Abstract
IgA nephropathy (IgAN) is a complex syndrome with high genetic heterogeneity. More recently, a genome-wide association study (GWAS) from Southern Han population revealed that variants within 8p23.1, where the DEFA genes encoding a-defensins assembled, were associated with susceptibility to IgAN. To replicate the association and fine-map the genetic variants, a case–control genetic study from an independent Northern Han cohort was conducted. A total of 60 single-nucleotide polymorphisms in a region spanning 350 kb encompassing the DEFA genes cluster were analyzed in 2096 individuals. Copy number variations of DEFA1A3 within the loci were also checked for the independent association. Functional significance of the associated variants was further examined by the in silico method as well as by cis-acting expression quantitative trait loci analysis with mRNA. It showed that 17 out of 60 (28.3%) variants were associated with susceptibility to IgAN. Two independent signals with functional potentials were discovered (rs2738058, P=4.64 × 10−5, odds ratio (OR)=0.76, 95% confidence interval (CI) 0.66–0.87 and rs9644778, P=4.78 × 10−3, OR=1.21, 95% CI 1.06–1.39). Besides, marginally significant association of rs9644778 risk genotype with lower proportion of gross hematuria (CC+CA vs AA 35.2% vs 30.2%, P=0.073) was observed. In conclusion, DEFA gene polymorphisms have potentially pathogenic roles in IgAN, and the role of mucosal immunity in the pathogenesis of IgAN has to be emphasized.
Similar content being viewed by others
Introduction
IgA nephropathy (IgAN), the most common cause of end-stage renal disease in Asian population, is a genetically complex and clinically heterogeneous renal disease.1 It is characterized as immune complexes containing IgA deposited in the glomerular mesangium, in renal biopsy. Although the pathogenesis of IgAN is not clear, the four-hit model (glycosylation deficiency of IgA1, synthesis of antiglycan IgG antibodies, formation of IgA1-containing immune complex and mesangial deposition of immune complex) suggesting abnormality in the innate and adaptive immunity may be involved in its pathogenesis.2 The familial aggregation, different prevalence among different ethnic populations and variable disease courses in individual patients support that genetic predisposing factors contribute to IgAN susceptibility and development.3, 4, 5, 6, 7, 8
The IgAN susceptibility loci discovered in the genome-wide association studies (GWASs) were mainly from eight genomic regions, including the MHC region at 6p21, DEFA locus at 8p23, TNFSF13 locus at 17p23, HORMAD2 locus at 22q12, CFH/CFHR locus at 1q32, ITGAM-ITGAX locus at 16p11, VAV3 locus at 1p13 and CARD9 locus at 9q34.5, 6, 9 Genetic studies have provided a glimpse into IgAN pathogenesis and identified the involvement of defects in adaptive immunity, innate immunity, mucosal immunity and the alternative complement pathway.2 Although the role of mucosal immunity in IgAN is still controversial, the loci including TNFSF13, DEFA and HORMAD2 discovered in the GWAS of IgAN were all likely implicated in the response of mucosal immunity, suggesting a deregulation role due to genetic defects in mucosal immunity. More recently, GWAS conducted in 4 discovery cohorts and 10 replication cohorts covering European and East Asian, the two major ancestries, confirmed that DEFA locus is strongly associated with IgAN susceptibility.9 DEFA locus, in which six human a-defensin peptides are encoded by only five DEFA genes, encodes α-defensins, which are antimicrobial peptides involved in maintenance of the intestinal mucosal barrier or regulation of the mucosal immune response.10 The DEFA1 and DEFA3 genes are interchangeable occupants of a 19-kb copy-variable unit. Both single-nucleotide polymorphisms (SNPs) and copy number variations (CNVs) can associate with diseases and influence gene expression. It has been reported that they have an effect on several diseases.11, 12, 13, 14, 15, 16, 17, 18 Two GWASs discovered variants in the association of DEFA locus with IgAN susceptibility. However, the top SNPs were inconsistent.6, 9 Replication by others can ensure that the original findings are robust and can provide a more accurate estimate of the likely effect size. Besides, variants discovered by previous GWASs were within the noncoding sequences of a gene or intergenic regions. To integrate expression data from diverse tissue types, genome and experimental-based annotation data covering noncoding and clinical phenotype association analysis are prominent. From a more important aspect for disease pathogenesis, the underlying functional significance needs to be further addressed.
In this study, we focus on exploring independent susceptibility variants in the DEFA region in a bid to provide evidence DEFA gene involvement in the pathogenesis of IgAN.
Results
The significant SNPs in the DEFA region associated with IgAN susceptibility
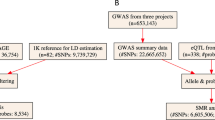
A total of 60 SNPs in the DEFA region were analyzed (Figure 1). Seventeen (28.3%) of them were significantly associated with the susceptibility to IgAN (P<0.05; Table 1). Among them, rs2738058 located in 14 kb 3' of the DEFA1 intergenic region (GRCh37/hg19) showed the strongest association signal (P=4.64 × 10−5, odds ratio (OR)=0.76, 95% confidence interval (CI) 0.66–0.87). The association of top SNP rs2738048 identified in the previous GWAS with susceptibility to IgAN was also confirmed (P=2.13 × 10−3, OR=0.81, 95% CI 0.71–0.93).6 Power calculations indicated that we had at least 98.6% power to detect loci with allelic frequencies >0.10 and relative risk >1.5 assuming an a-level of 0.05 (P<0.05) in the current study. Detailed power calculation data for the identified 17 variants are shown in Supplementary Table 1.
Further linkage disequilibrium (LD) analysis in the control groups revealed that these 17 SNPs can be grouped into four clusters (Figure 2). The most strongly associated SNPs from each block were chosen as tag SNPs, including rs2738058 (a variant at 14 kb in 3' of DEFA1), rs2702910 (a variant 11 kb 3' of DEFA1), rs4300027 (a variant 4.8 kb 5' of DEFA3) and rs9644778 (a variant 3.2 kb 3' of DEFA5), and together with rs2738048 (a variant 12 kb 3' of DEFA1), which was the top signal from the GWAS from Southern Han, for identifying the independent susceptibility variant associated with IgAN.
The LD maps of the 17 identified disease-associated SNPs in 902 healthy controls. The figure was generated using the Haploview software, and the LD was estimated using the r2 and the blocks defined according to CIs.38 The SNP marked with cross was the previous GWAS identified, rs2738048. The most strongly associated SNPs from each block, which were chosen as tag SNPs, are marked in the boxes.
To test the independence of the identified five SNPs, a stepwise logistic regression analysis was performed using the forward method. rs2738058 showed the strongest association with IgAN (OR=1.25(1.08–1.43); P=5.82 × 10−5). The second independent locus was rs9644778 (OR=1.15(1.00–1.32); P=0.015; Supplementary Table 2). Conditional logistic regression analysis was also performed to investigate the independent effects of the identified five SNPs, which showed the same results (Supplementary Table 3). Accordant with the data from the stepwise logistic regression analysis and conditional logistic regression analysis, rs2738058 and rs9644778 were in low LD (r2=0.16). These suggest that rs2738058 and rs9644778 were independent variants associated with susceptibility to IgAN.
Association analysis between DEFA1A 3 CNVs and SNPs
To determine whether the association of the SNPs within DEFA loci with IgAN is independent or is due to their LD with DEFA1A3 CNVs, we conducted a two-stage association analysis considering DEFA1A3 copy numbers (CNs).
In the first stage, we correlated the two different variants (SNP and CNV) in 45 HapMap Chinese Han Beijing (CHB) individuals by χ2 test and calculated the Pearson’s correlation coefficients for LD estimation. The previously GWAS-reported variant rs2738048, which was located at 12 kb in 3' of DEFA1, was significantly associated with CNs of DEFA1A3, with the risk allele correlating with lower DEFA1A3 CNs (P=0.012, r=−0.370). Another marginal significance was observed between rs2702910 risk genotypes and lower CNs (P=0.124, r=−0.233). For validation, we further checked DEFA1A3 CNs of 70 healthy controls with Northern Chinese ethnicity and their associations with the two tag SNPs. Similar to data from HapMap CHB, the correlation coefficients observed in the current Beijing controls were also approximately −0.3 (P=0.012 and r=−0.298 for rs2738048, P=0.033 and r=−0.255 for rs2702910; Figure 3). However, none of rs2738058, rs9644778 and rs4300027 genotypes was observed to correlate with CNs of DEFA1A3 in any of the cohorts (Supplementary Figure 1).
Regulatory variant annotation and cis-eQTL (expression quantitative trait loci) analysis exploring possible functional loci
As the majority of associated variants mapped to noncoding intergenic regions, we first checked the functionality of lead SNPs, including rs2738048, rs2738058 and rs9644778, by rSNPBase. In the rSNPBase, all SNPs were annotated with references to experimentally supported regulatory elements, which improved the reliability.19
SNP annotations indicated that both rs2738058 and rs9644778 rather than rs2738048 were regulatory SNPs. Considering regulation types, by ChIP-seq technology, it was observed that rs2738058 affects proximal transcriptional regulation, whereas rs9644778 affects distal transcriptional regulation;20 rs2738058 was located within different histone-marked regions in multiple cell lines, which was experimentally supported by the Broad Institute using ChIP-seq technology from ENCODE data. In T-lymphocyte and leukemia cell lines, rs2738058 was located within H3K4me1, H3K9ac, H3K9ac and H3K27ac histone-marked regions. In lung epithelial cells, B lymphocytes, embryonic stem cells, liver carcinoma cells and myoblast cells, rs2738058 was located within H4K20me1 histone-marked regions. rs2738058 was located within H3K9me1 histone-marked regions in leukemia cells and keratinocyte cells. rs9644778 affects distal transcriptional regulation in breast adenocarcinoma MCF-7 cell lines using ChIA-PET (Genome Institute of Singapore, Singapore) tools.20
It was reported that the functional SNP most strongly supported by experimental evidence was possibly an SNP in LD with the reported association rather than the reported SNP itself.21 We further located the set of SNPs that were in strong LD (r2⩾0.8) with leading SNPs in all HapMap populations, and annotated the SNPs using a scoring scheme integrated in Regulome DB. A total of 4 out of 7 SNPs and 6 out of 13 SNPs were in strong LD with regulatory SNPs rs2738058 and rs9644778. This was supported by more than one single experimental modality (for example, motif-based predictions, DNase I hypersensitivity peaks and ChIP-seq peaks).20 Among the many predicted regulatory SNPs, rs2615787 (17 kb 5' of DEFA4, in LD with rs2738058, r2=0.89 in Chinese Hans living in Beijing) had the highest Regulome DB score 2b (namely, containing predicted transcription factor-binding motifs supported by ChIP-seq, having protein-binding sequences as demonstrated by ChIP-seq, or located in the chromatin structures or the histone modification regions). rs2615787 overlapped with a region of predicted transcription factor-binding motifs (STAT,22 HSF222, 23), protein (CTCF,20 TAL124) binding, ChIP-seq peak (in 13 types of cell lines, that is, B lymphocyte–Gm12878 and Monocyte–Monocd14ro1746), and a DNase I hypersensitivity peak20 (see Supplementary Table 4).
Supporting eQTL data surely will support a higher functional significance of the associated variants in addition to TF binding, matched TF motif, matched DNase footprint and DNase peak. Gene expression in Epstein–Barr virus-transformed lymphoblastoid B-cell lines from 210 unrelated HapMap samples was utilized to study the correlation between SNP genotypes and gene expression. Risk genotypes of both rs2738058 and rs4300027 showed significant correlation with low DEFA3 mRNA expression (P=1.63 × 10−4, r=0.265 and P=8 × 10−3, r=0.187, respectively).
Genetic association of the variants with specific subphenotypes
Associations of the independent variants with clinical and demographic features of patients with IgAN were further analyzed in our cohort. As presented in Table 2, rs9644778 risk genotypes (CC+CA) showed significant association with early-onset (age⩽18) IgAN (17.5% vs 9.4%, P=4.69 × 10−5, OR=0.49, 95% CI 0.34–0.69) and marginally significant association with higher proportions of gross hematuria (CC+CA vs AA 35.2 vs 30.2%, P=7.3 × 10−2).
Discussion
DEFA gene cluster codes for small peptides that are important effector molecules in innate and adaptive immunity. In humans, there are two families of defensins, α-defensins and β-defensins. α-Defensins are mainly expressed in the neutrophils and in the paneth cells of the intestine, whereas β-defensins are expressed by the epithelial tissue.10 Elevated level of α-defensins was found in type 1 diabetic patients with nephropathy,11 rheumatoid arthritis,12 systemic lupus erythematosus13 and Sjögren’s syndrome.14 CNVs and gene polymorphisms of DEFA1 or/and DEFA3 are found associated with multiple immune-related diseases such as Crohn’s disease.17 All the above discoveries highlighted the critical role of α-defensins in autoimmunity, including mucosal innate immunity. SNPs together with CNVs may contribute to the making up of the IgAN susceptibility background.
For IgAN, it was suggested that innate, adaptive and mucosal immunity were involved in inherited defects in IgAN.2 Recent GWASs have revealed that the DEFA locus at 8p23.1 was associated with susceptibility to IgAN in the Southern Chinese Han population.6 In the present study, we investigated 60 SNPs in 8p23.1 spanning 350 kb encompassing the DEFA gene cluster and its upstream region. A total of 17 (28.3%) identified SNPs were observed to significantly associate with the susceptibility to IgAN. Two independent signals, rs2738058 and rs9644778, discovered in DEFA locus were in low LD, and survived from stepwise analysis and conditional analysis. Multiple variants identified in DEFA locus suggested a likely true effect of DEFA SNP effects on IgAN susceptibility. Besides, our results also indicated that rs2738048 disease association effects may be partially inflated by DEFA1A3 CNVs. Just like SNPs, CNVs are also an important source of genetic variation in the human genome. CNVs are expected to have an important role in the genetic basis of complex diseases such as autoimmune diseases and autism.25, 26, 27 DEFA1A3 locus on human chromosome 8p23.1 is also a region that exhibits multiallelic CNVs. Future studies addressing the respective contributions of CNVs and SNPs in DEFA locus to IgAN are deserved. More recently, Black et al.28 suggested that different structural haplotypes in DEFA1A3 locus were associated with DEFA1A3 CNs. However, they mainly focused on a 4.1-kb region in high LD from a Caucasian population. The top-tagging SNP rs4300027 was also in low LD with ours, r2 being between rs4300027 and rs2738048 (r2=0.16), rs2738058 (r2=0.10), rs2702910 (r2=0.06) and rs9644778 (r2=0.04; LD relationships between SNPs were adopted based on the HapMap data using CHB samples). Thus, the structural haplotypes they observed may not be well tagged in our cohort and need further testing in multiple populations, because repeated data supported that CNVs of the region suffer from power of selection. In addition, both rs2738058 and rs9644778 are expected to be highly functional variants in DEFA gene cluster, which may act as regulatory factors for gene expression. From a GWAS of chronic periodontitis conducted in a general German population, rs2738058 was also observed to be one of the top association signals, further suggesting that rs2738058 may be one of the causal variants.29 The risk genotypes (CC+CA) of rs9644778 were associated with gross hematuria marginally, which indicates that the SNPs in DEFA loci may not only influence disease susceptibility but also impact IgAN severity. Hematuria, a typical clinical presentation secondary to upper respiratory or gastrointestinal illness in IgAN patients, was speculated to be caused by the synergistic effect of innate, mucosal and adaptive immunity.30, 31 Although limited by the small sample size, as more IgAN patients with detailed clinical data are needed to confirm the result, our findings support the notion that α-defensins are involved in the inherited defect in IgAN, via innate and mucosal immunity disorders. Cis-eQTL analysis showed that risk genotypes of both rs2738058 and rs4300027 showed significant correlations with lower gene expressions. When α-defensin concentration was low, human monocyte-derived dendritic cells elevated secretions of tumor-necrosis factor-α and interleukin-1β, contributing to aggregation of inflammatory medium.32 The risk genotypes correlated with lower gene expressions may lead to low serum α-defensin concentration, which would provide a proinflammatory environment in glomeruli mesangial cells to amplify proinflammatory loop by upregulation of IgA receptors in mesangial cells.33 In this case, more IgA may be deposited in mesangial cells, which is consistent with renal biopsy specimens. The data above showed the genetic, expression and clinical manifestation association of DEFAs and IgAN. However, the association in IgAN was relatively weak and precise mechanisms still need to be determined.
GWASs have identified numerous susceptibility loci for diseases or traits. One of the greatest challenges in the post-GWAS era is to clarify the functional variants within the identified loci. In this study, we investigated multiple SNPs spanning a 350-kb range in DEFA loci among 2096 CHB. We discovered two independent SNPs with potential regulatory functions and successfully replicated previously GWAS-reported rs2738048. Genetic, expression and clinical manifestation association suggested that DEFA gene polymorphisms have potentially pathogenic roles in IgAN. The role of mucosal immunity in the pathogenesis of IgAN should be emphasized.
Materials and methods
Patients and controls
The cases and controls analyzed in this study were from our previous GWAS cohort,5 including 1194 renal biopsy-proved IgA nephropathy patients and 902 healthy controls of Chinese Han ancestry from Northern China. Participants in this cohort were geographically and ethnically matched and statistically analyzed in a previous report.5 The study got the approval from the Medical Ethics Committee of the Peking University First Hospital and written informed consent was obtained from all the patients.
SNP selection and genotyping
For genotyping, a customized Illumina (San Diego, CA, USA) Human 610-Quad BeadChip platform was applied and genotyping yield was over 99%.5 A region spanning 350 kb (chromosome 8, 6 750 000–7 100 000, GRCh37/hg19) encompassing the DEFA gene cluster (132 kb, GRCh37/hg19) and its upstream (DEFA5, 5′ region, 200 kb, GRCh37/hg19) was selected, and a total of 60 SNPs within this region were included and analyzed in the current study.
Determination of DEFA1A3 CNVs
The relative standard curve method of quantitative real-time PCR (7500 Real-time PCR System, Applied Biosystems, Foster City, CA, USA) was used to determine DEFA1A3 CNVs, as described previously,34 without modifications of primers and PCR conditions. For quality control, an internal control from HapMap CHB population (NA18570) was selected and randomly assigned in every plate for calibration. Seven serial 1:2 dilutions of genomic DNA of the calibrator were used to generate standard curves on each PCR plate and the minimal DNA content was 0.5 ng in the reaction. The CN count of each sample was determined by the average of three times of repeated amplification. For methodological validations of the DEFA1A3 CNs determined by qPCR, we compared our results with the sequencing data in the database obtained from the Complete Genomics Institute (http://www.completegenomics.com/).35 Our qPCR results were totally concordant with sequencing data in the database from Complete Genomics Institute. The resulting CN between our qPCR and sequencing data was consistent in triplicated qPCR assays.
Cis expression quantative trait loci analysis
SNP–gene associations in eQTL were analyzed using data from Epstein–Barr virus-transformed lymphoblastoid cell lines of 210 unrelated HapMap samples, as previously reported.36 Normalized mRNA data were retrieved from the database of the Gene Expression Variation (GENEVAR) project at the Wellcome Trust Sanger Institute (http://www.sanger.ac.uk/humgen/genevar/).
Bioinformatic analysis
Regulatory SNPs were analyzed by rSNPBase (http://rsnp.psych.ac.cn).19 Annotations of variants, located in the transcription factor-binding motifs, protein-binding sequences, chromatin structures or the histone modification regions, were obtained from the Regulome DB database (http://www.regulomedb.org) in ENCODE project.37
Statistical analysis
The identified 60 SNPs within DEFA gene cluster were consistent with Hardy–Weinberg equilibrium genotype frequency expectations (P>0.05) using χ2 goodness-of-fit test. The degrees of LD were estimated by CI method using Haploview4.1 (Cambridge, MA, USA). Allelic and genotypic associations were accessed with PLINK. A stepwise logistic regression analysis using forward method was applied to determine the independence of the SNP. Univariate linear regression was used to determine the correlations between genotypes of associated SNPs and CNs. Power was calculated by Power and Sample Size Calculations Software (http://biostat.mc.vanderbilt.edu/). In cis-eQTL analysis, Spearman’s coefficient was calculated.
Means and s.d. were used to summarize normally distributed variables. Median and interquartile ranges were used for non-normally distributed variables. Categorical data were summarized by ratios and percentages. Student’s t-test, nonparametric Mann–Whitney U-test and one-way analysis of variance were performed for comparison of continuous variables. χ2 test or logistic regression analysis was used for categorical variables. A two-tailed P-value<0.05 was considered statistically significant. Statistical analysis was performed with the SPSS 16.0 software package (SPSS Inc., Chicago, IL, USA).
References
Tsukamoto Y, Wang H, Becker G, Chen HC, Han DS, Harris D et al. Report of the Asian Forum of Chronic Kidney Disease Initiative (AFCKDI) 2007. "Current status and perspective of CKD in Asia": diversity and specificity among Asian countries. Clin Exp Nephrol 2009; 13: 249–256.
Kiryluk K, Novak J, Gharavi AG . Pathogenesis of immunoglobulin A nephropathy: recent insight from genetic studies. Annu Rev Med 2013; 64: 339–356.
Kiryluk K, Julian BA, Wyatt RJ, Scolari F, Zhang H, Novak J et al. Genetic studies of IgA nephropathy: past, present, and future. Pediatr Nephrol 2010; 25: 2257–2268.
Feehally J, Farrall M, Boland A, Gale DP, Gut I, Heath S et al. HLA has strongest association with IgA nephropathy in genome-wide analysis. J Am Soc Nephrol 2010; 21: 1791–1797.
Gharavi AG, Kiryluk K, Choi M, Li Y, Hou P, Xie J et al. Genome-wide association study identifies susceptibility loci for IgA nephropathy. Nat Genet 2011; 43: 321–327.
Yu XQ, Li M, Zhang H, Low HQ, Wei X, Wang JQ et al. A genome-wide association study in Han Chinese identifies multiple susceptibility loci for IgA nephropathy. Nat Genet 2011; 44: 178–182.
Julian BA, Quiggins PA, Thompson JS, Woodford SY, Gleason K, Wyatt RJ . Familial IgA nephropathy. Evidence of an inherited mechanism of disease. N Engl J Med 1985; 312: 202–208.
Kiryluk K, Li Y, Sanna-Cherchi S, Rohanizadegan M, Suzuki H, Eitner F et al. Geographic differences in genetic susceptibility to IgA nephropathy: GWAS replication study and geospatial risk analysis. PLoS Genet 2012; 8: e1002765.
Kiryluk K, Li Y, Scolari F, Sanna-Cherchi S, Choi M, Verbitsky M et al. Discovery of new risk loci for IgA nephropathy implicates genes involved in immunity against intestinal pathogens. Nat Genet 2014; 46: 1187–1196.
Lehrer RI, Lu W . α-Defensins in human innate immunity. Immunol Rev 2012; 245: 84–112.
Saraheimo M, Forsblom C, Pettersson-Fernholm K, Flyvbjerg A, Groop PH, Frystyk J . Increased levels of alpha-defensin (-1, -2 and -3) in type 1 diabetic patients with nephropathy. Nephrol Dial Transplant 2008; 23: 914–918.
Bokarewa MI, Jin T, Tarkowski A . Intraarticular release and accumulation of defensins and bactericidal/permeability-increasing protein in patients with rheumatoid arthritis. J Rheumatol 2003; 30: 1719–1724.
Sthoeger ZM, Bezalel S, Chapnik N, Asher I, Froy O . High alpha-defensin levels in patients with systemic lupus erythematosus. Immunology 2009; 127: 116–122.
Peluso G, De Santis M, Inzitari R, Fanali C, Cabras T, Messana I et al. Proteomic study of salivary peptides and proteins in patients with Sjögren’s syndrome before and after pilocarpine treatment. Arthritis Rheum 2007; 56: 2216–2222.
Ahn JK, Cha HS, Lee J, Jeon CH, Koh EM . Correlation of DEFA1 gene copy number variation with intestinal involvement in Behcet’s disease. J Korean Med Sci 2012; 27: 107–109.
Chen Q, Hakimi M, Wu S, Jin Y, Cheng B, Wang H et al. Increased genomic copy number of DEFA1/DEFA3 is associated with susceptibility to severe sepsis in Chinese Han population. Anesthesiology 2010; 112: 1428–1434.
Jespersgaard C, Fode P, Dybdahl M, Vind I, Nielsen OH, Csillag C et al. Alpha-defensin DEFA1A3 gene copy number elevation in Danish Crohn’s disease patients. Dig Dis Sci 2011; 56: 3517–3524.
Zhou XJ, Cheng FJ, Lv JC, Luo H, Yu F, Chen M et al. Higher DEFB4 genomic copy number in SLE and ANCA-associated small vasculitis. Rheumatology (Oxford) 2012; 51: 992–995.
Guo L, Du Y, Chang S, Zhang K, Wang J . rSNPBase: a database for curated regulatory SNPs. Nucleic Acids Res 2014; 42: D1033–D1039.
Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C et al. Architecture of the human regulatory network derived from ENCODE data. Nature 2012; 489: 91–100.
Schaub MA, Boyle AP, Kundaje A, Batzoglou S, Snyder M . Linking disease associations with regulatory information in the human genome. Genome Res 2012; 22: 1748–1759.
Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res 2006; 34: D108–D110.
Pique-Regi R, Degner JF, Pai AA, Gaffney DJ, Gilad Y, Pritchard JK . Accurate inference of transcription factor binding from DNA sequence and chromatin accessibility data. Genome Res 2011; 21: 447–455.
Palii CG, Perez-Iratxeta C, Yao Z, Cao Y, Dai F, Davison J et al. Differential genomic targeting of the transcription factor TAL1 in alternate haematopoietic lineages. Embo J 2011; 30: 494–509.
Fanciulli M, Norsworthy PJ, Petretto E, Dong R, Harper L, Kamesh L et al. FCGR3B copy number variation is associated with susceptibility to systemic, but not organ-specific, autoimmunity. Nat Genet 2007; 39: 721–723.
Yang Y, Chung EK, Wu YL, Savelli SL, Nagaraja HN, Zhou B et al. Gene copy-number variation and associated polymorphisms of complement component C4 in human systemic lupus erythematosus (SLE): low copy number is a risk factor for and high copy number is a protective factor against SLE susceptibility in European Americans. Am J Hum Genet 2007; 80: 1037–1054.
Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T et al. Strong association of de novo copy number mutations with autism. Science 2007; 316: 445–449.
Black HA, Khan FF, Tyson J, Al AJ . Inferring mechanisms of copy number change from haplotype structures at the human DEFA1A3 locus. BMC Genomics 2014; 15: 614.
Teumer A, Holtfreter B, Volker U, Petersmann A, Nauck M, Biffar R et al. Genome-wide association study of chronic periodontitis in a general German population. J Clin Periodontol 2013; 40: 977–985.
Sanders JT, Wyatt RJ . IgA nephropathy and Henoch-Schonlein purpura nephritis. Curr Opin Pediatr 2008; 20: 163–170.
Wyatt RJ, Kritchevsky SB, Woodford SY, Miller PM, Roy SR, Holland NH et al. IgA nephropathy: long-term prognosis for pediatric patients. J Pediatr 1995; 127: 913–919.
Rodriguez-Garcia M, Oliva H, Climent N, Escribese MM, Garcia F, Moran TM et al. Impact of alpha-defensins1-3 on the maturation and differentiation of human monocyte-derived DCs. Concentration-dependent opposite dual effects. Clin Immunol 2009; 131: 374–384.
Barratt J, Feehally J . IgA nephropathy. J Am Soc Nephrol 2005; 16: 2088–2097.
Cheng FJ, Zhou XJ, Zhao YF, Zhao MH, Zhang H . Alpha-defensin DEFA1A3 gene copy number variation in Asians and its genetic association study in Chinese systemic lupus erythematosus patients. Gene 2013; 517: 158–163.
Drmanac R, Sparks AB, Callow MJ, Halpern AL, Burns NL, Kermani BG et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science 2010; 327: 78–81.
Zhou XJ, Lu XL, Lv JC, Yang HZ, Qin LX, Zhao MH et al. Genetic association of PRDM1-ATG5 intergenic region and autophagy with systemic lupus erythematosus in a Chinese population. Ann Rheum Dis 2011; 70: 1330–1337.
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012; 489: 57–74.
Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B et al. The structure of haplotype blocks in the human genome. Science 2002; 296: 2225–2229.
Acknowledgements
We thank our collaborators of Ali G Gharavi from the Department of Medicine, Columbia University College of Physicians and Surgeons, New York City, NY, USA, for genotyping sorting and assistance. We also thank all the members of the laboratory for technical assistance and the patients and their families for their cooperation and for giving consent to participate in this study. This work was supported by grants from the National Natural Science Foundation of China (No. 81200524), The Foundation of Ministry of Education of China (20120001120008), Major State Basic Research Development Program of China (973 program, No. 2012CB517700), Research Fund of Beijing Municipal Science and Technology for the Outstanding PhD Program (20121000110) and Natural Science Fund of China to the Innovation Research Group (81321064).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Disclaimer
The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Supplementary Information accompanies this paper on Genes and Immunity website
Supplementary information
Rights and permissions
About this article
Cite this article
Qi, Y., Zhou, X., Cheng, F. et al. DEFA gene variants associated with IgA nephropathy in a Chinese population. Genes Immun 16, 231–237 (2015). https://doi.org/10.1038/gene.2015.1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gene.2015.1
- Springer Nature Limited