Abstract
Most cervical cancer patients are prone to developing acquired cisplatin (DDP) resistance. Hsa_circ_0074269 (circ_0074269) plays a promoting role in cervical cancer, but whether circ_0074269 mediates cervical cancer resistance to DDP is unclear. Expression of circ_0074269 was detected by real-time quantitative polymerase chain reaction (RT-qPCR). The half-maximal inhibitory concentration (IC50) value, viability, proliferation, colony formation, migration, and apoptosis of DDP-resistant cervical cancer cells were determined. The molecular mechanisms associated with circ_0074269 were predicted by bioinformatics analysis and confirmed by dual-luciferase reporter and RIP assays. Xenograft assay was conducted to validate the effect of circ_0074269 on DDP resistance in vivo. Exosomes were isolated by ultracentrifugation. Circ_0074269 was overexpressed in DDP-resistant cervical cancer samples and cells. Silencing of circ_0074269 elevated DDP sensitivity, repressed DDP-resistant cervical cancer cell proliferation, and induced DDP-resistant cervical cancer cell apoptosis in vivo and in vitro and curbed DDP-resistant cervical cancer cell migration in vitro. And circ_0074269 could regulate DDP resistance via regulating TUFT1 expression via sponging miR-485-5p. More strikingly, circ_0074269 was also overexpressed in exosomes from DDP-resistant cervical cancer cells, and circ_0074269 could be delivered via exosomes. Circ_0074269 facilitated DDP resistance via elevating TUFT1 expression via sponging miR-485-5p, proving novel evidence to offer circ_0074269 as a target for cervical cancer treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cervical cancer, a common genital cancer among women, is caused by high-risk human papillomavirus in more than 99% of cases [1, 2]. HPV vaccination and Pap smear testing reduce its incidence and promote early detection. The treatment of cervical cancer includes surgery, radiotherapy, and chemotherapy, as well as adjuvant therapy such as targeted therapy and immunotherapy. Patients before stage IIB (excluding stage IIB) can be treated with surgery, and locally advanced (IIB-IVA) or relapsed patients are mainly treated with radiotherapy or concurrent chemo-radiation [3]. About 30% of patients relapse after undergoing surgery combined with radio-chemotherapy [4]. Cisplatin (DDP)-based chemotherapy often leads to drug resistance, while targeted therapy and immunotherapy have limited effects [5]. And most patients who relapse have experienced chemotherapy or radio-chemotherapy failure.
DDP, usually administered intravenously, is utilized as the first-line chemotherapy for the cancer patient, including cervical cancer. It exerts cytotoxic effects to inhibit DNA synthesis and cell growth through binding to DNA and forming intra-strand DNA adducts [6]. In addition, it can bind to tubulin, thioredoxin reductase (TrxR), and membrane-bound Na+/H+ exchanger (NHE) protein, impairing their function [7]. It is relatively common for tumors to develop drug resistance after cyclic treatment, and drug resistance is a challenge for DDP-based anti-cancer therapy [8].
Circular RNAs (circRNAs), new regulators of gene expression, are mostly formed by parental genes through reverse splicing and exon skipping or lasso formation [9]. Since circRNAs have no free ends, they are more stable than linear mRNAs. Changes in the expression of some circRNAs can reflect pathological conditions [10], and their biomarker potential has been demonstrated by various researches. In particular, circRNAs have been unmasked to intervene in tumor growth and resistance [11, 12]. Recent shreds of evidence have proved the significant role of circRNAs in cervical cancer resistance. For instance, circRNA-MTO1 and circ-0023404 decreased cervical cancer sensitivity toward DDP through upregulating S100A1 [13] and VEGFA [14], respectively. Hsa_circ_0074269 (circ_0074269), derived from the ANKHD1-EIF4EBP3 gene, has been uncovered to exert a promoting effect on cervical cancer progression [15]. Currently, whether circ_0074269 is related to the resistance of cervical cancer to DDP has not been reported yet.
CircRNAs can sequester microRNAs (miRNAs) by binding to miRNA response elements, thereby acting as competing endogenous RNAs (ceRNAs) to diminish the effect of miRNA-mediated regulatory activities [16]. miRNAs mediate target genes related to cancer progression. miR-485-5p is generally downregulated in different sorts of human tumors [17,18,19], and miR-485-5p can alleviate DDP resistance in ovarian cancer [20] and oral cancer [21]. Also, miR-485-5p exerts an anti-tumor role in cervical cancer [22, 23]. Tuftelin (TUFT1), encoding tuftelin protein, is thought to act on tooth enamel mineralization [24]. Recent studies have discovered the association of TUFT1 with various tumors, and TUFT1 promotes chemoresistance in cervical cancer [25] and breast cancer [26]. However, it is not clear how TUFT1 is deregulated in cervical cancer resistance to DDP.
Our results proved the promoting effect of circ_0074269 on DDP resistance in cervical cancer. Moreover, circ_0074269 increased TUFT1 expression via sequestering miR-485-5p, facilitating DDP resistance and tumor growth in cervical cancer.
Materials and Methods
Study Subjects
Patients with cervical cancer were recruited from Jingjiang People’s Hospital under permission of the Ethical Committee of Jingjiang People’s Hospital. All registered patients signed an informed consent form and received radiotherapy and chemotherapy combined with surgical resection. The samples collected during the operation were divided into DDP-resistant group (23 cases) and DDP-sensitive group (23 cases) according to the evaluation criteria for curative effect of solid tumors (RECIST).
Cell Culture and Treatment
The Ect1/E6E7 cells (CRL-2614, ATCC, Manassas, VA, USA) were cultured in Keratinocyte-Serum Free medium (Sigma-Aldrich (SA), St. Louis, MO, USA) with 0.1 ng/mL human recombinant EGF (SA), 0.05 mg/mL BPE (Cell Applications Inc., San Diego, CA, USA), and 44.1 mg/L CaCl2 (SA). Cervical cancer cell lines CaSki (CRM-CRL-1550, ATCC) and HeLa (CCL-2, ATCC) were respectively cultured in ATCC-formulated RPMI-1640 Medium (30–2001) and EMEM (30–2003) supplemented with 10% fetal bovine serum (Thermo, Waltham, MA, USA) and 1% pen/strep (SA). In an incubator with 37 °C, 95% air, and 5% CO2 were the growth conditions for these cells.
DDP-resistant cervical cancer cells CaSki/DDP-R and HeLa/DDP-R were constructed by exposing their parent cells to various concentrations of DDP (SA) continuously and incrementally.
Oligonucleotide Transfection
When grown to 40–50%, DDP-resistant cervical cancer cells were transfected with oligonucleotides synthesized by GenePharma (Shanghai, Chia) using Lipofectamine 3000 (Thermo) in Opti-MEM medium (Thermo). These oligonucleotide sequences were si-circ_0074269 (5′-TGGATGTACCTGCTATCAGGT-3′), si-NC (5′-UUCUCCGAACGUGUCACGUTT-3′), miR-485-5p inhibitor (anti-miR-485-5p: 5′-GAATTCATCACGGCCAGCCTCT-3′), anti-miR-NC (5′-GGUUCGUACGUACACUGUUCA-3′), miR-485-5p mimic (5′-AGAGGCUGGCCGUGAUGAAUUC-3′), and miR-NC (5′-CGGUACGAUCGCGGCGGGAUAUC-3′).
Plasmid Construction and Transfection
The TUFT1 overexpression plasmid (TUFT1) was constructed using the pcDNA3.1 vector (Thermo) with BamHI and XhoI restriction enzymes. A plasmid mixed with Lipofectamine 3000 in Opti-MEM medium (Thermo) was used to transfect DDP-resistant cervical cancer cells.
Real-Time Quantitative Polymerase Chain Reaction
RNA isolation was conducted using the TRIzol reagent (Thermo), followed by purification with the RNeasy kit (Qiagen, Valencia, CA, USA). Four hundred nanograms of total RNA were used in the complementary DNA synthesis reaction with an M-MLV Reverse Transcriptase Kit (Promega, Madison, WI, USA) or miScript II RT Kit (Qiagen). Quantitative PCR was done in triplicate using a ChamQ Universal SYBR® qPCR Master Mix (Vazyme, Nanjing, China) with specific primers (Table 1). The fold change was calculated by the equation 2−ΔΔCt, and the housekeeping genes β-actin and U6 were used for normalization.
In Vitro Drug-Sensitivity and Viability Analysis
About 1 × 104 DDP-resistant cells were cultured in the complete medium containing different concentrations of DDP (0, 2.5, 5, 10, 20, 40, 80, 160, 320 μg/mL) for 48 h, followed by incubation with 20 μL of MTT solution (5 mg/mL, SA). Four hours later, 150 μL of DMSO was used to dissolve the crystals and the absorbance at 490 nm was measured with an automatic enzyme-linked immunosorbent assay reader (BioTek, Winooski, VT, USA) for analysis of cell viability. The IC50 value was calculated from logarithmic sigmoidal dose–response curves generated using Graphpad Prism 8.0 software (Graphpad, San Diego, CA, USA).
5-ethynyl-2’-deoxyuridine Assay
Cell proliferation was analyzed using the Cell-Light EdU Apollo In Vitro Kit (Ribobio, Guangzhou, China). About 1 × 104 DDP-resistant cells were cultured for 24 h, followed by incubation with 100 μL of 50 μM EdU for 2 h. The cells were decolorized with 50 μL of 2 mg/mL glycine after immobilization with 4% paraformaldehyde. Then, 100 μL of 0.5% Triton X-100 was utilized to increase the permeability of the cell membrane. Afterward, the cells were incubated with 100 μL of 1 × Apollo-staining reaction solution, followed by staining with 100 μL of DAPI. A fluorescence microscope (Olympus, Tokyo, Japan) was used to capture images.
Colony Formation Assay
About 5 × 103 cells were seeded into 6-well plates and allowed to grow for 10 days. The cells were then stained with 0.1% crystal violet (Solarbio, Beijing, China), and the colonies were visualized using a microscope (Olympus).
Flow Cytometry Assay
Detection of cell apoptosis was carried out with the Annexin V-FITC/propidium iodide (PI) apoptosis detection kit (Beyotime, Shanghai, China) following the manufacturer’s instructions. In brief, the harvested cells were double-stained with FITC and PI, followed by analysis on a BD FACS flow cytometer (Beckman Coulter, Brea, CA, USA).
Wound-healing Assay
About 1 × 103 cells were seeded into 24-well plates and allowed to grow to 90% confluence, followed by creating wounds in the cell monolayer with a 200-μL sterile pipette tip. The plates were incubated for 24 h after removing the cell debris, followed by visualization and quantification of the scratch width.
Western Blotting
Proteins were extracted using RIPA lysis buffer (Thermo) and quantified using a BCA protein assay kit (Thermo). Western blotting was carried out as previously described [27]. Primary antibodies used for western blotting analysis were cleaved-caspase-3 (ab32042, 1:500, Abcam, Cambridge, MA, USA), MRP1 (ab260038, 1:1000, Abcam), TUFT1 (ab184949, 1:1000, Abcam), and GAPDH (ab181603, 1:10,000, Abcam). Bands were visualized using chemiluminescence (Thermo), and ImageJ software (NIH) was used for densitometry analysis.
Generation of Stable circ_0074269-knockdown HeLa/DDP-R Cells
The sh-circ_0074269 or sh-NC synthesized by GenePharma was cloned into lentiviral pLKO.1 vector (SA). The plasmids constructed above were respectively transfected into HEK293T cells along with the packaging plasmids psPAX2 and pMD2.G in accordance with the manufacturer’s guidelines. The collected viral supernatants from HEK293T cells were used to infect HeLa/DDP-R cells, followed by selection of the stable cell line with puromycin (2 μg/mL).
Animal Experiments
Xenograft experiments were conducted with the permission authorized by the Animal Care Committee of Jingjiang People’s Hospital. In short, 24 female BALB/c nude mice (5 weeks old, 15–20 g) (Vital River Laboratory, Beijing, China) were randomly divided into 4 groups (6 mice in each group) and processed as follows: (1) sh-NC + PBS; (2) sh-NC + DDP; (3) sh-circ_0074269 + PBS; (4) sh-circ_0074269 + DDP. Specifically, the HeLa/DDP-R cells (4 × 106) carrying sh-NC or sh-circ_0074269 were injected subcutaneously into the armpits of the mice. One week later, the mice were administered intraperitoneally with 200 μL of PBS or PBS containing DDP (7.5 mg/kg) every 3 days. Tumor volume was monitored in synchronization with the administration of PBS (volume = (length × width2)/2). Twenty-three days after the injection, the mice were killed, followed by excision of the tumor and measurement of its weight.
Immunohistochemistry Analysis and Tunel Assay
IHC analysis was performed as previously described [28]. An antibody against Ki67 (ab279653, 1:100, Abcam) was used for proliferation analysis in tissues. Tunel assay was performed to evaluate cell apoptosis in tissues using the Tunel kit (Promega, Madison, WI, USA) following the manufacturer’s instructions.
Dual-Luciferase Reporter Assay
Luciferase reporters were generated by ligating the fragments of WT-circ_0074269, MUT-circ_0074269, WT-TUFT1 3′UTR, or MUT-TUFT1 3′UTR into the psiCHECK-2 vector (Promega), respectively. DDP-resistant cells were co-transfected with miR-485-5p mimic or miR-NC and a luciferase reporter. The relative luciferase activity was normalized to Renilla luciferase activity after measuring the luciferase activity using the dual-luciferase assay system (Promega).
RNA Immunoprecipitation Assay
A Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore) was utilized for RIP analysis following the manufacturers’ guidelines. The antibodies used in RIP analysis were anti-Ago2 (Abcam) and anti-IgG (Abcam). The immunoprecipitated RNAs were detached and then purified for RT-qPCR analysis.
Isolation and Identification of Exosomes
In short, the collected cell culture supernatant was centrifuged as follows: 300 g for 15 min, 2000 g for 15 min, and 10,000 g for 40 min. The supernatant was filtered with a 0.22-μm filter after each centrifugation. The particles obtained after centrifugation (110,000 g, 75 min) of the filtrate were considered exosomes. The surface markers CD63 and CD9 of exosomes were analyzed by western blotting with antibodies against CD63 (ab134045, Abcam) and CD9 (ab236630, Abcam). The morphology of the isolated exosomes was observed using a transmission electron microscope (TEM) (JEM-2100; JEOL, Tokyo, Japan). The concentration and diameter distribution of the isolated exosomes were analyzed using NanoSight LM10 system (NanoSight, Amesbury, UK) with NTA 2.3 analysis software (NanoSight).
Statistical Analysis
All in vitro experiments included 3 biological replicates. Statistical analysis was performed using Graphpad Prism 8.0 software (GraphPad), in which the data was evaluated using the mean ± SD. Our data conformed to the normal distribution and had been tested using the Shapiro–Wilk normality test. Comparisons between 2 groups were made by Student’s t-test, whereas comparisons among more than 2 groups were made by one-way and two-way ANOVA. Statistical significance was ascribed when P < 0.05.
Results
Elevated circ_0074269 Expression in Cervical Cancer-resistant Samples and Cells
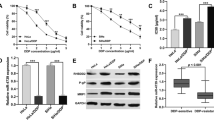
To address the role of circ_0074269 in DDP resistance of cervical cancer, we first validated the overexpression of circ_0074269 in cervical cancer-resistant samples (Fig. 1A). We established 2 DDP-resistant cervical cancer cell lines CaSki/DDP-R and HeLa/DDP-R, and the resistance to DDP in DDP-resistant cell lines was increased relative to their parental cells (Fig. 1B). Moreover, circ_0074269 expression was higher in cervical cancer cells (compared to Ect1/E6E7 cells) and DDP-resistant cells (compared to their parental cells) (Fig. 1C). Furthermore, circ_0074269 was resistant to RNase R exonuclease, while linear GAPDH was digested post-RNase R treatment (Fig. 1D and E). Compared with random primers, circ_0074269 expression in DDP-resistant cells was significantly reduced when using Oligo (dT)18 primers, but not linear GAPDH, indicating that circ_0074269 had no poly-A tail (Fig. 1F and G). In addition, RT-qPCR assay presented the predominant cytoplasmic distribution of circ_0074269 in DDP-resistant cells (Fig. 1H and I). In conclusion, circ_0074269 was overexpressed in DDP-resistant cervical cancer.
Circ_0074269 expression was elevated in DDP-resistant cervical cancer. A Relative levels of circ_0074269 in DDP-resistant samples and DDP-sensitive samples (unpaired Student’s t-test). B The IC50 of DDP-resistant cells (one-way ANOVA). C Relative levels of circ_0074269 in DDP-resistant cells and their parental cells (one-way ANOVA). D and E Relative levels of circ_0074269 and linear GAPDH in DDP-resistant cells treated with RNase R (two-way ANOVA). Both groups were normalized with the β-actin level of the mock group (as an internal control). F and G Analysis of circ_0074269 and linear GAPDH in DDP-resistant cells with random and oligo (dT)18 primers (two-way ANOVA). H and I The percentage of circ_0074269 in the cytoplasm and nucleus of DDP-resistant cells (two-way ANOVA). ***P < 0.001 and ****P < 0.0001
Circ_0074269 Silencing Increased Cervical Cancer Cell Sensitivity to DDP
To interrogate the potential significance of circ_0074269 in cervical cancer resistance toward DDP, we knocked down circ_0074269 in DDP-resistant cells with si-circ_0074269 (Fig. 2A). Moreover, a high sensitivity toward DDP was observed in circ_0074269-knockdown DDP-resistant cells (Fig. 2B). Also, circ_0074269 silencing caused a decrease in circ_0074269 expression in both HeLa and HeLa/DDP-R cell lines. However, circ_0074269 knockdown decreased the IC50 of DDP in the HeLa/DDP-R cell lines but not the HeLa cell line (supplementary Fig. 1). The MTT assay exhibited lower cell viability in circ_0074269-knockdown DDP-resistant cells than control cells (Fig. 2C). EdU and colony formation assays showed that circ_0074269 silencing resulted in an overt reduction in the number of EdU-positive cells and colony formation, manifesting that circ_0074269 knockdown could decrease DDP-resistant cell proliferation (Fig. 2D and E). Flow cytometry assay presented that circ_0074269 knockdown elevated cell apoptosis in DDP-resistant cells (Fig. 2F). Wound-healing assay indicated that silenced circ_0074269 expression decreased DDP-resistant cell migration (Fig. 2G). In addition, circ_0074269 inhibition decreased MRP1 protein levels and elevated cleaved-caspase-3 protein levels (Fig. 2F and I). Collectively, these results suggested the promoting effect of circ_0074269 on resistance toward DDP in cervical cancer.
Downregulation of circ_0074269 reduced cervical cancer cell resistance toward DDP. A Knockdown efficiency of circ_0074269 was measured by RT-qPCR (two-way ANOVA). B The IC50 value of circ_0074269-knockdown DDP-resistant cells (two-way ANOVA). C, D, E, F, G The viability (C), proliferation (D), colony formation (E), apoptosis (F), and migration (G) of circ_0074269-knockdown DDP-resistant cells were evaluated (two-way ANOVA). H and I Relative protein levels of cleaved-caspase-3 and MRP1 in circ_0074269-knockdown DDP-resistant cells (two-way ANOVA). **P < 0.01, ***P < 0.001, and ****P < 0.0001
Circ_0074269 Silencing Improved Cervical Cancer Cell Sensitivity Toward DDPIn Vivo
To corroborate our in vitro findings, we constructed a stable circ_0074269 knockdown HeLa/DDP-R cell line, and the knockdown efficiency was exhibited in Fig. 3A. And the effect of circ_0074269 knockdown produced anti-tumor activity was further verified by mouse models. Administration of sh-NC + DDP or sh-circ_0074269 + PBS decreased tumor volume and weight compared with the sh-NC + PBS control. Administration of sh-circ_0074269 + DDP also significantly decreased tumor volume and weight with respect to the sh-NC + DDP control (Fig. 3B and C). Furthermore, administration of sh-circ_0074269 + PBS overtly decreased circ_0074269 expression in tumor tissues relative the sh-NC + PBS control, and mice administrated with sh-circ_0074269 + DDP showed lower circ_0074269 levels compared to the sh-NC + DDP control (Fig. 3D). Also, mice administrated with sh-circ_0074269 + PBS and sh-circ_0074269 + DDP showed a decrease in the number of Ki67-positive cells, but an elevation in the number of Tunel-positive cells, manifesting circ_0074269 silencing repressed cell proliferation and induced cell apoptosis (Fig. 3E). In summary, these findings demonstrated that circ_0074269 knockdown reduced tumor growth and sensitized the response to DDP in vivo.
Inhibition of circ_0074269 reduced tumor growth and sensitized the response to DDP in vivo. A The knockdown efficiency of sh-circ_0074269 in HeLa/DDP-R cells (unpaired Student’s t-test). B and C Tumor volume and weight of mice administrated with sh-NC + PBS, sh-NC + DDP, sh-circ_0074269 + PBS, and sh-circ_0074269 + DDP (one-way ANOVA). D Relative levels of circ_0074269 in tumor tissues derived from mice (one-way ANOVA). E IHC analysis and Tunel assay showing the number of Ki67/Tunel-positive cells in tumor tissues. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001
Circ_0074269 was a ceRNA of miR-485-5p
Circ_0074269 was mainly in the cytoplasm, enabling it to act as a ceRNA. To clarify the specific miRNAs interrelating with circ_0074269, we used the online tool starbase to search for partner molecules that could bind to circ_0074269. And miR-485-5p had sequence complementarity with circ_0074269 on 11 bases (Fig. 4A). We then overexpressed miR-485-5p in DDP-resistant cells (Fig. 4B), and miR-485-5p overexpression restrained the luciferase activity in DDP-resistant cells with the WT-circ_0074269 reporter (Fig. 4C and D). Moreover, RIP assay showed that circ_0074269 and miR-485-5p were enriched in Ago2-RIPs compared with control IgG-RIPs (Fig. 4E and F). The Cancer Genome Atlas (TCGA) database download cervical squamous cell carcinoma (CESC) data showed lower levels of miR-485-5p in the primary tumor and individual cancer stages of CESC (Fig. 4G and H). Similar results were also observed in DDP-resistant cervical cancer tissues (Fig. 4I), and miR-485-5p expression was negatively correlated with circ_0074269 expression (Fig. 4J). Similarly, miR-485-5p expression was decreased in DDP-resistant cells compared to their parental cells (Fig. 4K). Together, these results indicated that circ_0074269 served as a miR-485-5p sponge.
Circ_0074269 acted as a miR-485-5p decoy. A A schematic drawing showing the putative miR-485-5p-binding sites with respect to circ_0074269. B Relative levels of miR-485-5p in DDP-resistant cells after transfection of miR-485-5p mimic or miR-NC (two-way ANOVA). C, D, E, F Dual-luciferase reporter and RIP assays were performed to verify the prediction relationship between circ_0074269 and miR-485-5p (two-way ANOVA). G and H Expression of miR-485-5p in the primary tumor and individual cancer stages of CESC in the TCGA database. I Relative levels of miR-485-5p in DDP-resistant samples and DDP-sensitive samples (unpaired Student’s t-test). J The correlation between circ_0074269 and miR-485-5p in DDP-resistant samples. K Relative levels of miR-485-5p in DDP-resistant cells and their parental cells (one-way ANOVA). **P < 0.01, ***P < 0.001, and ****P < 0.0001
Circ_0074269 Mediated Cervical Cancer Cell Sensitivity Toward DDP Through miR-485-5p
We further investigated the interaction between circ_0074269 and miR-485-5p in DDP-resistant cell resistance toward DDP. Transfection of miR-485-5p inhibitor reversed the elevated miR-485-5p expression in circ_0074269-silenced DDP-resistant cells (Fig. 5A). Moreover, miR-485-5p inhibitor weakened the decreased IC50 value of DDP-resistant cells mediated by circ_0074269 knockdown (Fig. 5B). In addition, the altered viability, proliferation, colony formation, apoptosis, and migration in DDP-resistant cells caused by circ_0074269 inhibition were impaired after miR-485-5p silencing (Fig. 5C-G). And the elevated cleaved-caspase-3 protein levels and the reduced MRP1 protein levels in circ_0074269-inhibiting DDP-resistant cells were overturned by miR-485-5p knockdown (Fig. 5H and I). In summary, circ_0074269 regulated cervical cancer cell sensitivity toward DDP via miR-485-5p.
circ_0074269 sponged miR-485-5p to mediate cervical cancer cell sensitivity toward DDP. A, B, C, D, E, F, G, H, I DDP-resistant cervical cancer cells were transfected with si-NC, si-circ_0074269, si-circ_0074269 + anti-miR-NC, or si-circ_0074269 + anti-miR-485-5p. A Relative levels of miR-485-5p in DDP-resistant cells (two-way ANOVA). B The IC50 value of DDP-resistant cells (two-way ANOVA). C, D, E, F, G Evaluation of cell viability, proliferation, colony formation, apoptosis, and migration in DDP-resistant cells was performed (two-way ANOVA). H and I Relative protein levels of MRP1 and cleaved-caspase-3 in DDP-resistant cells (two-way ANOVA). **P < 0.01, ***P < 0.001, and ****P < 0.0001
TUFT1 was a Direct Target of miR-485-5p
Bioinformatics analysis predicted that TUFT1 might be a target of miR-485-5p, and the predicted binding sites were presented in Fig. 6A. And miR-485-5p upregulation led to a decrease in the luciferase activity of the WT-TUFT1 3′UTR reporter (Fig. 6B and C). RT-qPCR revealed an over 37-fold enrichment of miR-485-5p and TUFT1 in purified RNAs derived from the Ago2 group (Fig. 6D and E). In addition, there was an apparent elevation in TUFT1 expression in the primary tumor and individual cancer stages of CESC (Fig. 6F–H). And the upregulation of TUFT1 mRNA was also obtained in DDP-resistant tumor samples (Fig. 6I). Moreover, a negative correlation between miR-485-5p and TUFT1 mRNA was observed in DDP-resistant tumor samples (Fig. 6J). Consistently, TUFT1 protein levels showed an upregulation trend in DDP-resistant tumor samples and cell lines compared to controls (Fig. 6K and L). Collectively, TUFT1 acted as a target of miR-485-5p.
Identification of the targeting relationship between miR-485-5p and TUFT1. A Sequence alignment of the miR-485-5p and TUFT1 3′-UTR-binding sites. B and C Relative luciferase activities in DDP-resistant cells co-transfected with miR-485-5p mimic or miR-NC and WT-TUFT1 3′UTR or MUT-TUFT1 3′UTR (two-way ANOVA). D and E RT-qPCR analysis of miR-485-5p and TUFT1 mRNA levels in DDP-resistant cells using an antibody against Ago2 or IgG (two-way ANOVA). F, G, H Expression of TUFT1 mRNA in the primary tumor and individual cancer stages of CESC in the TCGA database. I The TUFT1 mRNA levels in DDP-resistant/-sensitive cervical cancer samples (unpaired Student’s t-test). J Correlation of TUFT1 mRNA and miR-485-5p expression in DDP-resistant cervical cancer samples. K and L The TUFT1 protein levels in DDP-resistant cervical cancer samples (K, unpaired Student’s t-test) and DDP-resistant cells (L, one-way ANOVA). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001
miR-485-5p Elevated Cervical Cancer Cell Sensitivity to DDP Via Targeting TUFT1
To validate whether miR-485-5p-mediated functional effects depend specifically on TUFT1, DDP-resistant cells were co-transfected with miR-485-5p mimic and TUFT1 overexpression plasmid. miR-485-5p mimic repressed TUFT1 protein levels in DDP-resistant cells, but this effect was weakened after introduction of the TUFT1 overexpression plasmid (Fig. 7A). Additionally, miR-485-5p mimic caused a decrease in IC50 value, viability, proliferation, and colony formation, an increase in apoptosis rate, and a reduction in migration distance in DDP-resistant cells, but these changes were partially overturned after TUFT1 overexpression (Fig. 7B-G). Furthermore, miR-485-5p overexpression elevated cleaved-caspase-3 protein levels and reduced MRP1 protein levels, but these effects were impaired by TUFT1 upregulation (Fig. 7H and I). Collectively, miR-485-5p decreased cervical cancer cell sensitivity to DDP via targeting TUFT1.
miR-485-5p targeted TUFT1 to mediate cervical cancer cell sensitivity to DDP. A, B, C, D, E, F, G, H, I DDP-resistant cells were transfected with miR-NC, miR-485-5p, miR-485-5p + pcDNA, and miR-485-5p + TUFT1. A Analysis of TUFT1 protein levels in DDP-resistant cells (two-way ANOVA). B, C, D, E, F, G The IC50, viability, proliferation, colony formation, apoptosis, and migration of DDP-resistant cells were evaluated (two-way ANOVA). H and I Detection of cleaved-caspase-3 and MRP1 protein levels in DDP-resistant cells (two-way ANOVA). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001
Circ_0074269 Mediated TUFT1 Expression Through Sponging miR-485-5p
We then determined whether circ_0074269 could act as an endogenous miR-485-5p sponge to regulate TUFT1 expression. As shown in Fig. 8A and B, circ_0074269 knockdown resulted in a decrease in TUFT1 mRNA and protein levels in DDP-resistant cells, but the decrease in TUFT1 mRNA and protein levels caused by circ_0074269 knockdown was attenuated by miR-485-5p silencing, manifesting that circ_0074269 regulated TUFT1 expression through acting as an endogenous miR-485-5p sponge.
Circ_0074269 mediated TUFT1 expression through acting as a miR-485-5p sponge. A and B Relative levels of TUFT1 mRNA and protein in DDP-resistant cells with si-NC, si-circ_0074269, si-circ_0074269 + anti-miR-NC, or si-circ_0074269 + anti-miR-485-5p were detected (one-way ANOVA). **P < 0.01, ***P < 0.001, and ****P < 0.0001
Exosomes were Involved in the Transport of circ_0074269
Tumor cell–derived exosomes have been reported to participate in drug resistance [29]. Thus, we explored whether circ_0074269 could be transported via exosomes. Ultracentrifugation was carried out to isolate exosomes from the culture medium used to incubate DDP-resistant cervical cancer cells. TEM analysis showed that the separated exosomes had a round appearance (Fig. 9A). And the isolated pellets were determined to be exosomes by detecting the exosomal markers CD63 and CD9 (Fig. 9B). NTA showed that the size of the isolated exosomes ranged from 30 to 160 nm (Fig. 9C). Relative levels of circ_0074269 in exosomes from DDP-resistant cervical cancer cells and their parental cells were detected, and the results exhibited that circ_0074269 expression was higher in exosomes from cervical cancer cells (relative to Ect1/E6E7 cells) and DDP-resistant cervical cancer cells (relative to their parental cells) (Fig. 9D). We then incubated cervical cancer cells with exosomes derived from DDP-resistant cervical cancer cells, and an elevation in circ_0074269 expression was obtained in cervical cancer cells after incubation with exosomes (Fig. 9E). In addition, circ_0074269 expression in exosomes from DDP-resistant cervical cancer cells with GW4869 treatment was lower than that in exosomes from DDP-resistant cervical cancer cells with DMSO treatment, indicating that circ_0074269 existed in exosomes derived from DDP-resistant cervical cancer cells (Fig. 9F). These results manifested that circ_0074269 could be delivered in cervical cancer cells via exosomes.
Circ_0074269 could be delivered via exosomes. A TEM analysis of the morphology of separated exosomes. B The exosomal markers CD63 and CD9 were detected by western blotting. C NTA showing the size and number of isolated exosomes. D Relative levels of circ_0074269 in exosomes from cervical cancer cells (relative to Ect1/E6E7 cells) and DDP-resistant cervical cancer cells (relative to their parental cells) (one-way ANOVA). E Relative levels of circ_0074269 in cervical cancer cells treated with exosomes from DDP-resistant cervical cancer cells or PBS (two-way ANOVA). F Relative levels of circ_0074269 in exosomes from DDP-resistant cervical cancer cells with GW4869 or DMSO treatment (two-way ANOVA). **P < 0.01, ***P < 0.001, and ****P < 0.0001
Discussion
Most patients are prone to developing acquired DDP resistance, which is a huge obstacle encountered in cervical cancer treatment [30]. In recent years, several circRNAs have been confirmed to be associated with chemoresistance in different cancers, including cervical cancer [31]. So far, the mechanisms by which circRNAs mediate cervical cancer resistance to DDP resistance are unclear.
Our data verified the upregulation of circ_0074269 in DDP-resistant cervical cancer samples and cells. In xenograft models, circ_0074269 silencing decreased tumor growth and elevated DDP sensitivity. In vitro functional experiments uncovered that circ_0074269 knockdown reduced DDP resistance, repressed proliferation and migration, as well as induced apoptosis of DDP-resistant cervical cancer cells. These findings manifested that circ_0074269 might be a key node of cervical cancer intervention treatment. Our results were consistent with the published article that circ_0074269 silencing reduced cervical cancer cell proliferation and induced cervical cancer cell apoptosis [15]. In contrast, circ_0074269 was overtly low-expressed in hepatocellular cancer samples and cells, and circ_0074269 overexpression curbed cell growth in vitro and xenograft models, which might be related to the tissue-specific expression of circ_0074269.
The miRNA sponge function of circRNAs has been well demonstrated in tumorigenesis [32]. Herein, circ_0074269 preferred to locate in the cytoplasm of DDP-resistant cervical cancer cells, implying that circ_0074269 might function as a miRNA sponge. After bioinformatics analysis, circ_0074269 was identified as a potential sponge for miR-485-5p, which was reported to play a tumor-inhibiting role in cervical cancer [33,34,35]. Also, miR-485-5p silencing weakened circ_0074269 knockdown-mediated effects on DDP resistance, illustrating that circ_0074269 regulated DDP resistance via miR-485-5p in cervical cancer.
Furthermore, TUFT1 was validated as a miR-485-5p target. TUFT1 exhibits an oncogenic role in tumor progression [36,37,38]. Our results were consistent with Luo et al.’s report showing that TUFT1 was overexpressed in DDP-resistant cervical cancer cells [25]. Herein, TUFT1 upregulation impaired miR-485-5p mimic-mediated effects on DDP resistance, manifesting that miR-485-5p mediated DDP resistance through TUFT1 in cervical cancer. Notably, circ_0074269 could act as a ceRNA to regulate TUFT1 expression through adsorbing miR-485-5p. Accordingly, we inferred that circ_0074269 sponged miR-485-5p to elevate TUFT1 expression, thus promoting DDP resistance in cervical cancer.
Exosomes, tiny extracellular vesicles secreted by different types of cells, play a vital role in cell-to-cell communication [39]. Researchers found that circRNAs are enriched and stable in exosomes, and circRNAs can participate in tumor resistance through exosomes [40,41,42]. More strikingly, we discovered that circ_0074269 was also overexpressed in exosomes from DDP-resistant cervical cancer cells, and circ_0074269 expression was higher in cervical cancer cells after co-incubation with exosomes from DDP-resistant cervical cancer cells, indicating that circ_0074269 could be delivered via exosomes. Thus, we concluded that circ_0074269 could regulate DDP resistance in cervical cancer via exosomes. Unfortunately, we have not further explored whether exosomal circ_0074269 is involved in DDP resistance in cervical cancer, which can be explored in depth in the future.
In conclusion, circ_0074269 promoted DDP resistance via upregulating TUFT1 via sponging miR-485-5p in cervical cancer. We reported a new mechanism by which circ_0074269 decreased DDP sensitivity, contributing new evidence to support circ_0074269 as a treatment target for cervical cancer.
Data Availability
Not applicable.
References
de Martel C, Georges D, Bray F, Ferlay J, Clifford GM. Global burden of cancer attributable to infections in 2018: a worldwide incidence analysis. Lancet Glob Health. 2020;8(2):e180–90.
Crosbie EJ, Einstein MH, Franceschi S, Kitchener HC. Human papillomavirus and cervical cancer. Lancet. 2013;382(9895):889–99.
Cohen PA, Jhingran A, Oaknin A, Denny L. Cervical cancer. Lancet. 2019;393(10167):169–82.
Shieh KR, Huang A, Xu Y. Response to Immune checkpoint inhibitor treatment in advanced cervical cancer and biomarker study. Front Med. 2021;8:669587.
Chung HC, Ros W, Delord JP, Perets R, Italiano A, Shapira-Frommer R, Manzuk L, Piha-Paul SA, Xu L, Zeigenfuss S, et al. Efficacy and safety of pembrolizumab in previously treated advanced cervical cancer: results from the phase II KEYNOTE-158 study. J Clin Oncol. 2019;37(17):1470–8.
Shen D-W, Pouliot LM, Hall MD, Gottesman MM. Cisplatin resistance: a cellular self-defense mechanism resulting from multiple epigenetic and genetic changes. Pharmacol Rev. 2012;64(3):706–21.
Makovec T. Cisplatin and beyond: molecular mechanisms of action and drug resistance development in cancer chemotherapy. Radiol Oncol. 2019;53(2):148–58.
Tchounwou PB, Dasari S, Noubissi FK, Ray P, Kumar S. Advances in our understanding of the molecular mechanisms of action of cisplatin in cancer therapy. J Exp Pharmacol. 2021;13:303–28.
Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P, Wu M. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94.
Verduci L, Tarcitano E. CircRNAs: role in human diseases and potential use as biomarkers. Cell Death Dis. 2021;12(5):468.
Cui C, Yang J, Li X, Liu D, Fu L, Wang X. Functions and mechanisms of circular RNAs in cancer radiotherapy and chemotherapy resistance. Mol Cancer. 2020;19(1):58.
Jeyaraman S, Hanif EAM, Ab Mutalib NS, Jamal R, Abu N. Circular RNAs: potential regulators of treatment resistance in human cancers. Front Genet. 2019;10:1369.
Chen M, Ai G, Zhou J, Mao W, Li H, Guo J. circMTO1 promotes tumorigenesis and chemoresistance of cervical cancer via regulating miR-6893. Biomed Pharmacother. 2019;117:109064.
Guo J, Chen M, Ai G, Mao W, Li H, Zhou J. Hsa_circ_0023404 enhances cervical cancer metastasis and chemoresistance through VEGFA and autophagy signaling by sponging miR-5047. Biomed Pharmacother. 2019;115:108957.
Zhu Y, Jiang X, Zhang S. Hsa_circ_103973 acts as a sponge of miR-335 to promote cervical cancer progression. Onco Targets Ther. 2020;13:1777–86.
Verduci L, Strano S, Yarden Y, Blandino G. The circRNA-microRNA code: emerging implications for cancer diagnosis and treatment. Mol Oncol. 2019;13(4):669–80.
Lou C, Xiao M, Cheng S, Lu X, Jia S, Ren Y, Li Z. MiR-485–3p and miR-485–5p suppress breast cancer cell metastasis by inhibiting PGC-1α expression. Cell Death Dis. 2016;7(3):e2159.
Tu J, Zhao Z, Xu M, Chen M, Weng Q, Ji J. LINC00460 promotes hepatocellular carcinoma development through sponging miR-485–5p to up-regulate PAK1. Biomed Pharmacother. 2019;118:109213.
Liu H, Hu G, Wang Z, Liu Q, Zhang J, Chen Y, Huang Y, Xue W, Xu Y, Zhai W. circPTCH1 promotes invasion and metastasis in renal cell carcinoma via regulating miR-485-5p/MMP14 axis. Theranostics. 2020;10(23):10791–807.
Qiao HF, Liu YL, You J, Zheng YL, Chen LP, Lu XY, Du L, Shan F, Liu MH. G-5555 synergized miR-485–5p to alleviate cisplatin resistance in ovarian cancer cells via Pi3k/Akt signaling pathway. J Reprod Immunol. 2020;140:103129.
Lin XJ, He CL, Sun T, Duan XJ, Sun Y, Xiong SJ. hsa-miR-485-5p reverses epithelial to mesenchymal transition and promotes cisplatin-induced cell death by targeting PAK1 in oral tongue squamous cell carcinoma. Int J Mol Med. 2017;40(1):83–9.
Ou R, Lv J, Zhang Q, Lin F, Zhu L, Huang F, Li X, Li T, Zhao L, Ren Y, et al. circAMOTL1 motivates AMOTL1 expression to facilitate cervical cancer growth. Mol Ther Nucleic Acids. 2020;19:50–60.
Zhao D, Zhang H, Long J, Li M. LncRNA SNHG7 functions as an oncogene in cervical cancer by sponging miR-485–5p to modulate JUND Expression. Onco Targets Ther. 2020;13:1677–89.
Deutsch D, Palmon A, Fisher LW, Kolodny N, Termine JD, Young MF. Sequencing of bovine enamelin (“tuftelin”) a novel acidic enamel protein. J Biol Chem. 1991;266(24):16021–8.
Luo X, Wei J, Yang FL, Pang XX, Shi F, Wei YX, Liao BY, Wang JL. Exosomal lncRNA HNF1A-AS1 affects cisplatin resistance in cervical cancer cells through regulating microRNA-34b/TUFT1 axis. Cancer Cell Int. 2019;19:323.
Liu W, Han J, Shi S, Dai Y, He J. TUFT1 promotes metastasis and chemoresistance in triple negative breast cancer through the TUFT1/Rab5/Rac1 pathway. Cancer Cell Int. 2019;19:242.
Yang S, Shi F, Du Y, Wang Z, Feng Y, Song J, Liu Y. Long non-coding RNA CTBP1-AS2 enhances cervical cancer progression via up-regulation of ZNF217 through sponging miR-3163. Cancer Cell Int. 2020;20:343.
Luo KW, Zhu XH, Zhao T, Zhong J, Gao HC, Luo XL, Huang WR. EGCG enhanced the anti-tumor effect of doxorubicine in bladder cancer via NF-κB/MDM2/p53 Pathway. Front Cell Dev Biol. 2020;8:606123.
Mashouri L, Yousefi H, Aref AR, Ahadi AM, Molaei F, Alahari SK. Exosomes: composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol Cancer. 2019;18(1):75.
Zhu H, Luo H, Zhang W, Shen Z, Hu X, Zhu X. Molecular mechanisms of cisplatin resistance in cervical cancer. Drug Des Devel Ther. 2016;10:1885–95.
Xu T, Wang M, Jiang L, Ma L, Wan L, Chen Q, Wei C, Wang Z. CircRNAs in anticancer drug resistance: recent advances and future potential. Mol Cancer. 2020;19(1):127.
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–8.
Dai Y, Xie F, Chen Y. Reduced levels of miR-485–5p in HPV-infected cervical cancer promote cell proliferation and enhance invasion ability. FEBS Open Bio. 2020;10(7):1348–61.
Ou R, Lv J, Zhang Q, Lin F, Zhu L, Huang F, Li X, Li T, Zhao L, Ren Y, et al. circAMOTL1 motivates AMOTL1 expression to facilitate cervical cancer growth. Onco Targets Ther. 2020;19:50–60.
Zhao D, Zhang H, Long J, Li M. LncRNA SNHG7 functions as an oncogene in cervical cancer by sponging miR-485-5p to modulate JUND expression. FEBS Open Bio. 2020;13:1677–89.
Zhou B, Zhan H, Tin L, Liu S, Xu J, Dong Y, Li X, Wu L, Guo W. TUFT1 regulates metastasis of pancreatic cancer through HIF1-Snail pathway induced epithelial-mesenchymal transition. Cancer Lett. 2016;382(1):11–20.
Liu H, Zhu J, Mao Z, Zhang G, Hu X, Chen F. Tuft1 promotes thyroid carcinoma cell invasion and proliferation and suppresses apoptosis through the Akt-mTOR/GSK3β signaling pathway. Am J Transl Res. 2018;10(12):4376–84.
Wu MN, Zheng WJ, Ye WX, Wang L, Chen Y, Yang J, Yao DF, Yao M. Oncogenic tuftelin 1 as a potential molecular-targeted for inhibiting hepatocellular carcinoma growth. World J Gastroenterol. 2021;27(23):3327–41.
Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367(6478).
Luo Y, Gui R. Circulating exosomal circFoxp1 confers cisplatin resistance in epithelial ovarian cancer cells. J Gynecol Oncol. 2020;31(5):e75.
Zhang PF, Gao C, Huang XY, Lu JC, Guo XJ, Shi GM, Cai JB, Ke AW. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. J Gynecol Oncol. 2020;19(1):110.
Ding C, Yi X, Chen X, Wu Z, You H, Chen X, Zhang G, Sun Y, Bu X, Wu X, et al. Warburg effect-promoted exosomal circ_0072083 releasing up-regulates NANGO expression through multiple pathways and enhances temozolomide resistance in glioma. J Exp Clin Cancer Res. 2021;40(1):164.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
Written informed consents were obtained from all participants and this study was permitted by the Ethics Committee of Jingjiang People’s Hospital.
Consent for Publication
Not applicable.
Competing Interests
The authors declare no competing interests.
Supplementary Information

Rights and permissions
About this article
Cite this article
Chen, J., Wu, S., Wang, J. et al. Hsa_circ_0074269-mediated Upregulation of TUFT1 Through miR-485-5p Increases Cisplatin Resistance in Cervical Cancer. Reprod. Sci. 29, 2236–2250 (2022). https://doi.org/10.1007/s43032-022-00855-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43032-022-00855-9