Abstract
DNA double-strand breaks (DSBs) are highly cytotoxic DNA lesions that trigger non-proteolytic ubiquitylation of adjacent chromatin areas to generate binding sites for DNA repair factors. This depends on the sequential actions of the E3 ubiquitin ligases RNF8 and RNF168 (refs 1, 2, 3, 4, 5, 6), and UBC13 (also known as UBE2N), an E2 ubiquitin-conjugating enzyme that specifically generates K63-linked ubiquitin chains7. Whereas RNF168 is known to catalyse ubiquitylation of H2A-type histones, leading to the recruitment of repair factors such as 53BP1 (refs 8, 9, 10), the critical substrates of RNF8 and K63-linked ubiquitylation remain elusive. Here we elucidate how RNF8 and UBC13 promote recruitment of RNF168 and downstream factors to DSB sites in human cells. We establish that UBC13-dependent K63-linked ubiquitylation at DSB sites is predominantly mediated by RNF8 but not RNF168, and that H1-type linker histones, but not core histones, represent major chromatin-associated targets of this modification. The RNF168 module (UDM1) recognizing RNF8-generated ubiquitylations11 is a high-affinity reader of K63-ubiquitylated H1, mechanistically explaining the essential roles of RNF8 and UBC13 in recruiting RNF168 to DSBs. Consistently, reduced expression or chromatin association of linker histones impair accumulation of K63-linked ubiquitin conjugates and repair factors at DSB-flanking chromatin. These results identify histone H1 as a key target of RNF8–UBC13 in DSB signalling and expand the concept of the histone code12,13 by showing that posttranslational modifications of linker histones can serve as important marks for recognition by factors involved in genome stability maintenance, and possibly beyond.
Similar content being viewed by others
Main
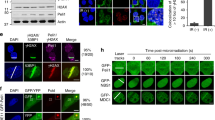
To explain mechanistically the critical role of UBC13 in the RNF8/RNF168 pathway, we generated human UBC13 knockout cells using CRISPR–Cas9 technology14,15 (Fig. 1a). As expected, loss of UBC13 abrogated the accumulation of RNF168, RNF168-dependent ubiquitin conjugates, and 53BP1 and other readers of these marks at DSB sites, while RNF8 recruitment was normal (Fig. 1b, c and Extended Data Fig. 1a, b). Reintroducing UBC13 into these cells restored 53BP1 focus formation, as did a fusion protein mimicking an RNF8–UBC13 E3–E2 complex16,17, independently of endogenous RNF8 (Fig. 1d and Extended Data Fig. 1c, d). In contrast, expression of RNF8 alone or other RNF8–E2 chimaeras failed to support 53BP1 accumulation at DSBs in these cells (Fig. 1d and Extended Data Fig. 1c). Unlike RNF8, overexpression of RNF168 was sufficient to restore 53BP1 recruitment to damaged DNA in UBC13-knockout cells (Extended Data Fig. 1e). These data suggest that UBC13 mainly cooperates with RNF8 but not RNF168 to catalyse K63 ubiquitylation of DSB-flanking chromatin to promote recruitment of repair factors.
a, Immunoblot analysis of HCT116 wild-type (WT) and UBC13-knockout (KO) cells. MCM6 was a loading control. WCE, whole-cell extract. b–d, Representative images of HCT116 wild-type and UBC13-knockout cells exposed to IR or laser micro-irradiation (n = 3 experiments). Cells in d were transfected with the indicated expression constructs (Extended Data Fig. 1c). HA, haemagglutinin. e, f, Binding of recombinant wild-type and mutant (MUT) K63-Super-UIM to di-ubiquitin (Ub2) linkages. The migration of molecular weight markers (kDa) is indicated on the left. g, Quantification of GFP–K63-Super-UIM and 53BP1 accumulation at DSBs in U2OS/GFP–K63-Super-UIM cells transfected with the indicated siRNAs. CTRL, control. Data are mean ± standard deviation (s.d.) from three independent experiments. Representative images are shown in Extended Data Fig. 2b. Scale bars, 10 μm. a, f, Uncropped blots are shown in Supplementary Fig. 1.
To understand the molecular basis of this process, we used a tandem ubiquitin-binding entity (‘K63-Super-UIM’)18 that showed remarkable specificity for binding to K63 linkages but not to other ubiquitin chain types (Fig. 1e, f). Indeed, green fluorescent protein (GFP)-tagged K63-Super-UIM, but not a ubiquitin-binding-deficient mutant (Fig. 1e, f), was efficiently recruited to microlaser- and ionizing radiation (IR)-generated DSBs (Extended Data Fig. 2a–c), thus providing an efficient sensor of DSB-associated K63 ubiquitylation19. Depletion of UBC13 or RNF8, but not RNF168, abrogated recruitment of the K63-Super-UIM, as well as an independently derived high-affinity binder of K63-linked ubiquitin chains, to DSB sites, and auto-ubiquitylation-generated K63 ubiquitin chains were abundantly present on RNF8 but not RNF168 (Fig. 1g and Extended Data Fig. 2b–f). This suggests that the roles of RNF8 and RNF168 in promoting DSB-associated chromatin ubiquitylation can be functionally uncoupled and that RNF8, but not RNF168, is a primary mediator of UBC13-dependent K63 ubiquitylation at DSB sites.
To identify the RNF8–UBC13-dependent ubiquitylation processes underlying RNF168 recruitment to DSBs, we first characterized the impact of UBC13 loss on global K63-linked ubiquitylation under steady-state conditions. While K63-linked ubiquitylation is thought to be mainly catalysed by UBC13 (ref. 7), the extent to which this E2 contributes to the total pool of K63 ubiquitin linkages in human cells is not known. To address this, we quantified the relative abundance of different ubiquitin chains in HCT116 wild-type and UBC13-knockout cells using stable isotope labelling by amino acids in cell culture (SILAC)-based mass spectrometry. Ablation of UBC13 decreased the global level of K63 ubiquitin linkages by ∼50%, while the abundance of other chain types was not markedly affected (Fig. 2a). Using the di-glycine approach20,21, we quantified over 3,000 ubiquitylation sites, of which fewer than 1% showed a >2-fold decrease in UBC13-knockout cells (Extended Data Fig. 3a–d and Supplementary Table 1). Thus, UBC13 has little impact on the conjugation of the initial ubiquitin moiety to substrates, consistent with previous reports that UBC13 primarily acts at the ubiquitin chain elongation step22,23,24. To identify UBC13-dependent K63-ubiquitylated proteins, we analysed K63-Super-UIM pull-downs from wild-type and UBC13-knockout cells under stringent buffer conditions by mass spectrometry. We identified 371 proteins that showed >2-fold enrichment in wild-type cells, several of which are known targets of K63 ubiquitylation (Extended Data Figs 3e–h, 4a, b and Supplementary Table 2).
a, Tukey boxplot showing levels of ubiquitin linkages in HCT116 UBC13-knockout (KO) cells relative to wild-type (WT) cells, quantified by SILAC-based proteomics. Data from two independent experiments are shown. **P < 0.01; NS, not significant (Welch’s t-test). b–d, Pull-down analysis of IR-induced K63-linked ubiquitylation of histone H1.2 in U2OS cells (b, d) or derivative cell lines expressing RNF8 or RNF168 short hairpin RNAs (shRNAs) in a doxycycline (DOX)-inducible manner (c) using recombinant wild-type or mutant (MUT) K63-Super-UIM. Where indicated, ubiquitin conjugates were digested with recombinant USP2 catalytic domain (USP2cc) before SDS–polyacrylamide gel electrophoresis (SDS–PAGE). Asterisk indicates a non-specific band. e, In vitro ubiquitylation of purified H1-containing oligonucleosomes by RNF8351–485 and indicated factors. WCE, whole-cell extract. The migration of molecular weight markers (kDa) is indicated on the left. b–e, Uncropped blots are shown in Supplementary Fig. 1.
We extended this proteomic strategy to search for the chromatin-bound substrate(s), whose K63 ubiquitylation by RNF8–UBC13 promotes recruitment of RNF168 and downstream factors to DSB sites (Extended Data Fig. 4c). Surprisingly, only few cellular proteins reproducibly displayed elevated K63-linked ubiquitylation upon IR-induced DSBs, and H1 linker histones, but not core histones, were major chromatin-associated factors showing such behaviour (Extended Data Fig. 4d). Using the K63-Super-UIM we confirmed biochemically for two endogenous histone H1 isoforms (H1.2 and H1x) that they were modified with K63-linked ubiquitin chains and, more importantly, that these ubiquitylations were markedly upregulated after DSBs (Fig. 2b and Extended Data Fig. 5a, b). Basal levels of H1 K63 polyubiquitylation in non-irradiated cells dropped substantially upon serum starvation (Extended Data Fig. 5a), suggesting that they result mostly from endogenous DNA damage generated by cell-cycle-dependent processes. Little if any K63-linked ubiquitylation of core histones was detectable, regardless of whether DSBs had been inflicted or not (Extended Data Fig. 5c), thus H1 appears unique among histones in undergoing robust DSB-stimulated K63 ubiquitylation. Importantly, the DNA-damage-induced increase in K63-linked polyubiquitylation of H1 was dependent on RNF8 and UBC13 but not RNF168 (Fig. 2c, d), whereas none of these enzymes appreciably affected H1 monoubiquitylation (Extended Data Fig. 5d). Moreover, mass spectrometry analysis showed that H1 was a major factor co-purifying with endogenous RNF8 (Extended Data Fig. 5e). Finally, recombinant RNF8 catalysed robust ubiquitylation of nucleosome-associated H1 with UBC13 in vitro (Fig. 2e). These data suggest that histone H1 is a major chromatin-associated substrate of RNF8–UBC13.
To address whether linker histones represent critical RNF8 substrates in DSB signalling, we downregulated overall H1 levels using a short interfering RNA (siRNA) cocktail targeting different isoforms (Extended Data Fig. 6a, b). Under these conditions, accumulation of K63-linked ubiquitin chains, RNF168, 53BP1 and BRCA1, but not MDC1, at DSB sites was clearly diminished (Fig. 3a–d and Extended Data Fig. 6c–g), suggesting that loss of H1 uncouples ubiquitin-dependent DSB signalling at the level of RNF168 accrual. Consistently, like RNF8 or RNF168 knockdown, H1 downregulation suppressed DSB-induced H2A ubiquitylation catalysed by RNF168 (Fig. 3e and Extended Data Fig. 6h)8,10. As an alternative strategy to interfere with H1 functionality, we overexpressed the high-mobility group protein HMGB1, which competes with H1 for chromatin binding in a non-site-specific manner25. Elevated HMGB1 levels impaired K63 ubiquitylation and 53BP1 accumulation at DSB sites despite enhancing γ-H2AX formation, and this could be rescued by co-overexpression of H1 or by wild-type but not catalytically inactive RNF168 (Fig. 3f, g and Extended Data Fig. 7a–c). A similar strong increase in γ-H2AX formation accompanied by only mildly elevated 53BP1 accumulation at DSBs, indicating a partial uncoupling of these events, was reported for murine cells lacking three of six H1 isoforms26. Together, these data suggest that H1-type histones are functionally critical targets of RNF8- and UBC13-dependent K63 ubiquitylation at DSB-flanking chromatin areas.
a, b, Representative images of GFP–K63-Super-UIM-expressing cells transfected with siRNAs. CTRL, control; DAPI, 4′,6-diamidino-2-phenylindole. c, Proportion of cells with visible accumulation of GFP–K63-Super-UIM at laser-induced DSBs (a). d, Intensity of GFP–K63-Super-UIM or 53BP1 accumulation at laser-induced DSBs normalized to γ-H2AX signal (a). e, Analysis of IR-induced γ-H2AX ubiquitylation (Ub) by RNF168 (marked by arrow) in chromatin fractions of U2OS cells transfected with the indicated siRNAs. f, Representative images of U2OS cells expressing the indicated constructs and exposed to IR. HMGB1 overexpression impairs 53BP1 recruitment to DSBs (marked by arrows). g, Quantification of data in f. Data in c and d are mean ± s.d. from three independent experiments. Data in g are mean ± standard error of the mean (s.e.m.) from two independent experiments. Scale bars, 10 μm. e, Uncropped blots are shown in Supplementary Fig. 1.
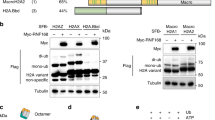
We asked whether K63-ubiquitylated H1 provides a platform for initial, RNF8–UBC13-dependent RNF168 recruitment to DSBs. RNF168 contains two DSB-targeting modules, both of which contain ubiquitin-binding domains4,5,27 (Fig. 4a). While the carboxy-terminal LRM2–MIU2 region, which we termed ubiquitin-dependent DSB recruitment module 2 (UDM2), recognizes RNF168-catalysed forms of ubiquitylated H2A, thereby effectively enabling RNF168 to autonomously propagate H2A ubiquitylation at DSB sites once recruited, its LRM1–UMI–MIU1 region (UDM1) has been suggested to bind as yet undefined RNF8-generated ubiquitylation products required for initial relocalization of RNF168 to break sites (Fig. 4a)11. Therefore, if K63-ubiquitylated H1 functions as a recruitment platform for RNF168 at DSB-modified chromatin, the UDM1 module should be capable of recognizing this modified form of H1. Consistent with this idea, we found that UDM1 bound to K63-linked ubiquitin but no other chain types (Fig. 4b). RNF168 UDM2 also interacted with K63 linkages, albeit with much lower affinity, and unlike UDM1 it also recognized other ubiquitin chains (Extended Data Fig. 8a, b). This might reflect the fact that UDM1, but not UDM2, has two adjacent ubiquitin-binding domains (UMI and MIU1), which together may confer specific binding to K63-linked chains. We also found that UDM1 interacted with histone H1 but not H2A in vitro, whereas UDM2 displayed the inverse preference, binding only to H2A, as previously shown11 (Fig. 4c). The LRM1 part of UDM1 has been hypothesized to impart target specificity to the ubiquitin-binding affinity of this module11. Consistently, the H1-binding ability of UDM1 was at least partially mediated by the LRM1 motif (Fig. 4a), which interacted strongly with H1 in vitro, unlike LRM2 (Extended Data Fig. 8c). However, additional sequence elements within UDM1 were also able to bind H1 (data not shown), possibly owing to the very acidic nature of the UDM1 region (Extended Data Fig. 8d), which could facilitate robust interaction with the highly basic H1 proteins28. Unlike UDM1, UDM2 interacted with neither modified nor unmodified forms of H1 (Fig. 4c, d). When expressed in cells, the UDM1 domain efficiently bound to high-molecular-weight forms of endogenous H1 isoforms in an IR-stimulated manner, suggesting that they correspond to K63-ubiquitylated H1 (Fig. 4d and Extended Data Fig. 8e). Supporting this, point mutations in the UMI or MIU1 domains that impair their ubiquitin-binding activity and RNF168 recruitment to DSBs11,27 markedly reduced the binding of UDM1 to modified H1 (Extended Data Fig. 8f). Moreover, UDM1 did not interact with ubiquitylated forms of core histones, in sharp contrast to its binding to ubiquitylated H1 (Extended Data Fig. 9a). Finally, in-frame deletion of UDM1 but not UDM2 impaired the ability of ectopically expressed RNF168 to promote IR-induced 53BP1 foci in cells lacking endogenous RNF168 (Fig. 4e). We conclude that the RNF168 UDM1 is a high-affinity recognition module for K63-ubiquitylated histone H1. While the isolated UDM1 domain was distributed pan-cellularly and interacted with a range of proteins when overexpressed in cells, the only such protein showing DSB-stimulated K63 polyubiquitylation was an H1 isoform (Extended Data Fig. 9b, c), further suggesting that ubiquitylated H1 is a major receptor for RNF168 at DSB-modified chromatin.
a, Composition and reported functions of ubiquitin-dependent DSB recruitment modules (UDMs) in human RNF168. b, Binding of recombinant UDM1 to di-ubiquitin (Ub2) linkages. c, Binding of recombinant UDM1 and UDM2 to purified histones (H1.0 and H2A). d, Pull-down assays of Strep-tagged UDM1 and UDM2 modules expressed in U2OS cells. The migration of molecular weight markers (kDa) is indicated on the left. WCE, whole-cell extract. e, Representative images of shRNF168-expressing cells transfected with GFP–RNF168 constructs and exposed to IR (n = 2 experiments). Deletion of UDM1 (ΔUDM1) impairs restoration of 53BP1 foci by GFP–RNF168 (indicated by arrows). Scale bar, 10 μm. f, Model of RNF8–UBC13 function in ubiquitin-dependent signalling after DSBs. Me, methyl group; P, phosphate. b–d, Uncropped blots are shown in Supplementary Fig. 1.
Collectively, our findings suggest an integrated model for how RNF8 and UBC13 promote ubiquitin-dependent protein recruitment to DSB sites. We propose that H1-type linker histones represent key chromatin-associated RNF8 substrates whose UBC13-dependent K63-linked ubiquitylation at DSB-containing chromatin provide an initial binding platform for RNF168 via the UDM1 (Fig. 4f). RNF168 then ubiquitylates H2A at K13/K15 and possibly other proteins to trigger recruitment of DSB repair factors. The notable absence of K63-linked ubiquitin chains on H2A suggests that RNF168 does not efficiently modify H2A in conjunction with UBC13, but may instead catalyse formation of other ubiquitin chains or monoubiquitylation of the H2A K13/K15 mark. Indeed, RNF168 was recently shown to catalyse the formation of K27-linked ubiquitin chains, at least when overexpressed29. It is possible, however, that RNF168 cooperates with UBC13 in K63-linked ubiquitylation of other factors at damaged chromatin. While we observed no marked DSB-induced change in overall nuclear H1 mobility and no overt enrichment or depletion of H1 isoforms at DSB sites, the K63-ubiquitylated forms of H1 proteins were more loosely associated with chromatin than their unmodified counterparts (Extended Data Fig. 10a, b; data not shown). This suggests that in addition to triggering RNF168 recruitment, the DSB-associated K63 ubiquitylation of H1 may play a part in facilitating chromatin remodelling to allow efficient repair.
H1-type histones consist of almost 30% lysine residues28, and proteomic studies have shown that many of these can be ubiquitylated20,21,30. Because UBC13 only generates few de novo ubiquitylation marks, it is conceivable that RNF8–UBC13 mainly extends pre-existing H1 ubiquitylations in response to DNA damage, rendering identification of the key H1 sites targeted by RNF8–UBC13-dependent K63 ubiquitylation challenging. Although numerous post-translational modifications (PTMs) have been mapped on different H1 isoforms28, these marks have not so far been connected directly to the ‘histone code’ specifying protein recruitment to different chromatin states12,13. In addition to providing the first insights into the functional role of H1 ubiquitylation, our findings also show that linker histones can indeed form part of a dynamic histone code for DNA repair, wherein RNF8 and RNF168 function as writer and reader, respectively, of K63-ubiquitylated H1. The multitude of site-specific H1 PTMs raises the possibility that some of these marks may have an integral role in the histone code, an area of study warranting further investigation.
Methods
Plasmids
cDNAs encoding K63-Super-UIM (wild type and mutant) and the Vps27-based K63 binder18 containing C-terminal His6 tags were produced as synthetic genes (Eurofins) and inserted into pDONR221 by BP reactions (Invitrogen). By means of LR reactions (Invitrogen) the inserts were then transferred to the Champion pET104 BioEase Gateway Biotinylation System (Invitrogen) for recombinant protein production or pcDNA-DEST53 (Invitrogen) for GFP-tagged constitutive mammalian expression. For inducible expression of GFP-tagged K63-Super-UIM, the GFP-K63-Super-UIM complementary DNA was inserted into pcDNA4/TO (Invitrogen). Plasmids encoding HA-tagged wild-type and catalytically inactive (CI) RNF8 (C403S), wild-type and CI (C16S/C19S) forms of RNF168, and UBC13, as well as chimaeras between RNF8 and different E2 enzymes were described previously1,4,16. The *FHA mutation (R42A) in HA–RNF8ΔR–UBC13 was generated by site-directed mutagenesis. RNF8 constructs were made resistant to RNF8-siRNA by introducing three silent mutations (bold) in the siRNA targeting sequence (5′-TGCGGAGTATGAGTACGAG-3′) in the plasmids by site-directed mutagenesis. The RNF168 UDM1 (amino acids 110–201) and UDM2 (amino acids 419–487) fragments were amplified by PCR and inserted into either pTriEx-5 (Novagen) for Strep- and His-tagged expression in Escherichia coli and mammalian cells, or pEGFP-C1 (Clontech) for expression of GFP-tagged versions. The Strep–RNF168 UDM1 mutants used in this study (*UMI (L149A) and *MIU1 (A179G)) were generated using the QuikChange site-directed mutagenesis kit (Stratagene). Constructs encoding GFP–H1 isoforms were cloned by inserting the respective cDNAs into the BglII and BamHI sites of pEGFP-C1 (Clontech). A plasmid encoding HMGB1–GFP was provided by M. Bianchi. A Flag–HMGB1 expression construct was generated by inserting the HMGB1 open reading frame (ORF) into pFlag–CMV2 (Sigma). All constructs were verified by sequencing. Plasmid transfections were done with FuGene 6 (Promega) or Genejuice (Novagene), siRNA transfections were done with Lipofectamine RNAiMAX (Invitrogen), according to the manufacturers’ instructions.
siRNA
siRNA sequences used in this study were as follows. Non-targeting control (CTRL), 5′-GGGAUACCUAGACGUUCUATT-3′; UBC13, 5′-GAGCAUGGACUAGGCUAUATT-3′; RNF8, 5′-UGCGGAGUAUGAAUAUGAATT-3′; RNF168, 5′-GUGGAACUGUGGACGAUAATT-3′ or 5′-GGCGAAGAGCGAUGGAAGATT-3′; histone H1(#1), 5′-GCUACGACGUGGAGAAGAATT-3′; H1(#2), 5′-GCUCCUUUAAACUCAACAATT-3′; H1(#3), 5′-GAAGCCAAGCCCAAGGUUATT-3′; H1(#4), 5′-CCUUUAAACUCAACAAGAATT-3′; H1(#5), 5′-CCUUCAAACUCAACAAGAATT-3′; H1(#6), 5′-UCAAGAGCCUGGUGAGCAATT-3′; H1(#7), 5′-GGACCAAGAAAGUGGCCAATT-3′; H1(#8), 5′-GCAUCAAGCUGGGUCUCAATT-3′; H1(#9), 5′-CAGUGAAACCCAAAGCAAATT-3′; H1(#10) (specific for H1x), 5′-CCUUCAAGCUCAACCGCAATT-3′; 53BP1, 5′-GAACGAGGAGACGGUAAUATT-3′; USP7, 5′-GGCGAAGUUUUAAAUGUAUTT-3′; and USP9x, 5′-GCAGUGAGUGGCUGGAAGUTT-3′.
Cell culture
Human U2OS, HCT116 and RPE1 cells were obtained from ATCC. U2OS and HCT116 were cultured in DMEM containing 10% FBS and 1×penicillin–streptomycin, while RPE1 cells were grown in a 1:1 mixture of Ham’s F12 and DMEM supplemented with 10% FBS and 1×penicillin–streptomycin. Serum-starvation of RPE1 cells was done by incubating cells for 24 h in medium supplemented with 0.25% FBS. A HCT116 UBC13-knockout cell line was generated using CRISPR–Cas9 technology14,15. A donor plasmid bearing a splice acceptor site and a puromycin resistance marker, flanked by homology arms, was co-transfected with pX300 (ref. 14) targeting the GGCGCGCGGGAATCGCGGCG sequence within the first intron of the UBC13 gene. To generate cell lines capable of doxycycline-induced expression of GFP-tagged K63-Super-UIM, U2OS cells were transfected with GFP–K63-Super-UIM plasmid and pcDNA6/TR and positive clones were selected with Zeocin (Invitrogen) and Blasticidin S (Invitrogen). Stable U2OS cell lines expressing RNF8 or RNF168 shRNA in a doxycycline-inducible manner or Strep–HA–ubiquitin were described previously1,4,31. All cell lines were regularly tested for mycoplasma infection. Unless otherwise indicated, cells were exposed to DSBs using IR (4 Gy for microscopy experiments and 10 Gy for biochemical analyses) or laser micro-irradiation (as described previously32), and collected 1 h later.
Recombinant protein production
Purified biotinylated K63-Super-UIM wild-type and mutant proteins containing an N-terminal, biotinylated BioEase tag and a C-terminal His6-tag were obtained by expressing the proteins in an E. coli strain expressing the BirA biotin ligase. Bacteria were grown in LB medium containing 0.5 mM biotin, induced with 0.25 mM isopropyl-β-d-thiogalactoside (IPTG) for 3 h at 30 °C, and then lysed by French press. The K63-Super-UIM constructs were purified using immobilized metal affinity chromatography (IMAC) followed by size-exclusion chromatography (SEC). Purity and complete biotinylation of the proteins was verified by mass spectrometry. Recombinant Strep–His6–RNF168 UDM-1/2 was produced in Rosetta2(DE3)pLacI (Novagen) bacteria induced with 0.5 mM IPTG for 3 h at 30 °C, lysed using Bugbuster (Novagen) supplemented with Protease Inhibitor Cocktail without EDTA (Roche). The proteins were purified on Ni2+-NTA-agarose (Qiagen). Recombinant human UBA1, UBCH5c, UBC13, MMS2, RNF8 and ubiquitin used for in vitro ubiquitylation assays were purified as described8.
Antibodies
Antibodies used in this study included: UBC13 (#4919, Cell Signaling), MCM6 (sc-9843, Santa Cruz), 53BP1 (sc-22760, Santa Cruz), γ-H2A.X (05-636, Millipore; or 2577, Cell Signaling), H2A.X (2595, Cell Signaling), MDC1 (ab11171, Abcam), conjugated ubiquitin (FK2) (BML-PW8810-0500, Enzo Life Sciences), HA (11867423991, Roche; and sc-7392, Santa Cruz), Myc (sc-40, Santa Cruz), His6 (631212, Clontech), GFP (sc-9996, Santa Cruz; 11814460001, Roche), ubiquitin (sc-8017, Santa Cruz), histone H1.2 (ab17677, Abcam), histone H1x (A304-604A, Bethyl Labs), histone H1 (pan, #AE-4 clone) (ab7789, Abcam), histone H2A (07-146, Millipore), histone H2B (ab1790, Abcam), histone H3 (ab1791, Abcam), histone H4 (ab7311, Abcam), cyclin A (sc-751, Santa Cruz), actin (MAB1501, Millipore), BRCA1 (sc-6954, Santa Cruz), RNF168 for immunofluorescence (06-1130, Millipore) and antibody to RNF168 (ref. 5) used for immunoblots were gifts from D. Durocher. Antibody to RNF8 has been described previously1.
Immunochemical methods
For pull-down of K63-ubiquitylated proteins, cells were lysed in high-stringency buffer (50 mM Tris, pH 7.5; 500 mM NaCl; 5 mM EDTA; 1% NP40; 1 mM dithiothreitol (DTT); 0.1% SDS) containing 1.25 mg ml−1 N-ethylmaleimide, 50 μM DUB inhibitor PR619 (LifeSensors), and protease inhibitor cocktail (Roche). Recombinant biotionylated K63-Super-UIM (25 μg ml−1) was added immediately upon lysis, followed by sonication and centrifugation. Streptavidin M-280 Dynabeads (Invitrogen) was added to immobilize the K63-Super-UIM, and bound material was washed extensively in high-stringency buffer. A Benzonase (Sigma) and MNase (NEB) treatment step was included to remove any contaminating nucleotides. Proteins were resolved by SDS–PAGE and analysed by immunoblotting. Where indicated, bound complexes were subjected to deubiquitylation by incubation with USP2cc (1 μM, Boston Biochem) in DUB buffer (50 mM HEPES, pH 7.5; 100 mM NaCl; 1 mM MnCl2; 0.01% Brij-35; 2 mM DTT) overnight at 30 °C before boiling in Laemmli Sample Buffer. Immunoblotting, Strep-Tactin pull-downs, and chromatin enrichment were done essentially as described32. Briefly, Strep–RNF168 UDM pull-down experiments from cells were performed after lysing cells in EBC buffer (50 mM Tris, pH 7.4; 150 mM NaCl; 0.5% NP-40; 1 mM EDTA) containing 1.25 mg ml−1 NEM, 50 μM PR619 (LifeSensors) and protease inhibitor cocktail (Roche). The soluble fraction was subsequently used for immunoprecipitation using Strep-Tactin sepharose (IBA). After washing in EBC buffer, proteins were eluted and analysed by immunoblotting. To isolate Strep–HA–ubiquitin-conjugated proteins, cells were lysed in denaturing buffer (20 mM Tris, pH 7.5; 50 mM NaCl; 1 mM EDTA; 1 mM DTT; 0.5% NP-40; 0.5% sodium deoxycholate; 0.5% SDS) containing 1.25 mg ml−1 NEM, 50 μM PR619 (LifeSensors) and protease inhibitor cocktail (Roche). After sonication and centrifugation, Strep–HA–ubiquitin-conjugated proteins were immobilized on Strep-Tactin sepharose (IBA). After extensive washing in denaturing buffer, proteins were eluted and analysed by immunoblotting. For chromatin fractionation, cells were first lysed in buffer 1 (100 mM NaCl; 300 mM sucrose; 3 mM MgCl2; 10 mM PIPES, pH 6.8; 1 mM EGTA; 0.2% Triton X-100) containing protease, phosphatase and DUB inhibitors and incubated on ice for 5 min. After centrifugation, the soluble proteins were removed and the pellet was resuspended in buffer 2 (50 mM Tris-HCl, pH 7.5; 150 mM NaCl; 5 mM EDTA; 1% Triton X-100; 0.1% SDS) containing protease, phosphatase and DUB inhibitors. Lysates were then incubated 10 min on ice, sonicated, and solubilized chromatin-enriched fractions were collected after centrifugation.
Immunofluorescence staining and microscopy
For immunofluorescence staining, cells were fixed in 4% paraformaldehyde for 15 min, permeabilized with PBS containing 0.2% Triton X-100 for 5 min, and incubated with primary antibodies diluted in DMEM for 1 h at room temperature. After staining with secondary antibodies (Alexa Fluor; Life Technologies) for 1 h, coverslips were mounted in Vectashield mounting medium (Vector Laboratories) containing nuclear stain DAPI. Images of GFP–K63-Super-UIM were all obtained from a stable cell line where GFP–K63-Super-UIM was induced by incubating with 1 μg ml−1 doxycycline for approximately 24 h unless otherwise stated. Images were acquired with an LSM 780 confocal microscope (Carl Zeiss Microimaging) mounted on Zeiss-Axiovert 100M equipped with Plan-Apochromat 40×/1.3 oil immersion objective, using standard settings. Image acquisition and analysis was carried out with ZEN2010 software. For ImageJ-based image analysis, images were acquired with an AF6000 wide-field microscope (Leica Microsystems) equipped with a Plan-Apochromat 40×/0.85 CORR objective, using the same microscopic settings. Fluorescence intensities of the micro-irradiated region (demarcated by γ-H2AX positivity) and the nucleus were first corrected for the general image background. Using these values, relative recruitment to DNA damage sites (relative fluorescence units (RFUs)) was calculated by normalizing the nuclear-background-corrected signal at the micro-irradiated region to that of the nuclear background. Finally, the RFU of the protein of interest was normalized to the RFU of the γ-H2AX signal and plotted as the average of biological triplicates. Fluorescense recovery after photobleaching (FRAP) was performed essentially as described33. Briefly, U2OS cells stably expressing GFP–H1 were grown in glass-bottom dishes (LabTek) in the presence of CO2-independent medium. A 2-μm-wide rectangular strip spanning the entire width of the cell was bleached by excitation with the maximal intensity of a 488 nm laser line, after which 95 frames of the bleached region were acquired at 4 s intervals. Mean fluorescence intensities were processed, normalized and analysed as described33.
In vitro binding and ubiquitylation assays
Binding of K63-Super-UIM to di-ubiquitin (Ub2) linkages (Boston Biochem) was done by incubating 100 ng Ub2 with 2.5 μg K63-Super-UIM immobilized on Streptavidin M-280 Dynabeads (Invitrogen) in buffer A (50 mM Tris, pH 7.5; 10% glycerol; 400 mM NaCl; 0.5% NP40; 2 mM DTT; 0.1 mg ml−1 BSA). After extensive washing, bound complexes were resolved by SDS–PAGE and analysed by immunoblotting. Binding of RNF168 UDM1/2 to di-ubiquitin (Ub2) linkages was analysed by incubating 100 ng Ub2 with 5 μg Strep–RNF168–UDM1/2 immobilized on Strep-Tactin sepharose (IBA BioTAGnology) in buffer B (50 mM Tris, pH 8; 5% glycerol; 0.5% NP40; 2 mM DTT; 0.1 mg ml−1 BSA; 2 mM MgCl2, supplemented with 250 mM KCl for UDM1 binding and 100 mM KCl for UDM2 binding). After extensive washing, bound complexes were resolved by SDS–PAGE and analysed by immunoblotting. Where indicated, UDM1/2 binding to K63-linked Ub2 was analysed in the presence of increasing KCl concentrations (75 mM, 150 mM and 250 mM). To analyse binding of RNF168 UDM1/2 to recombinant histones, purified Strep–RNF168 UDM1/2 (10 μg) was pre-bound to Strep-Tactin sepharose in buffer C (for binding to H1.0) (50 mM, Tris pH 8; 5% glycerol; 150 mM KCl; 0.5% NP40; 2 mM DTT; 0.1 mg ml−1 BSA) or D (for binding to H2A) (50 mM, Tris pH 8; 5% glycerol; 75 mM KCl; 0.05% NP40; 2 mM DTT; 0.1 mg ml−1 BSA), and incubated with 500 ng recombinant histone H1.0 or H2A (New England Biolabs). Bound complexes were washed and analysed by immunoblotting. To analyse binding of LRM1 and LRM2 peptides to histone H1.0 or H2A, magnetic Streptavidin beads were incubated with buffer E (25 mM, Tris pH 8.5; 5% glycerol; 50 mM KCl; 0.5% TX-100; 1 mM DTT; 0.1 mg ml−1 BSA) in the absence (control) or presence of 1.5 μg purified, biotinylated RNF168 LRM1 (amino acids 110–133) or LRM2 (amino acids 463–485) peptide. Samples were then incubated with 250 ng recombinant H2A or H1.0 for 2 h at 4 °C, and immobilized complexes were washed and analysed by SDS–PAGE and Colloidal Blue staining (Invitrogen).
For in vitro ubiquitylation assays, histone-H1-containing oligonucleosomes (10 µM) were purified in the presence of 55 mM iodoacetamide, essentially as described previously34, with the exception that micrococcal nuclease digestion was stopped with 20 mM EGTA and dialysis was started right after the second homogenization in buffer containing 50 mM Tris, pH 7.5; 150 mM NaCl; 1 mM TCEP; and 340 mM sucrose. Dialysed samples were then incubated with DUB inhibitor (Ubiquitin-PA35, 20 μM) for 20 min at room temperature. Nuclesomes were incubated with 0.5 µM human UBA1, 5 µM UBCH5c, 1 µM UBC13–MMS2 complex, 5 µM RNF8351–485 fragment (purified as described previously8) and 75 µM ubiquitin in reaction buffer (50 mM Tris, pH 7.5; 100 mM NaCl; 3 mM ATP; 3 mM MgCl; 1 mM TCEP) at 31 °C. Samples were analysed by immunoblot analysis.
SILAC-based quantification of di-glycine-containing peptides
For SILAC experiments, U2OS or HCT116 cells were grown in medium containing unlabelled l-arginine and l-lysine (Arg0/Lys0) as the light condition, or isotope-labelled variants of l-arginine and l-lysine (Arg6/Lys4 or Arg10/Lys8) as the heavy condition36. SILAC-labelled HCT116 wild-type and UBC13-knockout cells were lysed in modified RIPA buffer (50 mM Tris-HCl, pH 7.5; 150 mM NaCl; 1% Nonidet P-40; 0.1% sodium-deoxycholate; 1 mM EDTA) supplemented with protease inhibitors (complete protease inhibitor mixture tablets, Roche Diagnostics) and N-ethylmaleimide (5 mM). Lysates were incubated for 10 min on ice and cleared by centrifugation at 16,000g. An equal amount of protein from the two SILAC states was mixed and precipitated by adding fivefold acetone and incubating at −20 °C overnight. Precipitated proteins were dissolved in denaturing buffer (6 M urea; 2 M thiourea; 10 mM HEPES, pH 8.0), reduced with DTT (1 mM) and alkylated with chloroacetamide (5.5 mM). Proteins were digested with lysyl endoproteinase C (Lys-C) for 6 h, diluted fourfold with water and digested overnight with trypsin. The digestion was stopped by addition of trifluoroacetic acid (0.5% final concentration), incubated at 4 °C for 2 h and centrifuged for 15 min at 4,000g. Peptides from the cleared solution were purified by reversed-phase Sep-Pak C18 cartridges (Waters Corporation). Diglycine-lysine modified peptides were enriched using the Ubiquitin Remnant Motif Kit (Cell Signaling Technology), according to the manufacturer’s intructions. Briefly, peptides were eluted from the Sep-Pak C18 cartridges with 50% acetonitrile, which was subsequently removed by centrifugal evaporation. Peptides were incubated with 40 μl of anti-di-glycine-lysine antibody resin in immunoaffinity purification (IAP) buffer for 4 h at 4 °C. Beads were washed three times with IAP buffer, two times with water and peptides eluted with 0.15% trifluoroacetic acid. Eluted peptides were fractionated by microcolumn-based strong cation exchange chromatography (SCX) and cleaned by reversed-phase C18 stage-tips.
SILAC K63-Super-UIM pull-down and in-gel digestion
SILAC-labelled cells were lysed in high-stringency RIPA buffer (50 mM Tris-HCl, pH 7.5; 500 mM NaCl; 1% Nonidet P-40; 0.1% sodium-deoxycholate; 1 mM EDTA) containing 1.25 mg ml−1 N-ethylmaleimide, 50 μM DUB inhibitor PR619 (LifeSensors), and protease inhibitor cocktail (Roche). Lysates from different SILAC states were separately incubated for 10 min on ice and cleared by centrifugation at 16,000g. Extracts (5 mg) were incubated for 4 h at 4 °C with K63-Super-UIM immobilized to Streptavidin beads (approximately 5 μg K63-Super-UIM per experiment). Beads were washed three times with high-stringency RIPA, beads from the different SILAC conditions were mixed, and proteins were eluted with SDS sample buffer, incubated with DTT (10 mM) for 10 min at 70 °C and alkylated with chloroacetamide (5.5 mM) for 60 min at 25 °C. Proteins were separated by SDS–PAGE using a 4–12% gradient gel and visualized with colloidal blue stain. Gel lanes were sliced into six pieces, and proteins were digested in-gel using standard methods37.
Mass spectrometry and data analysis
Peptides were analysed on a quadrupole Orbitrap (Q Exactive, Thermo Scientific) mass spectrometer equipped with a nanoflow HPLC system (Thermo Scientific). Peptide samples were loaded onto C18 reversed-phase columns and eluted with a linear gradient (1–2 h for in-gel samples, and 3–4 h for di-glycine-lysine enriched samples) from 8 to 40% acetonitrile containing 0.5% acetic acid. The mass spectrometer was operated in a data-dependent mode automatically switching between MS and MS/MS. Survey full scan MS spectra (m/z 300–1200) were acquired in the Orbitrap mass analyser. The 10 most intense ions were sequentially isolated and fragmented by higher-energy C-trap dissociation (HCD). Peptides with unassigned charge states, as well as peptides with charge state less than +2 for in-gel samples and +3 for di-glycine-lysine enriched samples were excluded from fragmentation. Fragment spectra were acquired in the Orbitrap mass analyser. Raw MS data were analysed using MaxQuant software (version 1.3.9.21). Parent ion and tandem mass spectra were searched against protein sequences from the UniProt knowledge database using the Andromeda search engine. Spectra were searched with a mass tolerance of 6 ppm in the MS mode, 20 ppm for MS/MS mode, strict trypsin specificity and allowing up to two missed cleavage sites. Cysteine carbamidomethylation was searched as a fixed modification, whereas amino-terminal protein acetylation, methionine oxidation and N-ethylmaleimide modification of cysteines, and di-glycine-lysine were searched as variable modifications. Di-glycine-lysines were required to be located internally in the peptide sequence. Site localization probabilities were determined using MaxQuant (PTM scoring algorithm) as described previously38. A false discovery rate of less than 1% was achieved using the target-decoy search strategy39 and a posterior error probability filter. Information about previously known protein–protein interactions among putative UBC13-dependent K63-Super-UIM interacting proteins was extracted using the HIPPIE database40 (version 1.6), and interactions were visualized in Cytoscape41. The Gene Ontology (GO) biological process term analysis for UBC13-dependent K63-Super-UIM interacting proteins was filtered for categories annotated with at least 20 and not more than 300 genes. Redundant GO terms (less than 30% unique positive-scoring genes compared to more significant GO term) were removed and the five most significant (Fisher’s exact t-test) remaining GO term categories depicted. To determine the variation within the quantification of ubiquitin linkage types, an F-test was performed and the P values were adjusted using the Bonferroni method. A significant difference in the variances between K48 and K11, and K48 and K6 ubiquitin linkages was detected. To test the significance of the difference between the SILAC ratios measured for ubiquitin linkage types, the Welch two-sample t-test was performed and the obtained P values were adjusted using the Bonferroni method.
References
Mailand, N. et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131, 887–900 (2007)
Huen, M. S. et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131, 901–914 (2007)
Kolas, N. K. et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318, 1637–1640 (2007)
Doil, C. et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136, 435–446 (2009)
Stewart, G. S. et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136, 420–434 (2009)
Jackson, S. P. & Durocher, D. Regulation of DNA damage responses by ubiquitin and SUMO. Mol. Cell 49, 795–807 (2013)
Hofmann, R. M. & Pickart, C. M. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96, 645–653 (1999)
Mattiroli, F. et al. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell 150, 1182–1195 (2012)
Fradet-Turcotte, A. et al. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature 499, 50–54 (2013)
Gatti, M. et al. A novel ubiquitin mark at the N-terminal tail of histone H2As targeted by RNF168 ubiquitin ligase. Cell Cycle 11, 2538–2544 (2012)
Panier, S. et al. Tandem protein interaction modules organize the ubiquitin-dependent response to DNA double-strand breaks. Mol. Cell 47, 383–395 (2012)
Kouzarides, T. Chromatin modifications and their function. Cell 128, 693–705 (2007)
Jenuwein, T. & Allis, C. D. Translating the histone code. Science 293, 1074–1080 (2001)
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013)
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013)
Bekker-Jensen, S. et al. HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nature Cell Biol. 12, 80–86,–1–12 (2010)
Huen, M. S. et al. Noncanonical E2 variant-independent function of UBC13 in promoting checkpoint protein assembly. Mol. Cell. Biol. 28, 6104–6112 (2008)
Sims, J. J. et al. Polyubiquitin-sensor proteins reveal localization and linkage-type dependence of cellular ubiquitin signaling. Nature Methods 9, 303–309 (2012)
van Wijk, S. J. et al. Fluorescence-based sensors to monitor localization and functions of linear and K63-linked ubiquitin chains in cells. Mol. Cell 47, 797–809 (2012)
Wagner, S. A. et al. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell. Proteomics 10, M111.013284 (2011)
Kim, W. et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 (2011)
Petroski, M. D. et al. Substrate modification with lysine 63-linked ubiquitin chains through the UBC13-UEV1A ubiquitin-conjugating enzyme. J. Biol. Chem. 282, 29936–29945 (2007)
Christensen, D. E., Brzovic, P. S. & Klevit, R. E. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nature Struct. Mol. Biol. 14, 941–948 (2007)
Windheim, M., Peggie, M. & Cohen, P. Two different classes of E2 ubiquitin-conjugating enzymes are required for the mono-ubiquitination of proteins and elongation by polyubiquitin chains with a specific topology. Biochem. J. 409, 723–729 (2008)
Catez, F., Ueda, T. & Bustin, M. Determinants of histone H1 mobility and chromatin binding in living cells. Nature Struct. Mol. Biol. 13, 305–310 (2006)
Murga, M. et al. Global chromatin compaction limits the strength of the DNA damage response. J. Cell Biol. 178, 1101–1108 (2007)
Pinato, S., Gatti, M., Scandiuzzi, C., Confalonieri, S. & Penengo, L. UMI, a novel RNF168 ubiquitin binding domain involved in the DNA damage signaling pathway. Mol. Cell. Biol. 31, 118–126 (2011)
Harshman, S. W., Young, N. L., Parthun, M. R. & Freitas, M. A. H1 histones: current perspectives and challenges. Nucleic Acids Res. 41, 9593–9609 (2013)
Gatti, M. et al. RNF168 promotes noncanonical K27 ubiquitination to signal DNA damage. Cell Reports 10, 226–238 (2015)
Povlsen, L. K. et al. Systems-wide analysis of ubiquitylation dynamics reveals a key role for PAF15 ubiquitylation in DNA-damage bypass. Nature Cell Biol. 14, 1089–1098 (2012)
Danielsen, J. M. et al. Mass spectrometric analysis of lysine ubiquitylation reveals promiscuity at site level. Mol. Cell. Proteomics 10, M110.003590 (2011)
Poulsen, M., Lukas, C., Lukas, J., Bekker-Jensen, S. & Mailand, N. Human RNF169 is a negative regulator of the ubiquitin-dependent response to DNA double-strand breaks. J. Cell Biol. 197, 189–199 (2012)
Bekker-Jensen, S., Lukas, C., Melander, F., Bartek, J. & Lukas, J. Dynamic assembly and sustained retention of 53BP1 at the sites of DNA damage are controlled by Mdc1/NFBD1. J. Cell Biol. 170, 201–211 (2005)
Hernández-Muñoz, I. et al. Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc. Natl Acad. Sci. USA 102, 7635–7640 (2005)
Ekkebus, R. et al. On terminal alkynes that can react with active-site cysteine nucleophiles in proteases. J. Am. Chem. Soc. 135, 2867–2870 (2013)
Ong, S. E. et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 (2002)
Jensen, O. N., Wilm, M., Shevchenko, A. & Mann, M. Sample preparation methods for mass spectrometric peptide mapping directly from 2-DE gels. Methods Mol. Biol. 112, 513–530 (1999)
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature Biotechnol. 26, 1367–1372 (2008)
Elias, J. E. & Gygi, S. P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nature Methods 4, 207–214 (2007)
Schaefer, M. H. et al. HIPPIE: Integrating protein interaction networks with experiment based quality scores. PLoS ONE 7, e31826 (2012)
Cline, M. S. et al. Integration of biological networks and gene expression data using Cytoscape. Nature Protocols 2, 2366–2382 (2007)
Acknowledgements
We thank D. Durocher and M. Bianchi for providing reagents and J. Lukas for helpful discussions. This work was supported by grants from the Novo Nordisk Foundation (grants NNF14CC0001 and NNF12OC0002114), European Research Council, Nederlandse Organisatie voor Wetenschappelijk Onderzoek- Chemische Wetenschappen (NWO-CW), The Danish Cancer Society, and The Danish Council for Independent Research.
Author information
Authors and Affiliations
Contributions
T.T. initiated the project, designed and performed cell biological and biochemical experiments, and analysed data; A.R. and S.H. performed cell biological and biochemical experiments and analysed data; T.W. generated UBC13-knockout cells, designed and performed mass spectrometry experiments and analysed the data; M.U. performed in vitro ubiquitylation assays with purified nucleosomes; B.V. helped T.T. with biochemical experiments; T.N. analysed mass spectrometry data; T.K.S. supervised M.U.; C.C. supervised T.W. and T.N., designed mass spectrometry experiments and analysed the data; S.B.-J. performed and designed experiments and analysed data; N.M. conceived and supervised the project, designed experiments, analysed data and wrote the manuscript with input from the other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Protein recruitment to DSB sites in UBC13-knockout cells.
a, b, Representative images of HCT116 wild-type (WT) or UBC13-knockout (KO) cells exposed to IR. Where indicated, cells were transfected with HA–RNF8 plasmid before IR (n = 2 experiments). c, Constructs used in Fig. 1d and their ability to restore IR-induced 53BP1 foci in UBC13-knockout cells. d, Representative images of HCT116 UBC13-knockout cells transfected with non-targeting control (CTRL) or RNF8 siRNAs and subsequently with plasmids encoding UBC13 or siRNA-resistant UBC13–RNF8 fusion constructs (c) (n = 2). e, Representative images of UBC13-knockout cells transfected with plasmids encoding wild-type or catalytically inactive (CI) HA–RNF168 (n = 2). Expression of HA–RNF168 wild type restores IR-induced 53BP1 foci formation (arrows). Scale bars, 10 μm.
Extended Data Figure 2 RNF8- and UBC13-dependent K63-linked ubiquitylation at DSB sites.
a–c, Representative images of U2OS cells stably expressing GFP–K63-Super-UIM, transfected with siRNAs where indicated, and exposed to laser micro-irradiation or IR (n = 3). d, Representative images of U2OS cells stably expressing GFP- and nuclear localization signal (NLS)-tagged Vps27-UIM (with high affinity for binding to K63-linked ubiquitin18) transfected with the indicated siRNAs and exposed to laser micro-irradiation (n = 2). e, Loss of RNF8 or UBC13 has no impact on 53BP1 abundance. Immunoblot analysis of whole-cell extracts (WCEs) of U2OS cells transfected with indicated siRNAs. f, RNF8, but not RNF168, is modified by K63-linked ubiquitin chains. K63-Super-UIM pull-downs from U2OS cells transfected with empty vector (−), HA-tagged RNF8 (wild type (WT) or catalytically inactive (CI)) or HA–RNF168 plasmids were immunoblotted (IB) with indicated antibodies. Scale bars, 10 μm. e, f, Uncropped blots are shown in Supplementary Fig. 1.
Extended Data Figure 3 Experimental replicates of SILAC-based quantification of di-glycine-containing peptides and K63-Super-UIM pull-downs from wild-type and UBC13-knockout cells.
a, Schematic outline of SILAC-based mass spectrometry approach to quantify di-glycine-containing peptides in HCT116 wild-type (WT) and UBC13-knockout (KO) cells. b, c, Proportional Venn diagrams showing overlap between all identified di-glycine-containing peptides (b) and those with a SILAC ratio (UBC13-knockout/wild-type cells) <0.5 (c) in two independent experiments (Exp.) performed as shown in a (Supplementary Table 1). d, Scatter plot showing correlation between SILAC ratios of di-glycine-containing peptides. The Pearson’s correlation coefficient (R) is indicated. e, Schematic outline of SILAC-based mass spectrometry approach to identify UBC13-dependent K63-ubiquitylation targets in unperturbed HCT116 wild-type and UBC13-knockout cells. f, g, Proportional Venn diagrams showing overlap between all proteins identified in K63-Super-UIM pull-downs (f) and those with a SILAC ratio (UBC13-knockout/wild-type cells) <0.5 (g) in two independent experiments performed as shown in e (Supplementary Table 2). h, Scatter plot showing correlation between SILAC ratios of proteins identified in two experiments. The Pearson’s correlation coefficient (R) is indicated.
Extended Data Figure 4 Analysis of UBC13-dependent K63-ubiquitylated proteins in unperturbed cells and in response to DNA damage.
a, K63 linkages from extracts of HCT116 wild-type (WT) and UBC13-knockout (KO) cells were enriched by K63-Super-UIM pull-down (Supplementary Table 2). Interaction network shows proteins enriched at least twofold in wild-type cells. Proteins involved in UBC13-dependent activation of NF-κB signalling are highlighted in red. b, Functional annotation of potential UBC13-dependent K63-ubiquitylated proteins (a), showing enriched Gene Ontology (GO) biological process terms. c, Schematic outline of SILAC-based mass spectrometry approach to identify targets of K63 ubiquitylation in response to IR-induced DSBs. d, SILAC ratios of selected proteins from U2OS cells treated as in c. Data from a representative experiment are shown (n = 3).
Extended Data Figure 5 DSB-induced K63 ubiquitylation of H1-type linker histones.
a, Analysis of K63-linked ubiquitylation of histone H1.2 in RPE1 cells growing exponentially or kept quiescent by serum starvation. b, c, K63-Super-UIM pull-downs from U2OS cells exposed or not to IR were immunoblotted (IB) with the indicated antibodies. d, U2OS cells or U2OS cells stably expressing Strep–HA–ubiquitin (U2OS/Strep–Ub) were transfected with the indicated siRNAs and exposed or not to IR. Whole-cell extracts (WCEs) and Strep–ubiquitin-conjugated proteins immobilized on Strep-Tactin beads under denaturing conditions were analysed by immunoblotting. e, Proteins interacting with endogenous RNF8. U2OS cells stably expressing RNF8 shRNA in a doxycycline (DOX)-inducible manner1 was grown in light (L) or heavy (H) SILAC medium. Cells growing in light medium were induced to express RNF8 shRNA by treatment with DOX. Both cultures were then exposed to IR and processed for immunoprecipitation with RNF8 antibody. Bound proteins were analysed by mass spectrometry. Proteins displaying the highest H/L SILAC ratios are listed. a–d, The migration of molecular weight markers (kDa) is indicated on the left. Uncropped blots are shown in Supplementary Fig. 1.
Extended Data Figure 6 Knockdown of H1-type histones impairs accumulation of K63-linked ubiquitin conjugates, RNF168 and BRCA1 at DSB sites.
a, Immunoblot analysis of U2OS cells transfected sequentially with plasmids encoding GFP-tagged histone H1 isoforms (H1.0–H1.5) and the indicated siRNAs. Knockdown efficiency of the siRNAs (#1, #4, #5 and #9) used in the pan-H1 siRNA cocktail to reduce global H1 expression level is highlighted in red boxes. b, Immunoblot analysis of U2OS cells transfected with H1 siRNA cocktail (a). c–g, Representative images of siRNA-transfected U2OS/GFP–K63-Super UIM (c) or U2OS cells (d–g) exposed to IR or laser micro-irradiation (n = 2). h, Analysis of IR-induced γ-H2AX ubiquitylation (Ub) by RNF168 (marked by arrow) in chromatin fractions of U2OS cells transfected with indicated siRNAs. Scale bars, 10 μm. a, b, h, Uncropped blots are shown in Supplementary Fig. 1.
Extended Data Figure 7 HMGB1 overexpression impairs the RNF8/RNF168-dependent signalling response at DSB sites at the level of K63 ubiquitylation and RNF168 recruitment.
a, b, Representative images of U2OS cells co-transfected with constructs encoding HMGB1–GFP and wild-type (WT) or catalytically inactive (CI) HA–RNF168 and exposed to IR (n = 3). Arrows indicate cells expressing HA–RNF168 CI, in which 53BP1 foci formation is not restored. c, Representative images of U2OS/GFP–K63-Super-UIM cells transfected with Flag–HMGB1 construct and subjected to laser micro-irradiation (n = 3). Flag–HMGB1-expressing cells show reduced K63 ubiquitylation at DSB sites (indicated by arrows). Scale bars, 10 μm.
Extended Data Figure 8 Interaction of RNF168 UDM1 with K63-ubiquitylated H1.
a, Immunoblot analysis of immobilized recombinant RNF168 UDM1 or UDM2 incubated with K63 linked di-ubiquitin (Ub2) in the presence of increasing salt concentrations (75 mM, 150 mM and 250 mM KCl, respectively). b, Binding of immobilized recombinant Strep–UDM2 or empty Strep-Tactin beads to indicated di-ubiquitin (Ub2) linkages was analysed by immunoblotting. c, Biotinylated peptides corresponding to the LRM1 and LRM2 motifs in human RNF168 were analysed for binding to recombinant H2A or H1.0 in vitro by Streptavidin pull-down followed by SDS–PAGE and Colloidal Blue staining. The migration of molecular weight markers (kDa) is indicated on the left. d, Sequence of the UDM1 region in human RNF168, showing the location of the LRM1, UMI and MIU1 motifs. Acidic amino acids are highlighted in red. The sequence corresponding to the LRM1 peptide (c) and mutations introduced to generate UDM1 *UMI and *MIU1 (f) are indicated. e, f, Pull-down assays of Strep-tagged UDM1 and UDM2 constructs expressed in U2OS cells. a–c, e, f, Uncropped blots are shown in Supplementary Fig. 1.
Extended Data Figure 9 RNF168 UDM1 recognizes ubiquitylated forms of H1 but not core histones.
a, Pull-downs of Strep-tagged RNF168 UDM1 expressed in U2OS cells were immunoblotted (IB) with antibodies to indicated histones. b, Localization pattern of GFP-tagged UDM1 expressed in U2OS cells. Scale bar, 10 μm. c, Venn diagram showing overlap between proteins displaying increased K63-linked ubiquitylation after IR (SILAC ratio (IR/mock) >1.5) and proteins showing potential interaction with overexpressed GFP–UDM1 (SILAC ratio (GFP–UDM1/mock) >1.5). Only one protein, histone H1x, was common to both of these subsets of cellular proteins. a, Uncropped blots are shown in Supplementary Fig. 1.
Extended Data Figure 10 Impact of DSBs on H1 chromatin association.
a, FRAP analysis of U2OS cells stably expressing GFP–H1.2 and exposed or not to IR (10 Gy). Individual data points represent mean values from ten independent measurements and error bars represent twice the s.d. b, U2OS cells left untreated or exposed to IR were lysed in EBC buffer. Soluble and resolubilized, EBC-insoluble fractions were incubated with recombinant K63-Super-UIM and washed thoroughly. Bound material and input fractions were analysed by immunoblotting with indicated antibodies. b, Uncropped blots are shown in Supplementary Fig. 1.
Supplementary information
Supplementary Figure 1
This file contains the uncropped scans of immunoblot data with size marker indications. (PDF 5793 kb)
Supplementary Table 1
This table contains the Mass spectrometry data for SILAC-based quantification of di-glycine containing peptides isolated from HCT116 WT and Ubc13 KO cells. See Extended Data Fig. 3a for experimental set-up. (XLSX 1569 kb)
Supplementary Table 2
This table contains the Quantitative mass spectrometry data for proteins identified in K63-Super-UIM pull-downs from HCT116 WT and Ubc13 KO cells. See Extended Data Fig. 3e for experimental set-up. (XLSX 780 kb)
Rights and permissions
About this article
Cite this article
Thorslund, T., Ripplinger, A., Hoffmann, S. et al. Histone H1 couples initiation and amplification of ubiquitin signalling after DNA damage. Nature 527, 389–393 (2015). https://doi.org/10.1038/nature15401
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature15401
- Springer Nature Limited
This article is cited by
-
Genes expression profiles in vascular cambium of Eucalyptus urophylla × Eucalyptus grandis at different ages
BMC Plant Biology (2023)
-
A neutrophil mimicking metal-porphyrin-based nanodevice loaded with porcine pancreatic elastase for cancer therapy
Nature Communications (2023)
-
CAMK2D serves as a molecular scaffold for RNF8-MAD2 complex to induce mitotic checkpoint in glioma
Cell Death & Differentiation (2023)
-
De-ubiquitination of SAMHD1 by USP7 promotes DNA damage repair to overcome oncogenic stress and affect chemotherapy sensitivity
Oncogene (2023)
-
Non-proteolytic ubiquitylation in cellular signaling and human disease
Communications Biology (2022)