Abstract
Glucose oxidase (GOD) has many practical applications, but its poor thermostability limits its broader use. In this research, three primary mutants of wild-type GOD were designed using rational mutagenesis, and the GODm mutant was constructed by combinatorial design. The expression, purification, and enzymatic properties of the mutants were studied. The specific enzyme activity of GODm was 2.10-fold higher than that of wild type, and the (kcat/Km) value was increased by 1.45-fold. After treatment at 55 ℃ for 3 h, GODm retained 37.5% of its enzymatic activity, and the half-life (t1/2) of GODm at 55 ℃ and 65 ℃ was 2.28-fold and 3.36-fold higher than that of wild type, respectively. By analyzing the three-dimensional structure of wild type and the GODm mutant, it was found that T30V formed a new hydrogen bond with FAD and strengthened the hydrophobic interaction, D315K optimized the surface electrostatic interaction, and A162T improved the efficiency of the electron pathway. Thus, a novel mutant with improved thermostability and catalytic efficiency was obtained in this research.
Graphic abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glucose oxidase (GOD, EC.1.1.3.4) is a bisubstrate enzyme that oxidizes β-D-glucose via molecular oxygen and produces d-glucono-δ-lactone and hydrogen peroxide (H2O2) [1]. The catalytic characteristics of GOD meet the requirements of different fields, and the enzyme has been widely used in food manufacturing [2], clinical diagnosis and multiple other fields, for example, decreasing the glucose concentration to produce a reduced-alcohol wine in the beverage industry [3]; producing an enzymatically modified electrode with high turnover rates, stability and shelf-life [4]; loading on reagent-free colorimetric biosensors for glucose detection [5]; elevating tomato antiherbivore defenses [6]; improving the growth performance, gut health and cecal microbiota of broiler chickens [7]; producing sodium gluconate [8] and so on. GOD plays an increasingly important role in various fields, especially research on substitutes for antibiotics and agricultural and veterinary drugs. In industrial applications, sterilization, granulation and other processes inactivate enzymes, and enzymes with the catalytic characteristics of high specificity, turnover, and stability are needed [9].
Currently, GOD is primarily produced in industrial-scale cultivation using Aspergillus niger and Penicillium [10]. GOD was first discovered in the cell-free extract of A. niger and named in 1928 [11], and then, in 1948, GOD was found in P. glaucum for the first time [12]. According to previous studies, GODs from P. amagasakiense possess much higher catalytic efficiency, but GODs from A. niger exhibit superior thermostability [13]. The biotechnological applications of enzymes require them to have high catalytic efficiency and to be thermostable at the same time, but enzymes generally have an activity-stability trade-off without affecting their own adaptive fitness in the environment [14]. Many studies have overcome this limitation via physical, chemical or genetic modifications of nonnatural enzymes. For instance, poly (ethylene glycol) (PEG) [15], polyethylenimine (PEI) [16], and polyacrylonitrile membranes [17] have been used to immobilize GOD to improve its stability in the field of physical and chemical modifications. In the field of genetic modifications, most researchers improve the stability of enzymes through rational (computer-aided design, sequence alignment with thermophilic homologs) and irrational (error-prone PCR, gene rearrangement, etc.) approaches [18]. For example, Tao [19] obtained a mutant M4 with improved catalytic efficiency and thermostability using error-prone PCR, increasing noncovalent interactions and optimizing the surface charge distribution of GOD. Ge [20] constructed GOD mutants by introducing disulfide bonds, pi–pi bonds, hydrogen bonds, salt bridges, and surface charge interactions. These studies confirmed that the higher structural rigidity of thermophilic homologs is usually accompanied by more noncovalent (hydrophobic, hydrogen bonding, ionic, etc.) and covalent (disulfide bridges) interactions, reducing surface hydrophobicity, and amino acid compositional bias [21].
Due to the increasing applications of GOD, this paper improved the thermostability and catalytic efficiency of GOD from A. niger through rational mutagenesis. Based on bioinformatics analysis and homologous sequence alignment, four mutants were constructed and expressed in Pichia pastoris. Meanwhile, the enzymatic properties of the mutants were characterized, and the potential factors that may affect the thermostability and catalytic efficiency of GOD were identified.
Materials and methods
Strains, plasmids, and chemicals
The pPUC57 vector and Escherichia coli JM109 were preserved in our laboratory and used for construction and amplification, respectively. The expression host Pichia pastoris GS115 and plasmid pPIC9K were also preserved in our laboratory and used for heterologous expression. Primestar Max DNA polymerase, Premix Taq (Takara Taq version 2.0 plus dye), the restriction enzyme Sac I were purchased from Takara Biomedical Technology Co., Ltd (Beijing, China), and ClonExpress II One Step Cloning Kit was purchased from Vazyme Biotech Co., Ltd (Nanjing, China). Bradford Protein Concentration Determination Kit and SDS-PAGE Gel Parparation Kit were purchased from Beyotime Biotechnology Co., Ltd (Shanghai, China). All chemicals used in this study were analytical grade.
Media
Luria–Bertani (LB) medium, yeast peptone dextrose (YPD) medium, minimal dextrose (MD) medium, buffered methanol-complex (BMMY) medium were prepared according to the instructions of the Pichia pastoris expression kit (Invitrogen, Carlsbad, CA, USA). Minimum methanol medium: tryptone 20.0 g/L, yeast extract 10.0 g/L, agar 20.0 g/L, 1% methanol.
Gene cloning and mutagenesis
The gene encoding opanGOD from A. niger glucose oxidase gene (GenBank accession number CAC27315.1) was codon optimized and synthesized by Talen-bio Co., Ltd. (Shanghai, China). E. coli JM109 and pUC57 vector were used for cloning. The opanGOD gene was obtained by PCR amplification with pUC57-opanGOD as a template. The primers used for cloning opanGOD were opanGOD-F and opanGOD-R (Table S1). Then, the linearized vector pPIC9K was amplified by primers 9 K-F and 9 K-R (Table S1). The purified opanGOD gene was ligated into the pPIC9K expression vector using the CloneExpress™ One Step Cloning Kit, resulting in the plasmid pPIC9K-opanGOD. Sequences of mutated primers are listed in Supplementary Table S1. The operation of cloning the mutant gene into pPIC9K is similar to the described above. The verification of the expression plasmids was confirmed by DNA sequencing.
Heterologous expression in P. pastoris and positive transformants screening
Constructed plasmids were linearized with Sac I according to the instruction (TAKARA) and individually transformed into competent P. pastoris GS115 cells via electroporation. Transformants were cultivated on MD plates added 150 μg/mL G418 and incubated for 36 h at 30 ℃.
Preliminary screening: Solid plates were used for screening P. pastoris transformants expressing high levels of recombinant GOD activity according to the research with some modifications [19, 22]. The transformants were selected from MD plates and seeded on YPD resistant plate with 350 μg/mL G418. After 24 h of cultivation at 30 ℃, the transformants were seeded on the minimum methanol plate and incubated for 36 h at 28 °C. Then, the plate with transformants was placed at 55 ℃ for 10 min and subsequently covered with 15 mL of reaction mixture (1% o-dianisidine solution, 86% phosphate buffered saline (PBS, pH 6.0), 3% 90 U/mL horseradish peroxidase, 10% 18%(w/v) glucose solution) in 2% agar. After a 2 h reaction time at 28 ℃, residual enzyme activity following thermal treatment was detected by formation of a brown area around the colony. Confirmed positive transformants were picked up from the corresponding YPD resistant plate and rescreened by fermentation.
Rescreening: The residual enzyme activity of GOD at different temperature and time was calculated for quantitative screening. The specific method is as follows: Individual positive transformants were placed in YPD medium and then incubated at 30 ℃ for 24 h with 220 r/min of shaking. The transformants were cultivated further in BMMY medium at 28 ℃ for 96 h after the YPD medium was discarded by centrifugation. After the fermentation, the crude enzyme liquid was harvested by centrifugation. The crude enzyme liquid of the wild-type and the mutants were kept at 55 ℃, 65 ℃ and 75 ℃ for 10 min, 10 min, and 5 min, respectively, for determining the enzyme activity, then the residual enzyme activity was calculated according to the enzyme activity.
GOD activity assay and protein content assay
GOD activity assay was measured following the method described by the researches [19, 23] with some modifications. 2.5 mL of 0.21 mM o-dianisidine (TCI, Japan) solution (dissolved in 100 mM PBS, pH 6.0), 0.3 mL of 18% (w/v) glucose solution, and 0.1 mL of 90 U/mL horseradish peroxidase solution were mixed well in a tube and incubated for 2 min at 35 ℃. Then, 0.1 mL of sample solution was added and immediately kept at 35 ℃ incubating for 3 min. The reaction was stopped by adding 2 mL of 2 M sulfuric acid. Absorbance at 540 nm was measured and quantitated against a standard curve of different concentrations of glucose oxidase standard. All reactions were performed in triplicate. Protein content was measured according to the Bradford Protein Concentration Determination Kit.
Enzyme purification
The fermentation supernatant was collected by centrifugation. The crude enzyme was desalted and enriched using 10 kDa ultrafiltration membrane with 20 mM PBS (pH 6.5). The sample was purified by AKTA protein purification system (AKTA avant; GE Healthcare Life Science) and loaded onto Hitrap™ Q FP (5 mL) anion exchange chromatographic column for further purification. All purification steps were performed at 4 °C and PBS buffer (A: 20 mM PBS (pH 6.5); B: 20 mM PBS (pH 6.5) with 1 M NaCl) was used for dialysis. The protein purity was evaluated by SDS-PAGE.
Thermostability assay
The optimal temperature for recombinant GOD was determined based on the standard enzyme assays in the range of 20–65 ℃ in 100 mM PBS (pH 5.5) reaction system.
The purified recombinant GOD was diluted to 50 μg/mL and kept at 55 ℃ for 0 min, 10 min, 30 min, 60 min, 120 min, 180 min; 65 ℃ for 0 min, 0.5 min, 1 min, 3 min, 5 min, 10 min, 15 min, 30 min, and 75 ℃ for 0 s, 15 s, 30 s, 60 s, respectively. Samples were incubated for various times, and then placed immediately on ice. The activity was measured under standard conditions. The activity of the untreated enzyme was considered as 100%.
The half-lives (t1/2) of the wild-type GOD and its mutants were determined by incubating at 55 ℃ and 65 ℃ for various times. The half-lives (t1/2) were calculated by the following formula: t1/2 = ln2/kd. The rate constant, kd is the inactivation constant of GOD at 55 ℃, 65 ℃, respectively. The ln (residual enzyme activity) was used to plot the incubation time, and the slope obtained by linear fitting was the inactivation constant of GOD at the temperature. Tm values were measured by differential scanning calorimetry (DSC; GE Healthcare Life Science).
Enzymatic kinetic assays
The kinetic parameters, Km and Vmax, of wild-type GOD and its mutants were analyzed in the presence of 5–200 mM of glucose under standard assay conditions. Kinetic values were estimated using OriginPro 8.5 software to fit Michaelis–Menten plot using non-linear regression.
GOD protein structure analysis
The three-dimensional structure of GOD is obtained by SWISS MODEL (https://swissmodel.expasy.org/) and analyzed by PyMOL (Delano Scientific, San Carlos, CA, USA).
Results and discussion
Selection of mutant sites
The objective of the current study was to improve the thermostability and catalytic efficiency of GOD. Based on bioinformatics analysis and homologous sequence alignment, two rounds of mutations were designed. In the first round of mutation, a rational design was applied according to the previous research and the theory of hydrophobic interaction and surface modification, to investigate the changes of enzymatic properties after mutation. Then, a new mutant with thermostability and catalytic efficiency improved was obtained by integrating the sites screened in the first round of mutation, and in which modifications related to electron transport chain were introduced into the new mutants. The specific process of selecting mutation sites is described below.
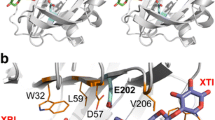
Sequence hydrophobicity is critical for folding stability, and the primary thermodynamic driver of protein folding is the formation of a hydrophobic interior [24]. Previous research has shown a high degree of conservation in the FAD- and substrate-binding domains among GODs. Mutating the nonconservative sites in the first region of the FAD-binding domain, which is the most conserved region of the FAD-binding site, may improve stability [25]. Based on conjecture and sequence analysis, T30 is located in this region (Fig. 3a) and was selected for mutation in the present study; the polar threonine was mutated into hydrophobic valine, T30V, and the mutant was named GOD1.
Surface modification is a strategy that can improve the thermostability of proteins and minimize damage to their activity. By optimizing electrostatic interactions on the surface away from the active site, stabilization can be achieved without compromising protein function [14]. Based on this theory and previous research [19], the D70 and D315 sites, which are far from the active site (Fig. 3a), were selected by protein sequence alignment and mutated into lysine, which is positively charged. By optimizing charge-charge interactions, the second mutant with the D70K and D315K mutations was obtained and named GOD2.
In addition, according to previous research [26], the thermostability and catalytic efficiency of mutant F91 (with mutations at T30V, R37K, I94V, V106I, A162T) were significantly improved after mutation. Therefore, the mutant F91 was selected for research and analysis in the present study.
In the second round of mutation, based on the above four mutants, it was found that the thermostability of GOD1 and GOD2 was significantly improved, while the catalytic efficiency and affinity of F91 were substantially improved compared with those of wild type. Some studies have shown that there may be an electron pathway, Thr110–Trp111–Glu144–His165–Cys164 in GOD [27], and the mutation A162T is close to the pathway (Fig. 3c). Mutation of hydrophobic alanine to polar threonine may improve the transfer efficiency of the electron transfer chain. Accordingly, T30V in GOD1, D70K, and D315K in GOD2 and A162T in F91 were combinatorially mutated to form a new mutant, which was named GODm.
Recombinant positive transformant screening assay
P. pastoris is a widely used eukaryotic expression system, and its convenient fermentation with fewer byproducts simplifies downstream purification. However, there may be differences in expression levels among transformants in P. pastoris, and screening for positive transformants seems to be necessary. Typically, screening of strains is time-consuming and inefficient. Establishing a rational high-throughput screening method can effectively reduce the workload of screening. Under the catalysis of horseradish peroxidase, H2O2 which is produced from glucose catalyzed by glucose oxidase can oxidize colorless o-anisidine to brown. It has been reported that the color reaction of o-anisidine can be used for high-throughput screening [23]. With modifications to this method, screening of G418 resistance and a heat treatment were added to the process of solid screening, and transformants with improved thermostability could be obtained by adding a heat treatment in the plate color screening method. Transformants with dark brown and large chromogenic rings on the plate were selected for fermentation (Fig. 1a).
After 96 h of methanol induction, the specific activity and thermostability of each strain were determined. The results are shown in Fig. 1b, and compared with wild type, the specific activity of the mutants decreased to varying degrees, which indicates that more heteroprotein was produced after mutation. The residual enzyme activity of the mutants and wild-type GOD were not obviously different after treatment at 55 °C, while after treatment at 65 °C or 75 °C, the thermostability of the mutants was significantly higher than that of wild type. In particular, the mutants GOD1, GOD2, and GODm retained 21.7%, 38.6%, and 41.2% of their residual enzyme activity, while wild type lost its initial activity following incubation at 75 °C for 5 min.
Thermostability of recombinant GOD
Diluting the purified wild-type GOD and mutants to the same protein concentration, their thermostability was studied at 55 ℃, 65 ℃, and 75 ℃ for different times. The activity of untreated enzyme was considered to be 100%, and the experimental results are shown in Fig. 2. The thermostability of most of the mutants was higher than that of wild-type GOD, especially GOD1, GOD2, and GODm, which retained 30.5%, 34.2%, and 37.5% of their residual enzyme activity after 3 h of treatment at 55 °C (Fig. 2a). As shown in Table 1, the half-life (t1/2) at 55 ℃ of wild type was 43.4 min, while that of GOD2 was 115.2 min, 1.65-fold higher than that of wild type, and that of GODm was 142.3 min, 2.28-fold higher than that of wild type. From the results, it was found that the thermostability of the GOD1, GOD2, and GODm mutants at 65 ℃ and 75 ℃ was significantly higher than that of wild type (Fig. 2b, c). The F91 mutant and wild-type GOD lost their activity after 15 min of treatment at 65 °C, but the GOD1, GOD2, and GODm mutants retained 3.6%, 6.0%, and 9.9% of their residual enzyme activity after 30 min of treatment. At 65 ℃, the half-life (t1/2) of the GOD1, GOD2, and GODm mutants was 1.79-, 3.14-, and 3.36-fold higher than that of wild type, respectively (Table 1). Meanwhile, the Tm of the GOD1, GOD2, and GODm mutants increased by 2.0 ℃, 2.8 ℃, and 3.1 ℃, respectively, compared to the wild type. The thermostability of the purified GODs was not as high as that of the crude enzyme liquid, which indicated that the heteroprotein in the crude enzyme liquid had a protective effect on the enzyme at high temperature. In addition, as shown in Fig. 2d that the optimum temperature of these mutants was not significantly different from that of wild-type GOD.
Thermostability of wild type and the mutant GODs. a Residual activity of wild type and the mutant GODs after treatment at 55 °C for different times. The data points correspond to the mean values of three independent experiments. The activity of the untreated enzyme was considered 100%. b Residual activity of wild type and the mutant GODs after treatment at 65 °C for different times. c Residual activity of wild type and the mutant GODs after treatment at 75 °C for different times. d The optimum temperature of wild type and the mutant GODs at 20 °C–65 °C
Kinetic analysis and specific activity of recombinant GODs
The kinetic constants and specific activity of the recombinant enzymes were measured using β-d-glucose as the substrate, and the results are summarized in Table 2. After purification, the specific enzyme activity of the mutants was higher than that of wild type. The GODm mutant had the highest specific enzyme activity, 2.10-fold higher than that of wild-type. Compared with that of wild-type GOD, the Km value of the mutants was lower, which is related to the substrate affinity. The Km values of the F91 and GODm mutants were only 35.7% and 37.8% of wild type, respectively, which indicated that their substrate affinity was enhanced. Otherwise, the (kcat/Km) value of the F91 and GODm mutants was increased by 1.69- and 1.45-fold, respectively, and their catalytic efficiency was significantly improved.
Protein structure analysis of the GODm mutant
To study the changes in the properties of the GODm mutant, the crystal structure of glucose oxidase was identified by comparison using SWISS MODEL, and the protein tertiary structure was analyzed by PyMOL software. According to analysis of the T30V site in the mutant sequence, as shown in Fig. 3b, when threonine was mutated into valine, a new hydrogen bond was formed with FAD. Meanwhile, the locations of hydrogen bonds have been found to make an important contribution to thermostability [28]. According to previous research, there is a hydrophobic core region near FAD [19], and mutating the polar Thr at position 30 into hydrophobic Val would strengthen the hydrophobic core. As previous researchers improved the thermostability of GOD by 26% by enhancing the hydrophobic core network [29].
Protein structure of the GODm mutant. a The location of the mutation sites and FAD in the protein. b Hydrogen bonding of T30V with other sites and FAD. c The positional relationship between A162 and the electron transport chain. d Comparison of D70K and D315K before and after charge-charge optimization
Protein stability can be modulated via surface charge-charge interactions [30]. In this research, the GOD2 mutant with D70K and D315K mutation suggested that the thermostability and catalytic efficiency could be improved at the same time. The catalytic efficiency did not decrease when the mutation sites were far from the active center, as shown in Table 2. GOD is a FAD-containing homodimeric protein with a large excess of negatively charged groups (66 Asp + Glu, 38 Arg + Lys, 20 His/His+) [31]. When negatively charged aspartic acid is transformed into positively charged lysine, the magnetic intensity of the electrostatic interaction is increased (Fig. 3d), and thermostability is improved by optimizing the surface charge distribution.
GOD is a flavoprotein that contains the compact noncovalently bound coenzyme flavin adenine dinucleotide (FAD). According to the following reaction formula for the catalysis of glucose by GOD, GOD uses molecular oxygen as an electron acceptor to catalyze the oxidation of the first hydroxyl group on β-d-glucose to d-gluconic-δ-lactone and produce H2O2. The electrons released from the oxidation of glucose are received by FAD to reduce FADH2. The electrons carried by FADH2 are transferred to molecular oxygen through the electron transport chain. Previous research has predicted that there may be an electron pathway, Thr110–Trp111–Glu144–His165–Cys164, within GOD; the pathway ends at C164, and electrons are released by N161 and N168 when the enzyme is glycosylated [27]. The A162T mutation is near the electron pathway (Fig. 3c). Mutation of hydrophobic alanine to polar threonine may improve the electron transfer efficiency of this pathway, which may result in a potentially significant improvement of the catalytic efficiency of the GODm and F91 mutants.
The whole reaction of GOD-catalyzed glucose oxidation is as follows [32]:
GOD(FAD) + β-d-glucose → GOD(FADH2) + d-gluconic-δ-lactone,
GOD(FADH2) + O2 → GOD(FAD) + H2O2,
β-d-glucose + GOD(FAD) + O2 → GA + GOD(FADH2) + H2O2.
Comparison of thermostability with commercial GOD
GOD can produce gluconic acid and H2O2 through specific catalysis which can inhibit pathogenic microorganisms, but it is conducive to the growth of probiotics. Thus, GOD is used as a feed additive to replace antibiotics, improve animal immunity and promote animal growth. In the process of feed granulation, high temperature is required, therefore, the thermostability of GOD is particularly important. Three different commercialized GODs named CG1, CG2, and CG3 which are applied in the feed industry were selected to study the difference of thermostability with the wild type and GODm mutant in this research. The crude enzyme liquid of GOD was treated at 55 ℃, 65 ℃, and 75 ℃ for 10 min, 10 min, and 5 min, respectively, to study their thermostability, the result is shown in Fig. 4. The thermostability of wild-type GOD reflects small divergence with CG2 and CG3, but it is far less than CG1. After optimizing the thermostability of GOD, the GODm mutant has higher stability at 65 ℃ and 75 ℃, similar to CG1, it can better meet the requirements of industrial application, especially in the process of processing.
Comparison of thermostability between wild type, mutant GODm, and commercial GOD. The crude enzyme liquid of GOD was treated at 55 ℃, 65 ℃, and 75 ℃ for 10 min, 10 min, and 5 min, respectively. The enzyme activity was compared with that of untreated enzyme liquid, and the relative activity was calculated
Conclusions
In the past, many studies used protein engineering and genetic engineering technology to modify GOD to achieve high catalytic efficiency and stability at the same time. In the present research, five GOD mutants were constructed via rational mutagenesis, and their enzymatic properties were evaluated. The GODm mutant screened in this study had a simultaneous enhancement of thermostability and catalytic efficiency, which accounted for the changes in the enzymatic properties of the mutant. The GODm mutant provides a reference for future research on GOD.
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
Zhang Y, Tsitkov S, Hess H. Proximity does not contribute to activity enhancement in the glucose oxidase-horseradish peroxidase cascade. Nat Commun. 2016;7:13982. https://doi.org/10.1038/ncomms13982.
Pang CP, Yin XX, Zhang GQ, Liu S, Zhou JW, Li JH, Du GC. Current progress and prospects of enzyme technologies in future foods. Syst Microbiol Biomanuf. 2021;1:24–32. https://doi.org/10.1007/s43393-020-00008-6.
Valencia P, Espinoza K, Ramirez C, Franco W, Urtubia A. Technical feasibility of glucose oxidase as a prefermentation treatment for lowering the alcoholic degree of red wine. Am J Enol Vitic. 2017;68:386–9. https://doi.org/10.5344/ajev.2017.16005.
Suroviec AH. Layer-by-layer assembly of glucose oxidase on carbon nanotube modified electrodes. Methods Mol Biol. 2017;1504:203–13. https://doi.org/10.1007/978-1-4939-6499-4_16.
Min SK, Kim DH, Lee J, Ahn HT, Kim MI, Lee J. Self color-changing ordered mesoporous ceria for reagent-free colorimetric biosensing. Nanoscale. 2019;12:1419–24. https://doi.org/10.1039/C9NR09182C.
Wang J, Peiffer M, Hoover K, Rosa C, Zeng R, Felton GW. Helicoverpa zea gut-associated bacteria indirectly induce defenses in tomato by triggering a salivary elicitor(s). New Phytol. 2017;214:1294–306. https://doi.org/10.1111/nph.14429.
Wu S, Li T, Niu H, Zhu Y, Liu Y, Duan Y, Sun Q, Yang X. Effects of glucose oxidase on growth performance, gut function, and cecal microbiota of broiler chickens. Poult Sci. 2018;98:828–41. https://doi.org/10.3382/ps/pey393.
Han XL, Liu GD, Song WX, Qu YB. Production of sodium gluconate from delignified corn cob residue by on-site produced cellulase and co-immobilized glucose oxidase and catalase. Bioresour Technol. 2018;248:248–57. https://doi.org/10.1016/j.biortech.2017.06.109.
Wijma HJ, Floor RJ, Janssen DB. Structure- and sequence-analysis inspired engineering of proteins for enhanced thermostability. Curr Opin Struct Biol. 2013;23:588–94. https://doi.org/10.1016/j.sbi.2013.04.008.
Fakhry B, Asghar KA, Jamshid R. Expression, characterization and one step purification of heterologous glucose oxidase gene from Aspergillus niger ATCC 9029 in Pichia pastoris. EuPA Open Proteom. 2018;19:1–5. https://doi.org/10.1016/j.euprot.2018.09.001.
Muller D. Oxidation von glukose mit extrakten aus Aspergillus niger. Biochem Z. 1928;199:136–70.
Keilin D, Hartree EF. Properties of glucose oxidase (notatin): Addendum. Sedimentation and diffusion of glucose oxidase (notatin). Biochem J. 1948;42:221–9. https://doi.org/10.1042/bj0420221.
Ning X, Zhang Y, Yuan T, Li Q, Tian J, Guan W, Bo L, Wei Z, Xu X, Zhang Y. Enhanced thermostability of glucose oxidase through computer-aided molecular design. Int J Mol Sci. 2018;19:425. https://doi.org/10.3390/ijms19020425.
Siddiqui KS. Defying the activity-stability trade-off in enzymes: taking advantage of entropy to enhance activity and thermostability. Crit Rev Biotechnol. 2016;37:309–22. https://doi.org/10.3109/07388551.2016.1144045.
Vardar G, Altikatoglu M, Basaran Y, Işıldak İ. Synthesis of glucose oxidase-PEG aldehyde conjugates and improvement of enzymatic stability. Artif Cells Nanomed Biotechnol. 2017;46:788–94. https://doi.org/10.1080/21691401.2017.1345920.
Padilla-Martínez S, Martínez-Jothar L, Sampedro JG, Tristan F, Pérez E. Enhanced thermal stability and pH behavior of glucose oxidase on electrostatic interaction with polyethylenimine. Int J Biol Macromol. 2015;75:453–9. https://doi.org/10.1016/j.ijbiomac.2015.02.005.
Bayramoğlu G, Metin AÜ, Altıntas B, Arıca MY. Reversible immobilization of glucose oxidase on polyaniline grafted polyacrylonitrile conductive composite membrane. Bioresour Technol. 2010;101:6881–7. https://doi.org/10.1016/j.biortech.2010.04.025.
Tanatarov D, Huang ZY, Liu YF, Lv XQ, Li JH, Du GC, Liu L. Current advances in design and engineering strategies of industrial enzymes. Syst Microbiol Biomanuf. 2021;1:15–23. https://doi.org/10.1007/s43393-020-00005-9.
Tu T, Wang Y, Huang H, Wang Y, Jiang X, Wang Z, Yao B, Luo H. Improving the thermostability and catalytic efficiency of glucose oxidase from Aspergillus niger by molecular evolution. Food chem. 2019;281:163–70. https://doi.org/10.1016/j.foodchem.2018.12.099.
Ge J, Jiang X, Liu W, Wang Y, Huang H, Bai Y, Su X, Yao B, Luo H. Characterization, stability improvement, and bread baking applications of a novel cold-adapted glucose oxidase from Cladosporium neopsychrotolerans SL16. Food Chem. 2020;310: 125970. https://doi.org/10.1016/j.foodchem.2019.125970.
Vieille C, Zeikus GJ. Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol Mol Biol Rev. 2001;65:1–43. https://doi.org/10.1128/MMBR.65.1.1-43.2001.
Hatzinikolaou DG, Macris BJ. Factors regulating production of glucose oxidase by Aspergillus niger. Enzyme Microb Technol. 1995;17:530–4. https://doi.org/10.1016/0141-0229(95)91708-7.
Valdivieso-Ugarte M, Ronchel C, Bauelos O, Velasco J, Ad Rio JL. Expression of an Aspergillus niger glucose oxidase in Saccharomyces cerevisiae and its use to optimize fructo-oligosaccharides synthesis. Biotechnol Prog. 2010;22:1096–101. https://doi.org/10.1021/bp060076k.
Stewart KL, Rathore D, Dodds ED, Cordes MJH. Increased sequence hydrophobicity reduces conformational specificity: a mutational case study of the Arc repressor protein. Proteins. 2019;87:23–33. https://doi.org/10.1002/prot.25613.
Holland JT, Harper JC, Dolan PL, Manginell MM, Arango DC, Rawlings JA, Apblett CA, Brozik SM. Rational redesign of glucose oxidase for improved catalytic function and stability. PLoS ONE. 2012;7: e37924. https://doi.org/10.1371/journal.pone.0037924.
Ostafe R, Prodanovic R, Nazor J, Fischer R. Ultra-high-throughput screening method for the directed evolution of glucose oxidase. Chem Biol. 2014;21:414–21. https://doi.org/10.1016/j.chembiol.2014.01.010.
Mano N. Engineering glucose oxidase for bioelectrochemical applications. Bioelectrochem. 2019;128:218–40. https://doi.org/10.1016/j.bioelechem.2019.04.015.
Noorbatcha IA, Sultan AM, Salleh HM, Amid A. Understanding thermostability factors of Aspergillus niger PhyA Phytase: a molecular dynamics study. Protein J. 2013;32:309–16. https://doi.org/10.1007/s10930-013-9489-y.
Jiang X, Wang YR, Wang Y, Huang HQ, Bai YG, Su XY, Zhang J, Yao B, Tu T, Luo HY. Exploiting the activity-stability trade-off of glucose oxidase from Aspergillus niger using a simple approach to calculate thermostability of mutants. Food Chem. 2021;342: 128270. https://doi.org/10.1016/j.foodchem.2020.128270.
Schweiker KL, Zarrine-Afsar A, Davidson AR, Makhatadze GI. Computational design of the Fyn SH3 domain with increased stability through optimization of surface charge-charge interactions. Protein Sci. 2007;16:2694–702. https://doi.org/10.1110/ps.073091607.
Sedlák E, Sedláková D, Marek J, Hančár J, Žoldák G. Ion-specific protein/water interface determines the Hofmeister effect on the kinetic stability of glucose oxidase. J Phys Chem B. 2019;123:7965–73. https://doi.org/10.1021/acs.jpcb.9b05195.
Leskovac V, Trivic S, Wohlfahrt G, Kandrac J, Pericin D. Glucose oxidase from Aspergillus niger: the mechanism of action with molecular oxygen, quinones, and one-electron acceptors. Int J Biochem cell Biol. 2005;37:731–50. https://doi.org/10.1016/j.biocel.2004.10.014.
Acknowledgements
The authors are grateful for the financial support from the National First-class Discipline Program of Light Industry Technology and Engineering (Grant No. LITE2018-04), the Topnotch Academic Programs Project of Jiangsu Higher Education Institutions (TAPP), and the National Natural Science Foundation of China (NSFC) (32072162).
Author information
Authors and Affiliations
Contributions
HZ: conceptualization, methodology, formal analysis, investigation, data curation, writing—original draft, and writing—review and editing. DW: conceptualization, resources, project administration, funding acquisition, and writing—review and editing. PZ: resources and supervision. PC and XY: supervision and validation.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhou, H., Zheng, P., Chen, P. et al. Enhanced thermostability and catalytic efficiency of glucose oxidase in Pichia Pastoris. Syst Microbiol and Biomanuf 2, 296–304 (2022). https://doi.org/10.1007/s43393-021-00057-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43393-021-00057-5