Abstract
Activating transcription factor 3 (ATF3) responds to diverse cellular stresses, and regulates oncogenic activities (for example, proliferation, survival and migration) through direct transcriptional regulation or protein-protein interactions. Although aberrant ATF3 expression is frequently found in human cancers, the role of ATF3 in tumorigenesis is poorly understood. Here, we demonstrate that ATF3 suppresses the development of prostate cancer induced by knockout of the tumor suppressor Pten in mouse prostates. Whereas the oncogenic stress elicited by Pten loss induced ATF3 expression in prostate epithelium, we found that ATF3 deficiency increased cell proliferation and promoted cell survival, leading to early onset of mouse prostatic intraepithelial neoplasia and the progression of prostate lesions to invasive adenocarcinoma. Importantly, the loss of ATF3 promoted activation of the oncogenic AKT signaling evidenced by high levels of phosphorylated AKT and S6 proteins in ATF3-null prostate lesions. In line with these in vivo results, knockdown of ATF3 expression in human prostate cancer cells by single guided RNA-mediated targeting activated AKT and increased matrix metalloproteinase-9 expression. Our results thus link ATF3 to the AKT signaling, and suggest that ATF3 is a tumor suppressor for the major subset of prostate cancers harboring dysfunctional Pten.
Similar content being viewed by others
Introduction
As a disease caused by genetic alterations successively occurring during its course, prostate cancer remains as one of the leading causes of death from cancer, partly because of the poor understanding of genetic factors that control the development of this deadly disease. Inactivation of the phosphatase and tension homolog protein (Pten) by gene mutations or deletion occurs in about 30% of prostate tumors and up to 60% of metastatic prostate cancers, and is often considered the driving force for prostate tumorigenesis.1 Pten inactivation results in loss of the enzymatic activity that catalyzes dephosphorylation of phosphatidylinositol (3,4,5)-triphosphate (PIP3), thereby activating the oncogenic phosphoinositide 3-kinase (PI3K)/AKT signaling crucial for the growth, survival and distal dissemination of prostate cancer cells.2 Moreover, Pten dysfunction confers advanced prostate cancer with resistance to conventional therapies that are mostly based on androgen deprivation. Indeed, deletion of Pten in mouse prostate epithelium not only recapitulates the progression of human disease from prostatic epithelial neoplasia to invasive adenocarcinoma,3 but results in malignant lesions that are intrinsically resistant to androgen deprivation or castration.4, 5 Given that Pten inactivation is one of the most common genetic alterations in prostate cancer, it would be of interest to identify other genetic alterations that may act in concert with Pten dysfunction to drive the development of prostate cancer.
The immediate early gene activating transcription factor 3 (ATF3) is an ATF/CREB family member whose expression is rapidly induced by a wide range of cellular stresses including DNA damage, cellular injury and oxidative stress.6 In response to cellular stresses, ATF3 regulates diverse cellular functions (for example, proliferation, survival and migration) through binding to the ATF/CREB cis-regulatory element,6 or interacting with other proteins (for example, p53 and NF-κB).7, 8 Accumulating evidence has linked ATF3 to several important cellular signaling pathways, including those mediated by p53, TGFβ and Toll-like receptor 4.7, 8, 9 Activation of p53 by ATF3, for instance, can regulate cellular responses to genotoxic stresses and prevent cellular transformation induced by oncogenic Ras expression.7 ATF3 can also interact with the NF-κB network to regulate cytokine expression thereby engaging in cellular immune responses.8 Although recent studies have revealed that ATF3 contributes to many important human diseases including secondary infections during sepsis-associated immunosuppression10 and skin cancer induced by immunosuppressants,11 the role of ATF3 in cancer, particularly prostate cancer, remains poorly understood.12 Whereas ATF3 appears to be proapoptotic in prostate cancer cells,13, 14 ATF3 also binds the androgen receptor (AR) and represses androgen signaling indispensable for sustaining prostate cancer cell proliferation and survival,1 indicating that ATF3 could be a putative tumor suppressor for prostate cancer. Indeed, several unbiased microarray results have revealed that ATF3 expression is downregulated in prostate cancers, particularly in metastatic prostate cancers.15, 16 In the similar vein, ATF3 has been shown to suppress tumor growth and metastasis in many other cancer types (for example, glioblastoma, colon, bladder and lung cancer).17, 18, 19, 20 However, ATF3 can also promote lung metastasis of mouse melanoma cells and rat prostate cancer cells.21, 22 Moreover, a recent report demonstrates that ATF3 expressed by stromal cells promotes breast cancer cells to disseminate into lungs.23 Therefore, the contributions of ATF3 to cancer remain elusive.
Here, we employed a Pten conditional knockout mouse model to determine the role of ATF3 in prostate cancer. Our results indicate that the loss of ATF3 promoted the development of prostate cancer through activating the AKT signaling. We thus provided the first genetic evidence arguing for that ATF3 is a tumor suppressor for the major subset of prostate cancer harboring Pten dysfunction.
Results
Loss of Pten induces ATF3 expression in prostate epithelium
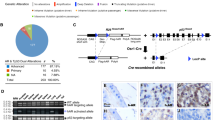
We previously reported that ATF3-knockout mice developed prostatic hyperplasia due to increased AR activity, but ATF3 deficiency alone was not sufficient to induce mouse prostatic intraepithelial neoplasm (mPIN) or carcinoma.24 To further explore the role of ATF3 in prostate cancer, we crossed ATF3−/−, PtenL/L and PB-Cre4 mice (all in C57BL/6 background), and generated offspring with a genotype of PtenL/L;ATF3+/+, PtenL/L;ATF3−/−, PtenL/L;ATF3+/+;Cre+ or PtenL/L;ATF3−/−;Cre+, referred to as WT, ΔATF3, ΔPten and ΔATF3ΔPten, respectively (Figure 1a). Loss of Pten expression in prostatic epithelial cells of Pten-knockout mice (that is, ΔPten and ΔATF3ΔPten) was confirmed by immunofluorescence staining (Figure 1b). Interestingly, although ATF3 was weakly expressed in mouse prostatic epithelial cells, ATF3 staining was significantly increased in Pten-knockout prostates (ΔPten vs WT, Figures 1b and c), arguing for the notion that ATF3 is a stress-inducible gene in prostates and can be induced by the oncogenic stress caused by Pten deficiency. Such oncogenic stress also induced expression of the tumor suppressor p53 as reported (Figure 1b, ΔPten vs WT).25 However, p53 induction was diminished in ATF3-null prostates (Figures 1b and c, ΔATF3ΔPten vs ΔPten,)—a result consistent with our previous report that ATF3 stabilizes p53 under stressed conditions.7
ATF3 expression is induced by the loss of Pten in mouse prostate epithelium. (a) Representative genotyping results of four groups of mice used in this study. (b, c) ATF3 and p53 levels were elevated in ΔPten mice that lost Pten expression. APs were dissected from indicated mice (9 weeks of age), and subjected to immunofluorescence or immunohistochemistry staining for Pten, ATF3 and p53 expression. Staining intensity was quantified and presented as integrated optical density (c).
Loss of ATF3 promotes the development of prostate cancer in mice
It was recently reported that deletion of Pten in prostate epithelium of albino (white) C57BL/6 mice, which contain a spontaneous mutation at the tyrosinase gene, leads to mPIN, but does not cause adenocarcinoma.26 Similar to that study, we found that the loss of Pten in our mutant mice also resulted in progressively enlarged prostates (Supplementary Figure S1). However, in addition to cribiform-like mPIN lesions, the loss of Pten in our black C57/BL6 mice resulted in apparent epithelial invasion into stromal tissues in anterior prostates (AP) and dorsal prostates (DP) (Figure 2a and Supplementary Figure S2, arrows) evidenced by the lack of α-smooth muscle actin staining in invasion regions (Figure 2b, arrows), suggesting the development of adenocarcinoma in these mice. Microinvasion was first seen in 6-week-old DP and 9-week-old AP, and 100% of mice older than 12 weeks developed carcinoma (Figure 2c). In contrast, only low-grade mPIN was seen in ventral prostates whereas no lesion other than hyperplasia was found in lateral prostates of ΔPten mice (Supplementary Figure S2). The cancerous cells were originated from luminal epithelial cells as they were positive for AR staining but negative for p63 expression (Supplementary Figure S3). Thus, the loss of Pten led to rapid development of adenocarcinoma in our mouse model. Interestingly, whereas ATF3 expression was initially induced by Pten loss (Figure 1b and Supplementary Figure S4b), the ATF3 expression level was decreased along with the progression of prostate lesions from mPIN to adenocarcinoma in ΔPten mice (Supplementary Figures S4b and c), suggesting that the loss or downregulation of ATF3 expression appeared to be required for the development of Pten-null prostate cancer.
Loss of ATF3 promotes the development of prostate cancer. (a) H&E-stained sections of AP from indicated mice at 4, 6, 9, 12 and 20 weeks of age. Arrows indicate invasion. (b) α-Smooth muscle actin stained sections of AP and DP at 9 and 12 weeks show more invasive lesions in ΔATF3ΔPten mice. Arrows indicate invasion sites marked by discontinuous α-smooth muscle actin staining. (c) Percentages of mice with hyperplasia, mPIN or adenocarcinoma in their APs. (d, e) Percentages of lesions with invasion are shown for AP (d) and DP (e). Invasive lesions were counted based on α-smooth muscle actin staining. All glands in each section (average 42 AP and 79 DP) were counted. *P<0.05; **P<0.01; ***P<0.001; ns, no significant difference; Mann-Whitney U-test.
Indeed, we found that the loss of ATF3 promoted the development of prostate cancer in Pten-knockout mice. In contrast to ΔPten mice, which developed mPIN at 6 weeks of age in 4 out of 9 mice, 10 out of 11 ΔATF3ΔPten mice developed mPIN at the same age (P<0.05, Fisher’s Exact test) (Figure 2c). Similarly, adenocarcinoma was found in 8 out of 9 ΔATF3ΔPten mice as compared to 4 out of 11 ΔPten mice at 9 weeks (P<0.05, Fisher’s Exact test) (Figure 2c). Moreover, mPIN in ΔATF3ΔPten prostates was often high-grade, and more prostate lesions in these compound-mutant mice were invasive (Figure 2a and Supplementary Figure 2a, arrows). Staining the prostates for α-smooth muscle actin expression (Figure 2b, arrows) confirmed that ΔATF3ΔPten mice had a significantly larger number of invasive adenocarcinoma in both AP (Figure 2d) and DP (Figure 2e). Taken together, these results indicate that the loss of ATF3 promoted the development of prostate cancer induced by Pten deletion.
Loss of ATF3 increases proliferation but decreased apoptosis of Pten-loss-induced tumor cells
To understand the mechanism by which ATF3 deficiency promoted the development of prostate cancer, we tested whether ATF3 affects proliferation and survival of prostate epithelial cells under the Pten-knockout condition. Towards this end, we stained the prostates for Ki67 expression (a proliferation marker) and cleaved caspase 3 expression (a apoptosis marker), and counted positively stained cells. As expected, the oncogenic stress conferred by Pten deletion promoted proliferation (Figure 3a) while inducing apoptosis of prostate cancer cells (Figure 3c). Importantly, the number of Ki67-positive cells was significantly increased in ΔATF3ΔPten lesions than ΔPten lesions in mice at 6 weeks and 9 weeks of age (Figures 3a and b). Conversely, ΔATF3ΔPten lesions contained a significantly lower number of apoptotic cells as compared with ΔPten prostates at all ages (Figures 3c and d). The decrease in the apoptotic cell number in ΔATF3ΔPten lesions at 12 weeks of age (Figure 3d) is noteworthy given that ATF3 deficiency did not promote proliferation under the same condition (Figure 3b). These results thus indicate that the loss of ATF3 likely promoted the development of prostate cancer by increasing cell proliferation while inhibiting apoptosis.
Loss of ATF3 promotes cell proliferation and survival in Pten-null prostate lesions. (a) APs were stained for Ki67 expression. Representative images are shown. (b) Ki67-positive cells were counted from random microscopic fields, and used to calculate positivity. (c) APs from 9-week-old mice were stained for cleaved caspase 3 expression. Representative images are shown. (d) Cells expressing cleaved caspase 3 were counted from random fields, and used to calculate positivity. *P<0.05; ***P<0.001; Student t-test.
Loss of ATF3 enhances AKT signaling in mouse prostatic epithelial cells
A major consequence of Pten inactivation is AKT phosphorylation, which in turn triggers a cascade of events that drive cell proliferation, sustain cell survival and also promote cell invasion.2 To gain a further insight of the mechanism by which ATF3 deficiency promoted cell proliferation, survival and subsequent development of prostate cancer, we examined AKT activation in prostate lesions by staining prostates for AKT phosphorylation. Indeed, the loss of Pten resulted in phosphorylation of AKT at S473 and T308 (Figure 4b, ΔPten), which otherwise was undetectable in Pten-wild-type prostates (Figure 4a). Loss of ATF3 alone was not sufficient to induce AKT phosphorylation (Figure 4a, ΔATF3). However, we found that ATF3 deficiency dramatically elevated the levels of S473- and T308-phosphorylated AKT under the Pten-knockout condition (Figures 4b–d, ΔATF3ΔPten vs ΔPten). Not only the overall AKT phosphorylation level, but the level of membrane-bound phosphorylated AKT was largely increased in ΔATF3ΔPten lesions as compared with ΔPten tumors (Figure 4b). Moreover, ATF3 deficiency also dramatically increased the phosphorylation level of S6 protein (Figures 4c and d)—a downstream effector of the mTOR pathway which is activated by AKT and also required for prostate tumorigenesis induced by Pten loss.27 Neither the total AKT level nor the total S6 level was altered by the loss of ATF3 (Figures 4b–d). These results indicate that the loss of ATF3 enhanced the AKT signaling in prostate cancer induced by Pten deficiency.
Loss of ATF3 promotes AKT activation and enhances AKT signaling in Pten-null prostate lesions. Prostate sections from indicated mice (9 weeks of age) were subjected to immunohistochemistry staining for expression of indicated proteins (a, b and c). Staining intensity of each protein was quantified using the Image-Pro Plus software, and presented as the integrated optical density (d).
Knockdown of ATF3 expression in prostate cancer cells activates AKT signaling
To corroborate the important finding that the loss of ATF3 promoted AKT signaling, we employed the emerging CRISPR-Cas9-based technology28 to knock down ATF3 expression in human prostate cancer cells (LNCaP, PC3 and DU145). This gene-targeting technology takes advantage of a short, single guided RNA (sgRNA) that specifically binds to a target genomic region (the region spanning the ATF3 start codon in our case, Figure 5a) and recruits a nuclease Cas9 to cleave DNA (Figure 5a, vertical arrow). The generated double-strand DNA break would then be repaired by the error-prone homology-directed repair mechanism, resulting in a deletion or insertion in a region immediate downstream of the start codon of the ATF3 gene thereby disrupting ATF3 gene expression. Employing this approach, we isolated several clones that expressed a significantly lower level of ATF3 (Figures 5c–e). The Surveyor-based mutagenesis detection assay confirmed that these clones carried at least one mutant ATF3 allele (Figure 5b). Western blotting showed dramatic elevation of the phosphorylation levels of AKT and S6 in these ATF3-low-expressing prostate cancer cells, although the expression level of total AKT and S6 was not altered (Figures 5c–e). These results thus demonstrate that downregulation of ATF3 expression in human prostate cancer cells enhances AKT signaling as well.
Downregulation of ATF3 expression by sgRNA-mediated targeting enhances AKT signaling in human prostate cancer cells. (a) Diagram depicts that the nuclease hCas9 recruited by a sgRNA (sgATF3) specifically recognizing a region spanning the ATF3 start codon cleaves the ATF3 gene. The vertical arrow shows the potential cleavage site. (b) The Surveyor nuclease cleaves the mismatch site of a heteroduplex DNA (580 bp) formed by the wild-type and mutated ATF3 gene fragments amplified by PCR using the primers indicated by arrows in a, which generates two cleaved products (340 and 240 bp). Genomic DNA from indicated cells was used as templates for PCR. (c–e) Western blotting results show elevated AKT signaling in sgATF3-targeting cells.
ATF3 deficiency/downregulation increases the NF-κB signaling and MMP-2/-9 expression
The AKT signaling can promote cancer progression by regulating the expression of matrix metalloproteinases (MMPs).29, 30, 31, 32, 33 To understand the mechanism by which the loss of ATF3 promotes invasion of prostate cancer cells (Figure 2b), we carried out qRT-PCR assays to examine expression of a range of MMPs in sgATF3-expressing and control DU145 cells. Whereas MMP-2, -8 and -12 expression were undetectable, downregulation of ATF3 significantly increased the expression of MMP-9, -10, -13, -14 and -15 in DU145 cells (Figure 6a). Using zymography, we confirmed the induction of MMP-9 expression in LNCaP, DU145 and PC3 cells (Figure 6b). Interestingly, sgATF3 also induced MMP-2 expression in LNCaP cells although the parental LNCaP cells did not express detectable MMP-2 (Figure 6b). As IKK phosphorylation by AKT can activate NF-κB29, 30 and the latter is well known to regulate MMP-9 transcription,30, 31 we explored a possibility that enhanced AKT signaling in ATF3-downregulated prostate cancer cells transactivates the MMP-9 promoter by activating the NF-κB signaling. Indeed, whereas the MMP-9 promoter activity was significantly higher in sgATF3-expressing prostate cancer cells, mutating the κB cis-regulatory element in the MMP-9 promoter31 completely abolished the sgATF3-mediated transactivation of the promoter. Consistent with these results, the NF-κB activity was increased in the ATF3-downregulating cells as the activity of a luciferase reporter driven by tandem repeats of κB elements (κB-luc), but not mutated κB elements (mκB-luc), was significantly higher in the sgATF3-expressing cells than that in control DU145 cells (Figure 6d). Moreover, the IKK and IκB phosphorylation levels as well as the nuclear p65 level were increased in sgATF3 cells (Figure 6e). Collectively, these results are in line with the notion that AKT activates the NF-κB signaling, leading to induction of MMP-9 expression under ATF3 downregulated or deficient conditions. To determine whether ATF3 affects MMP expression in mouse prostate lesions as well, we stained the prostate sections for MMP-2 and MMP-9 expression. We found that the loss of ATF3 indeed dramatically promoted MMP-2 and MMP-9 expression in Pten-knockout prostate lesions (Figures 6f and g, ΔATF3ΔPten vs ΔPten). Therefore, the loss of ATF3 likely enhances the AKT signaling, leading to increased MMP expression thereby promoting the invasion of Pten-null prostate cancer. Of note, ATF3 might directly repress MMP-2 transcription as previously reported.34, 35, 36 We thus conclude that the loss/downregulation of ATF3 expression likely contributes to the development of Pten-deficient prostate cancer by activating AKT and consequently promoting its downstream events including cell proliferation, resistance to apoptosis and expression of pro-invasive genes (Figure 6h).
Loss of ATF3 increases MMP-2 and MMP-9 expression in prostate cancer cells. (a) MMP expression was increased in ATF3-downregulated DU145 cells. Cells were subjected to qRT-PCR assays to measure MMP expression levels. (b) MMP-9 and MMP-2 expression was increased in ATF3-downregulated human prostate cancer cells. Conditional media from indicated cells were subjected to zymography after normalized to equal cell number. HT1080 cells expressing high levels of MMP-2 and MMP-9 were used as positive controls. (c) The MMP-9 promoter activity was increased in ATF3-downregulated cells. A luciferase reporter driven by a human MMP-9 promoter (−670~+34) or a κB-mutated (−600) promoter was transfected into DU145 or sgATF3-expressing cells along with pRL-CMV for dual-luciferase activity assays. (d) ATF3-downregulated cells harbored increased NF-κB reporter activity. A reporter driven by tandem repeats of κB cis-regulatory elements (κB-luc) or mutated κB elements (mκB-luc) were transfected into DU145 or sgATF3 cells for dual luciferase activity assays. (e) The NF-κB signaling was increased in sgATF3-expressing cells as detected by western blotting. WCL, whole-cell lysate; NE, nuclear extract. (f, g) Loss of ATF3 increases MMP-2/-9 expression in Pten-null prostate lesions. Prostate sections from 9-week-old mice were subjected to immunohistochemistry staining for MMP-2 and MMP-9 expression (f). Staining intensity was quantified and presented as integrated optical density (g). (h) A model indicates that ATF3 induced by Pten loss could block AKT activation, leading to decreased proliferation, survival and invasiveness of prostate cancer cells. ATF3 could also directly inhibit MMP-2 expression thereby preventing prostate cancer cells from invasion. *P<0.05; **P<0.01; ***P<0.001; ns, not significance; Student t-test.
Discussion
In line with an early report that ATF3 expression can be induced by oncogenic stresses elicited by oncogene expression (for example, Ras),37 we showed here that the oncogenic stress caused by Pten loss could also induce ATF3 expression in prostate epithelium (Figure 1b). This result suggests that ATF3 could serve as an anti-cancer barrier that functions to eliminate oncogenic stresses thereby preventing the development of prostate cancer. Indeed, whereas ATF3 expression was downregulated in Pten-null adenocarcinoma in ΔPten mice (Supplementary Figure S4), we have demonstrated that the lack of ATF3 induction in double-knockout mice promoted the development of prostate cancer. Thus, the downregulation of ATF3 expression, frequently found in human prostate cancer,15, 16 might promote the progression of the major subset of prostate cancer harboring dysfunctional Pten (Figure 6e). We therefore identified an additional genetic alteration that may act in concert with Pten inactivation to promote the development of human prostate cancer.
The tumor suppressor role of ATF3 in prostate cancer is also supported by the observations that ATF3 is a proapoptotic molecule in prostate cancer cells and that ATF3 represses AR signaling required for sustaining the growth and survival of prostate cancer cells.1, 13, 14 However, ATF3 expression was also shown to promote invasion of a human prostate cancer cell line and lung colonization of a rat prostate cancer cell line.22, 38 These seemingly controversial results well represent the fact that ATF3 often regulates gene expression in a context-dependent manner. For instance, ATF3 induces expression of the metastasis suppressor KAI1 when it interacts with JunB, but represses expression of the same gene when co-expressed with NF-κB.38 Nevertheless, our current study provides the first genetic evidence supporting that ATF3 suppresses prostate cancer harboring Pten dysfunction. Whereas it remains unclear whether ATF3 also suppresses prostate cancer carrying wild-type Pten, we recently showed that ATF3 can counteract the oncogenic activities of mutant p53 proteins thereby suppressing metastasis of TP53-mutated cancer.39 Given that TP53 mutation is a common genetic alteration in late-stage prostate cancer,1 ATF3 might function as a tumor suppressor for advanced prostate cancer in general. Indeed, downregulation of ATF3 expression is more often seen in metastatic prostate cancers (Supplementary Figure S5).40 It is worth noting that several other reports found elevated ATF3 expression in prostate cancer as compared with normal prostate tissues.40, 41 Such an elevation might be the result of acute ATF3 induction by oncogenic stresses in the early stage of tumorigenesis as we observed in this study (Figure 1b), and the ATF3 expression level in normal prostate tissues adjacent to tumor is indeed often higher than that in their counterparts obtained from organ donors free of prostate cancer (Supplementary Figure S5b). Elevated ATF3 expression might also be the result of upregulated expression of ATF3 splice variants, some of which can function as natural antagonists for the full-length ATF3.42 Alternatively, it might also be that downregulation of ATF3 expression is only required for the development of a subset of prostate cancer harboring Pten dysfunction. Therefore, it might be necessary to stratify prostate cancer patients for a better understanding of the relevance of ATF3 expression to clinical outcomes.
We previously showed that ATF3 can activate p53,7 the tumor suppressor which has been shown to block the development of prostate adenocarcinoma in Pten-knockout mice.25 In line with this, we found that p53 induced by Pten deletion appeared to depend on ATF3 expression (Figure 1b). However, the ATF3-mediated activation of p53 does not appear to be sufficient to cause the suppression of prostate cancer development in ΔPten mice as prostate lesions in these mice rapidly progressed to adenocarcinoma. Likewise, although ATF3 is an AR repressor,24 it is unlikely that ATF3 suppressed prostate cancer development in ΔPten mice through repressing androgen signaling, as it has been shown that Pten-null prostate cancer does not require AR signaling for their growth.4, 5 Indeed, castration of ΔPten mice at 7 weeks of age when mPIN emerges did not affect the prostate cancer progression, nor cause apparent tumor regression (Supplementary Figure S6).
A striking finding from this study is the link between ATF3 and the AKT signaling. Previously, ATF3 was shown to activate AKT in neurons43 and promote cytokine-induced AKT phosphorylation in mouse master cells.44 In this study, we found that ATF3 rather inhibited the oncogenic AKT signaling in Pten-null prostate lesions. Whereas knockdown of ATF3 expression in Pten-null (LNCaP and PC3) or -defective (DU145) human prostate cancer cells also activated AKT, it remains elusive whether Pten dysfunction, the transformation state, or the cell/tissue type, is the prerequisite for ATF3-mediated AKT inhibition. Given that ATF3 mainly localizes in the nuclei of prostate epithelial cells, it is more likely that ATF3 suppresses AKT activation through regulating transcription of other AKT regulators. Expression of CTMP, an AKT activator in breast cancer,45 for instance, can be repressed by ATF3.46 Regardless of which mechanism(s) may involve, activation of AKT in ATF3-null or downregulated prostate cancer cells would promote proliferation, prevent death and lead to a cascade of molecular events contributing to the development of cancer (Figure 6h). One such downstream event was the induction of pro-invasive MMP-9 expression, a likely consequence of transactivation of the MMP-9 promoter by the transcription factor NF-κB31 activated by AKT.47 Although ATF3 itself is a transcription factor, we previously showed that ATF3 does not directly regulate MMP-9 expression.34
It is well established that the genetic background of mouse strain determines the effect of Pten deletion on mouse prostate tumorigenesis.48 Whereas studies on mice with mixed 129 backgrounds often report highly penetrate invasive adenocarcinoma in all of the four prostate lobes,3, 49 knockout of Pten in a pure C57BL/6 background generated mixed results. Although monoallelic Pten knockout led to fully penetrant adenocarcinoma in APs,27 it was a surprise that biallelic Pten-knockout mice only developed mPIN in a recent report.26 It is important to note that the latter study employed an albino strain that contains a mutation at the tyrosinase gene. This strain appears to be different from the black C57BL/6 mice used in our study as we found that 100% of Pten-null mice developed invasive adenocarcinoma in AP and DP at 12 weeks of age. Whether other genetic alteration(s) accompanied by the tyrosinase gene mutation in the albino mice prevents the development of prostate adenocarcinoma is an intriguing question and worthy of further investigation. It is important to note that monoallelic knockout of Pten (Ptenpc(+/−)) in the prostates of our C57BL/6 mice only led to low-grade mPIN lesions even when the mice were older than 11 months (Supplementary Figure S7). Therefore, partial loss of Pten does not appear to be sufficient to cause prostate cancer in our mouse model.
Materials and Methods
Animals and genotyping
Animal experiments were carried out according to protocols approved by the Institutional Committee of Animal Care and Use (ICACU) of the Albany Medical College and the ICACU of Georgia Regents University. ATF3 knockout (ATF3−/−) mice (in C57BL/6 background) were described previously,24, 50 and we obtained B6.129S4-Ptentm1Hwu/J (PtenloxP/loxP, or PtenL/L) and PB-Cre4 (Cre+) mice from the Jackson Laboratory and the NCI Mouse Repository, respectively. To generate compound mutants, ATF3−/− mice were first crossed with PtenL/L and Cre+ mice, yielding PtenL/L;ATF3−/− and ATF3−/−;Cre+ mice, respectively. The cross of the latter two strains generated male PtenL/+;ATF3−/−;Cre+ mice, which were further bred with female PtenL/L;ATF3−/− mice to generate male PtenL/L;ATF3−/−;Cre+ (ΔATF3ΔPten) and PtenL/L;ATF3−/− (ΔATF3) for histolopathological examinations. Male PtenL/L;ATF3+/+;Cre+ (ΔPten) and PtenL/L;ATF3+/+(WT) mice were generated similarly. For genotyping, mouse tails were lysed in PBND buffer supplemented with 0.2 μg/ml Proteinase K at 55 °C overnight, and lysates were directly subjected to PCR following the protocols provided by the Jackson Laboratory. For castration, testes of 7-week-old male mice were surgically removed, and the mice were killed at 12 weeks of age for histopathological examinations.
Immunohistochemistry and immunofluorescence staining
Prostate lobes were separated by microdissection, and embedded in paraffin for sectioning.24 After antigen retrieval in hot citrate buffer, sections were blocked in 5% of normal horse serum and 1% of normal goat serum, and subjected to immunohistochemistry staining using the ABC Elite Kit and the DAB Kit (Vector Laboratories, Burlingame, CA, USA) according to the manufacturers’ recommendations. The following antibodies were used: Ki-67 (ab15580, 1:600) and MMP-9 (ab137867) from Abcam (Cambridge, MA, USA); ATF3 (sc-188, 1:200), AR (sc-816, 1:200), and p63 (sc-8430, 1:200) from Santa Cruz Biotechnology (Dallas, TX, USA); p-AKT S473 (#4060, 1:200), p-AKT T308 (#2965,1:200), AKT (#4691, 1:200), p-S6 (#2211, 1:400), S6 (#2217, 1:200), cleaved caspase-3 (#9661, 1:300), and Pten (#9188, 1:100) from Cell Signaling (Danvers, MA, USA); and MMP2 (NB200-193, 1:200) from Novus (Littleton, CO, USA). For α-smooth muscle actin staining, sections were incubated with alkaline phosphatase (AP)-conjugated anti-α-smooth muscle actin antibody (1:600, Sigma-Aldrich, St Louis, MO, USA) followed by detection of AP activity using the SIGMAFAST Fast Red TR/Naphthol AS-MX tablets (F4523, Sigma) according to the supplier’s protocol. For quantifying immunohistochemistry staining intensity, random microscopic fields were captured and digitized by a CCD camera (Olympus, Center Valley, PA, USA). Signal intensity was determined using the Image-Pro Plus software (MediaCybernetics, Rockville, MD, USA) and presented as integrated optical density.
Cell culture and sgRNA-mediated knockdown of ATF3 expression
LNCaP, DU145 and PC3 cells were cultured in DMEM, RPMI1640 and T-medium supplemented with 5–10% fetal bovine serum, respectively. The CRISPR-Cas9 system was used to generate ATF3 low-expressing cells as described.28 Essentially, a plasmid expressing a sgRNA targeting a region spanning the ATF3 start codon (5′-AAAATGATGCTTCAACACCCAGG-3′; the start codon was underlined) was constructed and co-transfected into cells with a hCas9-expressing plasmid.28 Single cells were plated into 96-well plates, and derived clones screened for ATF3 downregulation using western blotting. To confirm that obtained clones were derived from sgRNA-guided knockdown, genomic DNA was prepared and subjected to Surveyor mutation detection using a kit purchased from Transgenomic.
Western blotting and quantitative reverse transcription-PCR (qRT-PCR)
These were carried out as described previously.7, 51 Briefly, cells were lysed in RIPA buffer (50 mM Tris-HCl, pH 7.4, 1% Nonidet P-40, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA, 1 mM PMSF, and 1 mM NaF, 1 mM Na3VO4, and protease inhibitor cocktail (Roche, Indianapolis, IN, USA)), and subjected to SDS-polyacrylamide electrophoresis for western blotting. We purchased the following antibodies from Cell Signaling: IKKα (#11930), phospho-IKKα/β (#2697), IκBα (#4814), phospho-IκBα (#2859) and NF-κB p65 (#8242). For qRT-PCR assays, total RNA was extracted from cells using Trizol reagent (Life Technologies, Grand Island, NY, USA), and then reverse-transcribed, followed by real-time PCR assays for MMP expression.51 The primer sequences are available upon request.
Zymography
Zymography was performed as described previously.34 Briefly, condition media were collected, concentrated and loaded on a polyacrylamide gel containing 1 mg/ml gelatin after normalization to equal cell number. The gel was rinsed with 2.5% Triton X-100 at room temperature for 1 h, and then incubated in a buffer containing 50 mM Tris-HCl, pH 7.5, 10 mM CaCl2 and 150 mM NaCl at 37 °C overnight. The gel was stained with 0.2% Coomassie Blue, and MMPs were detected as transparent proteolytic bands against a dark blue background.
References
Shen MM, Abate-Shen C . Molecular genetics of prostate cancer: New prospectes for old challenges. Gene Dev 2010; 24: 1967–2000.
Song MS, Salmena L, Pandolfi PP . The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol 2013; 13: 283–296.
Wang S, Gao J, Lei Q, Rozengurt N, Pritchard C, Jiao J et al. Prostate-specific deletion of the muring Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell 2003; 4: 209–221.
Mulholland DJ, Tran LM, Li Y, Cai H, Morim A, Wang S et al. Cell autonomous role of PTEN in regulating castration-resistant prostate cancer growth. Cancer Cell 2011; 19: 792–804.
Carverm BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell 2011; 19: 575–586.
Hai T, Wolfgang CD, Marsee DK, Allen AE, Sivaprasad U . ATF3 and stress responses. Gene Expr 1999; 7: 321–325.
Yan C, Lu D, Hai T, Boyd DD . Activating transcription factor 3, a stress sensor, activates p53 by blocking its ubiquitination. EMBO J 2005; 24: 2425–2435.
Gilchrist M, Thorsson V, Li B, Rust AG, Korb M, Roach JC et al. Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature 2006; 441: 173–178.
Kang Y, Chen C, Massague J . A self-enabling TGFß response coupled to stress signaling: Smad engages stress response factor ATF3 for Id1 repression in epithelial Cells. Mol Cell 2003; 11: 915–926.
Hoetzenecker W, Echtenacher B, Guenova E, Hoetzenecker K, Woelbing F, Bruck J et al. ROS-induced ATF3 causes susceptibility to secondary infections during sepsis-associated immunosuppression. Nat Med 2011; 18: 128–134.
Wu X, Nguyen B, Dziunycz P, Chang S, Brooks Y, Lefort K et al. Opposing roles for calcineurin and ATF3 in squamous skin cancer. Nature 2010; 465: 368–372.
Hai T, Wolford CC, Chang Y-S . ATF3, a hub of the cellular adaptive-response network, in the pathogenesis of diseases: Is modulation of inflammation a unifying component? Gene Exp 2010; 15: 1–11.
Huang X, Li X, Guo B . KLF6 induced apoptosis in prostate cancer cells through upregulation of ATF3. J Biol Chem 2008; 283: 29795–29801.
Liu Y, Gao F, Jiang H, Niu L, Bi Y, Young CY et al. Induction of DNA damage and ATF3 by retigeric acid B, a novel topoisomeriase II inhibitor, promotes apoptosis in prostate cancer cells. Cancer Lett 2013; 337: 66–76.
Lapointe J, Li C, Higgins JP, Van De Rijn M, Bair E, Montgomery K et al. Gene expression profiling identifies clinically relevant subtypes of prostate cancer. Proc Natl Acad Sci USA 2004; 101: 811–816.
Tomlins SA, Mehra R, Rhodes DR, Cao X, Wang L, Dhanasekaran SM et al. Integrative molecular concept modeling of prostate cancer progression. Nat Genet 2011; 39: 41–51.
Gargiulo G, Cesaroni M, Serresi M, de Vries N, Hulsman D, Bruggeman SW et al. In vivo RNAi screen for BMI1 targets identifies TGF-β/BMP-ER stress pathways as key regulators of neural- and malignant glioma-stem cell homeostasis. Cancer Cell 2013; 23: 660–676.
Hackl C, Lang SA, Moser C, Mori A, Fichtner-Feigl S, Hellerbrand C et al. Activating transcription factor-3 (ATF3) functions as a tumor suppressor in colon cancer and is up-regulated upon heat-shock protein 90 (Hsp90) inhibition. BMC Cancer 2010; 10: 668.
Yuan X, Yu L, Li J, Xie G, Rong T, Zhang L et al. ATF3 suppresses metastasis of bladder cancer by regulating gelsolin-mediated remodeling of the actin cytoskeleton. Cancer Res 2013; 73: 3625–3637.
Jan Y-H, Tsai H-Y, Yang C-J, Huang M-S, Yang Y-F, Lai T-C et al. Adenylate kinase-4 is a marker of poor clinical outcomes that promotes metastasis of lung cancer by downregulating the transcription factor 3. Cancer Res 2012; 72: 5119–5129.
Ishiguro T, Nakajima M, Naito M, Muto T, Tsuruo T . Identification of genes differentially expressed in B16 murine melanoma sublines with different metastatic potentials. Cancer Res 1996; 56: 875–879.
Bandyopadhyay S, Wang Y ZR, Pai S, Watabe M, Iiizumi M, Furuta E et al. The tumor metastasis suppressor gene Drg-1 down-regulates the expression of activating transcription factor 3 in prostate cancer. Cancer Res 2006; 66: 11983–11990.
Wolford CC, McConoughey SJ, Jalgaonkar SP, Leon M, Merchant AS, Dominick JL et al. Transcription factor ATF3 links host adaptive response to breast cancer metastasis. J Clin Invest 2013; 123: 2893–2906.
Wang H, Jiang M, Cui H, Chen M, Buttyan R, Hayward SW et al. The stress response mediator ATF3 represses androgen signaling by binding the androgen receptor. Mol Cell Biol 2012; 32: 3190–3202.
Chen Z, Trotman LC, Shaffer D, Lin H-H, Dotan ZA, Niki M et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 2005; 436: 725–730.
Svensson RU, Haverkamp JM, Thedens DR, Cohen MB, Ratliff TL, Henry MD . Slow disease progression in a C57BL/6 Pten-deficient mouse model of prostate cancer. Am J Pathol 2012; 179: 502–512.
Blando J, Portis M, Benavides F, Alexander A, Mills G, Dave B et al. PTEN deficiency is fully penetrant for prostate adenocarcinoma in C57BL/6 mice via mTOR-dependent growth. Am J Pathol 2009; 174: 1869–1879.
Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE et al. RNA-guided human genome engineering via Cas9. Science 2013; 339: 823–826.
Romashkova J, Makarov SS . NF-κB is a target of AKT in anti-apoptotic PDGF signalling. Nature 1999; 401: 86–90.
Madrid LV, Mayo MW, Reuther JY, Baldwin AS . Akt stimulates the transactivation potential of the RelA/p65 subunit of NF-κB through utilization of the IκB kinase and activation of the mitogen-activated protein kinase p38. J Biol Chem 2001; 276: 18934–18940.
Yan C, Wang H, Boyd DD . KiSS-1 represses 92-kDa type IV collagenase expression by down-regulating NF-κB binding to the promoter as a consequence of IκBα-induced block of p65/p50 nuclear translocation. J Biol Chem 2001; 276: 1164–1172.
Yan C, Boyd DD . Regulation of matrix metalloproteinase gene expression. J Cell Physiol 2007; 211: 19–26.
Agarwal A, Das K, Lerner N, Sathe S, Cicek M, Casey G et al. The AKT/IκB kinase pathway promotes angiogenic/metastatic gene expression in colorectal cancer by activating nuclear factor-κB and β-catenin. Oncogene 2005; 24: 1021–1031.
Yan C, Wang H, Boyd DD . ATF3 represses 72-kDa type IV collagenase (MMP-2) expression by antagonizing p53-dependent trans-activation of the collagenase promoter. J Biol Chem 2002; 277: 10804–10812.
Chen HH, Wang DL . Nitric oxide inhibits matrix metalloproteinase-2 expression via the induction of activating transcription factor 3 in endothelial cells. Mol Pharmacol 2004; 65: 1130–1140.
Stearns ME, Kim G, Garcia F, Wang M . Interleukin-10 induced activating transcription factor 3 transcriptional suppression of matrix metalloproteinase-2 gene expression in human prostate CPTX-1532 cells. Mol Cancer Res 2004; 2: 403–416.
Lu D, Wolfgang C, Hai T . Activating transcription factor 3, a stress-inducible gene, suppresses Ras-stimulated tumorigenesis. J Biol Chem 2006; 281: 10473–10481.
Liu W, Liizumi-Gairani M, Okuda H, Kobayashi A, Watabe M, Pai SK et al. KAI1 gene is engaged in NDRG1 gene-mediated metastasis suppression through the ATF3-NFκB complex in human prostate cancer. J Biol Chem 2011; 286: 18949–18959.
Wei S, Wang H, Lu C, Malmut S, Zhang J, Ren S et al. The activating transcription factor 3 protein suppresses the oncogenic function of mutant p53 proteins. J Biol Chem 2014; 289: 8947–8959.
Varambally S, Yu J, Laxman B, Rhodes DR, Mehra R, Tomlins SA et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell 2012; 8: 393–406.
Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol 2004; 22: 2790–2799.
Chen BPC, Liang G, Whelan J, Hai T . ATF3 and ATF3ΔZip: Transcriptional repression versus activation by alternatively spliced isoforms. J Biol Chem 1994; 269: 15819–15826.
Nakagomi S, Suzuki Y, Namikawa K, Kiryu-Seo S, Kiyama H . Expression of the activating transcription factor 3 prevents c-Jun N-terminal kinase-induced neuronal death by promoting heat shock protein 27 expression and Akt activation. J Neurosci 2003; 23: 5187–5196.
Gilchrist M, Henderson WR Jr, Morotti A, Johnson CD, Nachman A, Schmitz F et al. A key role for ATF3 in regulating mast cell survival and mediator release. Blood 2010; 115: 4734–4741.
Liu YP, Liao WC, Ger LP, Chen JC, Hsu TI, Lee YC et al. Carboxyl-terminal modulator protein positively regulates Akt phosphorylation and acts as an oncogenic driver in breast cancer. Cancer Res 2013; 73: 6194–6205.
Huang CY, Chen JJ, Wu JS, Tsai HD, Lin H, Yan YT et al. Novel link of anti-apoptotic ATF3 with pro-apoptotic CTMP in the ischemic brain. Mol Neurobiol (e-pub ahead of print 26 April 2014; doi:10.1007/s12035-014-8710-0.
Manning BD, Cantley LC . AKT/PKB signaling: Navigating downstream. Cell 2007; 129: 1261–1274.
Freeman D, Lesche R, Kertesz N, Wang S, Li G, Gao J et al. Genetic background controls tumor development in Pten-deficient mice. Cancer Res 2006; 66: 6492–6496.
Zhang Q, Liu S, Ge D, Zhang Q, Xue Y, Xiong Z et al. Interleukin-17 promotes formation and growth of prostate adenocarcinoma in mouse models. Cancer Res 2012; 72: 2589–2599.
Hartman MG, Lu D, Kim ML, Kociba GJ, Shukri T, Buteau J et al. Role for activating transcription factor 3 in stress-induced β-cell apoptosis. Mol Cell Biol 2004; 24: 5721–5732.
Yan C, Boyd DD . Histone H3 acetylation and H3 K4 methylation define distinct chromatin regions permissive for transgene expression. Mol Cell Biol 2006; 26: 6357–6371.
Acknowledgements
This work was supported by NIH grants R01CA139107, R01CA164006 and a Department of Defense award W81XWH-07-1-0095 to CY. We thank Dr George Church for providing sgRNA targeting reagents and Dr Honglin Li for providing NF-κB reagents.
Author Contributions
ZY and DX bred the mice. ZY carried out the experiments with the help of HD and JZ, JK performed statistical analyses of clinical data. TH provided the ATF3−/− mice and analyzed the data. HD, JZ and TH edited the manuscript. CY conceived the study, analyzed the data and wrote the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Wang, Z., Xu, D., Ding, HF. et al. Loss of ATF3 promotes Akt activation and prostate cancer development in a Pten knockout mouse model. Oncogene 34, 4975–4984 (2015). https://doi.org/10.1038/onc.2014.426
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2014.426
- Springer Nature Limited
This article is cited by
-
ATF3 in atherosclerosis: a controversial transcription factor
Journal of Molecular Medicine (2022)
-
Induction of ferroptosis by ATF3 elevation alleviates cisplatin resistance in gastric cancer by restraining Nrf2/Keap1/xCT signaling
Cellular & Molecular Biology Letters (2021)
-
Extracellular vesicles-derived microRNA-222 promotes immune escape via interacting with ATF3 to regulate AKT1 transcription in colorectal cancer
BMC Cancer (2021)
-
Decreased expression of ATF3, orchestrated by β-catenin/TCF3, miR-17-5p and HOXA11-AS, promoted gastric cancer progression via increased β-catenin and CEMIP
Experimental & Molecular Medicine (2021)
-
Impaired AKT signaling and lung tumorigenesis by PIERCE1 ablation in KRAS-mutant non-small cell lung cancer
Oncogene (2020)