds could enhance the chemosensitivity of melanoma cells by both promoting the mitochondrial function of ATF2 and simultaneously derepressing IFNβ1 signaling.
In conclusion, our study has identified a molecular mechanism that underlies the oncogenic function of ATF2, as reflected in its ability to drive chemoresistance of melanomas. PKCɛ, which we previously found to be upregulated in metastatic melanoma,14 and which is one of the top 10 kinases that can confer resistance in melanoma to mutant BRAF inhibition,2 phosphorylates ATF2 and triggers the downregulation of IFNβ1 signaling that we have now shown determines the therapeutic responsiveness of melanoma cells by modulating cell-cycle dynamics and sensitivity to stress induced by chemotherapeutic therapeutic agents. Notably, the recent finding that ATF2 mediates sorafenib resistance in liver cancer suggests that the mechanism of therapeutic resistance that we have uncovered might represent a mode of resistance that pertains to other types of cancers beyond melanoma.
Materials and methods
Cell lines
All cell lines were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and antibiotics.
Antibodies and immunostaining reagents
Antibodies employed were purchased as follows: ATF2 (C-19 for immunoblotting and C-19X for chromatin immunoprecipitation assays) and PKCɛ (C15), IFNβ1 (E-20, for immunostaining of sections) from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA); IFNβ1 (AP18065PU-N, for intracellular fluorescence-activated cell sorting and immunoneutralization assays) from Acris (San Diego, CA, USA); β-tubulin (E7-s) from Developmental Studies Hybridoma Bank (University of Iowa, Iowa city, IA, USA); and pT52-ATF2 (Phosphosolutions, Aurora, CO, USA). For immunofluorescent staining of ATF2 in patient sections, we used a polyclonal antibody made by SDIX, LLC (Newark, DE, USA) against the amino-acid residues 129–278.
DNA constructs and transfection
DNA plasmids were all transfected using JetPrime (Polyplus, Illkirch, France) or Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturers’ protocols. The constitutively active HIS-tagged PKCɛ construct was a generous gift from Dr Jorge Moscat (Sanford-Burnham Medical Research Institute, La Jolla, CA, USA). Other plasmids have been previously described.14
Flow cytometric analyses
Cell-cycle analysis
Cells were seeded at 1 × 105 cells per well into 6-well tissue culture plates and treated the next day as indicated in the figure legend. Following treatment, cells were harvested by trypsinization and fixed in 70% EtOH in phosphate-buffered saline. After a single wash in phosphate-buffered saline, the cells were stained in cell cycle staining buffer (60 μg/ml propidium iodide/0.15 mg/ml RNAse A (Sigma, St Louis, MO, USA)). After incubation for 20 min, the cells were immediately analyzed by fluorescence-activated cell sorting, n=10 000 cells (within G1 to G2 gates) per replicate over 3 independent experiments. The fluorescence activated cell sorting data were subsequently analyzed using FlowJo software (TreeStar, Ashland, OR, USA).
Cell death analyses
Cells were seeded at 1 × 105 cells per well into 6-well tissue culture plates and treated the next day as indicated in the figure legend. After treatment, the cells were harvested and stained using the BioVision Annexin-V-FITC Apoptosis Detection Kit (BioVision, Milpitas, CA, USA), n=10 000 cells (within whole cell FSC:SSC gates) per replicate over 3 independent experiments. The fluorescence-activated cell sorting data were subsequently analyzed using FlowJo software (TreeStar).
References
Sun C, Wang L, Huang S, Heynen GJ, Prahallad A, Robert C et al. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature 2014; 508: 118–122.
Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature 2010; 468: 968–972.
Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 2010; 468: 973–977.
Van Allen EM, Wagle N, Sucker A, Treacy DJ, Johannessen CM, Goetz EM et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov 2014; 4: 94–109.
Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 2010; 363: 711–723.
Robert C, Thomas L, Bondarenko I, O'Day S, Weber J, Garbe C et al. Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N Engl J Med 2011; 364: 2517–2526.
Topalian SL, Sznol M, McDermott DF, Kluger HM, Carvajal RD, Sharfman WH et al. Survival, durable tumor remission, and long-term safety in patients with advanced melanoma receiving nivolumab. J Clin Oncol 2014; 32: 1020–1030.
Hamid O, Robert C, Daud A, Hodi FS, Hwu WJ, Kefford R et al. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N Engl J Med 2013; 369: 134–144.
Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 2012; 366: 2443–2454.
Ives NJ, Stowe RL, Lorigan P, Wheatley K . Chemotherapy compared with biochemotherapy for the treatment of metastatic melanoma: a meta-analysis of 18 trials involving 2,621 patients. J Clin Oncol 2007; 25: 5426–5434.
Garbe C, Eigentler TK, Keilholz U, Hauschild A, Kirkwood JM . Systematic review of medical treatment in melanoma: current status and future prospects. Oncologist 2011; 16: 5–24.
Rubin KM . Management of primary cutaneous and metastatic melanoma. Semin Oncol Nurs 2013; 29: 195–205.
Schuchter LM . Adjuvant interferon therapy for melanoma: high-dose, low-dose, no dose, which dose? J Clin Oncol 2004; 22: 7–10.
Lau E, Kluger H, Varsano T, Lee K, Scheffler I, Rimm DL et al. PKCepsilon promotes oncogenic functions of ATF2 in the nucleus while blocking its apoptotic function at mitochondria. Cell 2012; 148: 543–555.
Rudalska R, Dauch D, Longerich T, McJunkin K, Wuestefeld T, Kang TW et al. In vivo RNAi screening identifies a mechanism of sorafenib resistance in liver cancer. Nat Med 2014; 20: 1138–1146.
Bhoumik A, Jones N, Ronai Z . Transcriptional switch by activating transcription factor 2-derived peptide sensitizes melanoma cells to apoptosis and inhibits their tumorigenicity. Proc Natl Acad Sci USA 2004; 101: 4222–4227.
Varsano T, Lau E, Feng Y, Garrido M, Milan L, Heynen-Genel S et al. Inhibition of melanoma growth by small molecules that promote the mitochondrial localization of ATF2. Clin Cancer Res 2013; 19: 2710–2722.
Mirzoeva OK, Petrini JH . DNA replication-dependent nuclear dynamics of the Mre11 complex. Mol Cancer Res 2003; 1: 207–218.
Haferkamp S, Borst A, Adam C, Becker TM, Motschenbacher S, Windhovel S et al. Vemurafenib induces senescence features in melanoma cells. J Invest Dermatol 2013; 133: 1601–1609.
Pencheva N, Buss CG, Posada J, Merghoub T, Tavazoie SF . Broad-spectrum therapeutic suppression of metastatic melanoma through nuclear hormone receptor activation. Cell 2014; 156: 986–1001.
Koyanagi K, O'Day SJ, Gonzalez R, Lewis K, Robinson WA, Amatruda TT et al. Serial monitoring of circulating melanoma cells during neoadjuvant biochemotherapy for stage III melanoma: outcome prediction in a multicenter trial. J Clin Oncol 2005; 23: 8057–8064.
Koyanagi K, O'Day SJ, Boasberg P, Atkins MB, Wang HJ, Gonzalez R et al. Serial monitoring of circulating tumor cells predicts outcome of induction biochemotherapy plus maintenance biotherapy for metastatic melanoma. Clin Cancer Res 2010; 16: 2402–2408.
Gajewski TF, Fuertes MB, Woo SR . Innate immune sensing of cancer: clues from an identified role for type I IFNs. Cancer Immunol Immunother 2012; 61: 1343–1347.
Jacobs AT, Ignarro LJ . Lipopolysaccharide-induced expression of interferon-beta mediates the timing of inducible nitric-oxide synthase induction in RAW 264.7 macrophages. J Biol Chem 2001; 276: 47950–47957.
Malmgaard L, Salazar-Mather TP, Lewis CA, Biron CA . Promotion of alpha/beta interferon induction during in vivo viral infection through alpha/beta interferon receptor/STAT1 system-dependent and -independent pathways. J Virol 2002; 76: 4520–4525.
Wang J, Basagoudanavar SH, Wang X, Hopewell E, Albrecht R, Garcia-Sastre A et al. NF-kappa B RelA subunit is crucial for early IFN-beta expression and resistance to RNA virus replication. J Immunol 2010; 185: 1720–1729.
Middleton MR, Grob JJ, Aaronson N, Fierlbeck G, Tilgen W, Seiter S et al. Randomized phase III study of temozolomide versus dacarbazine in the treatment of patients with advanced metastatic malignant melanoma. J Clin Oncol 2000; 18: 158–166.
Rao RD, Holtan SG, Ingle JN, Croghan GA, Kottschade LA, Creagan ET et al. Combination of paclitaxel and carboplatin as second-line therapy for patients with metastatic melanoma. Cancer 2006; 106: 375–382.
Testori A, Rutkowski P, Marsden J, Bastholt L, Chiarion-Sileni V, Hauschild A et al. Surgery and radiotherapy in the treatment of cutaneous melanoma. Ann Oncol 2009; 20: vi22–vi29.
Bhoumik A, Takahashi S, Breitweiser W, Shiloh Y, Jones N, Ronai Z . ATM-dependent phosphorylation of ATF2 is required for the DNA damage response. Mol Cell 2005; 18: 577–587.
Eiro N, Bermudez-Fernandez S, Fernandez-Garcia B, Atienza S, Beridze N, Escaf S et al. Analysis of the expression of interleukins, interferon beta, and nuclear factor-kappa B in prostate cancer and their relationship with biochemical recurrence. J Immunother 2014; 37: 366–373.
Snijders AM, Langley S, Mao JH, Bhatnagar S, Bjornstad KA, Rosen CJ et al. An interferon signature identified by RNA-sequencing of mammary tissues varies across the estrous cycle and is predictive of metastasis-free survival. Oncotarget 2014; 5: 4011–4025.
Ryu H, Oh JE, Rhee KJ, Baik SK, Kim J, Kang SJ et al. Adipose tissue-derived mesenchymal stem cells cultured at high density express IFN-beta and suppress the growth of MCF-7 human breast cancer cells. Cancer Lett 2014; 352: 220–227.
Maeda S, Wada H, Naito Y, Nagano H, Simmons S, Kagawa Y et al. Interferon-alpha acts on the S/G2/M phases to induce apoptosis in the G1 phase of an IFNAR2-expressing hepatocellular carcinoma cell line. J Biol Chem 2014; 289: 23786–23795.
Yue C, Xu J, Tan Estioko MD, Kotredes KP, Lopez-Otalora Y, Hilliard BA et al. Host STAT2/type I interferon axis controls tumor growth. Int J Cancer 2014; 136: 117–126.
Acknowledgements
We thank Serge Fuchs (UPENN) and members of the Ronai laboratory for crucial scientific discussions and critical reading of this manuscript. Additional thanks to Mitch Levesque and Valerie Amann for their assistance in providing patient samples from the Department of Dermatology, University of Zurich. We are grateful to Jian-Liang Li of the SBMRI Bioinformatics Core, as well as the SBMRI Flow Cytometry and Histology Core, for technical support. Support from NCI P01 (CA128814), R01 (CA179170), Hervey Family Non-endowment Fund at The San Diego Foundation and a Melanoma Research Foundation grant to ZAR are gratefully acknowledged. Support from NCI Grants R01 (CA164679) and P01 (CA177322) to CW and P50 SPORE (CA121973) to JMK is also gratefully acknowledged. EL has been supported by K99 (CA172705) and T32 (CA121949) Grants.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Lau, E., Sedy, J., Sander, C. et al. Transcriptional repression of IFNβ1 by ATF2 confers melanoma resistance to therapy. Oncogene 34, 5739–5748 (2015). https://doi.org/10.1038/onc.2015.22
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2015.22
- Springer Nature Limited
This article is cited by
-
The two-faced role of ATF2 on cisplatin response in gastric cancer depends on p53 context
Cell & Bioscience (2022)
-
The interplay between ATF2 and NEAT1 contributes to lung adenocarcinoma progression
Cancer Cell International (2020)
-
Stress and interferon signalling-mediated apoptosis contributes to pleiotropic anticancer responses induced by targeting NGLY1
British Journal of Cancer (2018)
-
Essential role of HCMV deubiquitinase in promoting oncogenesis by targeting anti-viral innate immune signaling pathways
Cell Death & Disease (2017)
-
miR-218-5p restores sensitivity to gemcitabine through PRKCE/MDR1 axis in gallbladder cancer
Cell Death & Disease (2017)