Abstract
The world is heading towards an era of intractable and impending untreatable N. gonorrhoeae, thereby underlining the significance of rapid and accurate prediction of drug resistance as an indispensable need of the hour. In the present study, we optimized and evaluated a stable isotope labeling-based approach using the MALDI-TOF MS (Matrix-Assisted Laser Desorption/Ionization-Time of Flight Mass Spectrometry) for rapid and reliable detection of ciprofloxacin and azithromycin resistance in N. gonorrhoeae. All the isolates were cultured under three varied condition setups viz. medium supplemented with normal lysine, heavy lysine (isotope), and heavy lysine along with the antibiotics (ciprofloxacin/azithromycin), respectively. After incubation, spectra were acquired using the MALDI-TOF MS which were further screened for unique patterns (media-specific spectra) to differentiate drug-susceptible and resistant isolates. The results of the stable isotope labeling assay were comparable to the results of phenotypic methods used for susceptibility testing.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The extensive and injudicious use of antibiotics has led to an increase in the number of drug-resistant microorganisms, giving way to the origin of many superbugs that are multi-drug resistant [1]. Neisseria gonorrhoeae is also one of the members of this colossal group of drug-resistant microorganisms, which is proving as a menace to global reproductive health [2]. There have been reports of multidrug-resistant (MDR) and extensively drug-resistant (XDR) N. gonorrhoeae across the globe [2,3,4]. The current situation demands judicious use of antimicrobials, alongside delimiting the transmission of MDR/XDR strains and early diagnosis and management of such cases [5]. The currently accepted methods for the detection of drug resistance in N. gonorrhoeae include the agar dilution assay and E-test [6]. However, these are time-consuming and may lead to delays in treatment [7]. Besides the phenotypic methods, there are reports of a few genotypic methods that can be employed for the detection of drug resistance. However, the co-relation of genotypic methods with phenotypic methods is not reliable. Moreover, the methods fail to detect drug resistance due to newly acquired and novel mutations [8]. The Matrix-Assisted Laser Desorption/Ionization-Time of Flight Mass Spectrometry (MALDI-TOF MS) is an advanced technique used in various laboratories for rapid and reliable identification of clinically significant pathogenic organisms, without performing tedious and time-consuming biochemical assays [9]. The MALDI-TOF MS has also been exploited for the detection of drug resistance by identifying drug modifying enzymes and varied mutations in the target sites [10,11,12,13,14,15]. However, the identification of drug modifying enzymes limits the usage of MALDI-TOF MS as a tool to detect a plethora of MDR organisms and limits its potential. Recently, a novel technique of stable isotope labeling by amino acid-based assay using the MALDI-TOF MS has been employed for rapid detection of drug resistance in bacteria and fungi [16,17,18]. This assay is based on the detection of the metabolic profile, in the presence and absence of drugs that could discriminate between susceptible and resistant bacteria. In the present study, we optimized and evaluated a SILAC-based approach using the MALDI-TOF MS for rapid and reliable detection of ciprofloxacin and azithromycin resistance in N. gonorrhoeae.
Material and methods
N. gonorrhoeae isolates
Twenty-five recent clinical isolates of N. gonorrhoeae, available from the sexually transmitted disease laboratory repository of our tertiary care center, were included in this study, and reference isolates (WHO-M, WHO-P, and ATCC-49226) were employed for the initial standardization of the experiments. The isolates were grown on GC Agar plates (3.6% Difco GC medium base agar (BD Diagnostics, Sparks, MD, USA) supplemented with 1% hemoglobin (BD Diagnostics) and 1% IsoVitalex (BD Diagnostics)), incubated at 37 °C in a CO2 incubator (Binder GmBH, Tuttlingen, Germany) and fresh overnight cultures were used for further experiments.
Antibiotic susceptibility testing
Antibiotic susceptibility testing of the isolates for ciprofloxacin and azithromycin was performed using the E-test (bioMérieux, France) according to the manufacturer’s instructions. Briefly, 0.5 McFarland inoculum was prepared from overnight N. gonorrhoeae culture and the suspension was further used to seed the GC agar plates (3.6% Difco GC medium base agar (BD Diagnostics, Sparks, MD, USA) supplemented with 1% hemoglobin (BD Diagnostics) and 1% IsoVitalex (BD Diagnostics). The E-test strip was placed on the agar, and minimum inhibitory concentration (MIC) was determined after incubation of 18 h at 37 °C in a CO2 incubator.
Optimization of L-lysine concentration
The liquid medium for SILAC was prepared according to the protocol detailed by Wade and Graver (2007), using M199 cell culture medium supplemented with Earle’s salts without L-glutamine phenol red, sodium bicarbonate, and L-lysine (Himedia, Mumbai, India), in place of M199 cell culture medium with Earle’s salts but without L-glutamine, phenol red or sodium bicarbonate [19]. The liquid medium prepared for SILAC studies was evaluated for the growth of N. gonorrhoeae (1 × 104 cells) with varying concentrations of normal L-lysine (NL) (Invitrogen, USA) from 100 to 500 mg/L at different concentrations in a 96-well microtitre plate incubated in a CO2 incubator with shaking (300 rpm) (Binder-GmbH, Tuttlingen, Germany). The N. gonorrhoeae WHO-M, WHO-K, and ATCC-49226 reference isolates were employed for this experiment. The OD600 was observed at intervals of 2 h, and a growth curve was plotted to assess the ability of media to support the growth of N. gonorrhoeae.
Optimization of cell count and time to stable isotope incorporation
Two independent setups were prepared for each isolate the first setup with NL (Invitrogen, USA) and the second setup containing heavy lysine (HL) (13C6 15N2-L-lysine; Sigma-Aldrich, Germany). The best initial inoculum concentration for each setup was optimized by inoculating with various concentrations of cells (1 × 106, 5 × 106, and 1 × 107 cells/mL) in a 96-well microtiter plate and incubating at 37 °C in a CO2 incubator with shaking for varying time intervals. The N. gonorrhoeae culture from both the setups was observed by the Microflex LT Biotyper instrument (Bruker Daltonik, Bremen, Germany) every 2 h for determining the optimum incorporation of stable heavy isotopes.
Optimization of the drug concentrations
Three independent 300-µL setups were prepared for each isolate, the first setup containing media supplemented with NL, the second setup containing media with HL, and the third containing HL with ciprofloxacin/azithromycin (HL + Cip./HL + Azi.) at different concentrations (Cip. (0.015 to 8 µg/ml) and Azi. (1 to 16 µg/ml)).
Sample preparation and data acquisition
The cells of N. gonorrhoeae were transferred into a sterile 1.5-mL centrifuge tube and pelleted down. The supernatant was carefully aspirated without disturbing the pellet. 25 µl of milli-Q water was added to the cell pellet and vortexed vigorously, 2 µl of this suspension was spotted on the MALDI plate and air-dried. The dried spot was overlaid with 1 μl of HCCA matrix solution and dried at room temperature. Each spot was examined by using the Microflex LT Biotyper instrument (Bruker Daltonik, Bremen, Germany) with the default instrument settings as follows: laser frequency; 60 Hz, lens; 8.5 kV, ion source 1; 20 kV, and ion source 2; 18.1 kV. Spectra were recorded in the positive linear mode in the mass range of 1000 to 12,000 Da with 25–35% laser intensity, and spectra produced by a sum of 240 laser shots were considered for each sample to analyze the data [18]. The MALDI Biotyper 3.1 was used to analyze the acquired data.
Analysis of acquired spectra
The FlexAnalysis 3.3 program (Bruker Daltonics GmbH, Germany) was employed for the visual analysis of the spectra. Briefly, the spectra were smoothened and baseline subtracted, and this data was subjected to visual analysis. The results of the visual analysis were reconfirmed by virtual gel analysis and cluster analysis. The similarity in the spectra obtained from the N. gonorrhoeae isolates grown in different setups was observed by using the virtual gel analysis module present in the MALDI Biotyper software, which represents all the significantly visible peaks as bands on the virtual gel created by the module. Clustering of the spectra obtained from different setups was accomplished by constructing the PCA dendrogram. The PCA dendrogram determined the similarity and differences between the spectra acquired from different conditions [20].
Results
N. gonorrhoeae isolates and antimicrobial susceptibility profile
Antimicrobial susceptibility testing of the clinical isolates demonstrated 3 isolates as susceptible and 22 isolates to be resistant to ciprofloxacin. On the contrary, 22 isolates were noted to be susceptible while 3 were found resistant to azithromycin (Fig. S1).
Optimization of media components and inoculum concentration
The growth curves for WHO-M, WHO-P, and ATCC49226 were plotted using OD600 for time 0 to 24 h at time intervals of 2 h. The isolates displayed no noticeable variations in the growth curves with varying concentrations of lysine, 200 mg/L of lysine was used for the further experiments to flush in a high concentration of lysine (Supplementary Fig. S2). It was observed that the cells harvested at the time point of 6 h were consistently demonstrating the incorporation of the stable heavy isotope with a cell number of 1 × 107 cells/mL (Supplementary Fig. S3). Thus, a count of 1 × 107 cells/ml was used for all the further experiments.
Assessment of acquired spectra using existing database
The acquired spectra of the isolates grown in NL and HL were compared with the existing MALDI-TOF MS database to assess the identification status. None of the isolates growing in HL had a log score < 1.4 and were not identified as N. gonorrhoeae during the MALDI-TOF MS analysis, while all the isolates growing in NL were correctly identified as N. gonorrhoeae with a log score of > 1.9 (Supplementary Table S1 & S2).
Assessment of the spectral shift
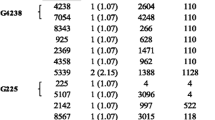
Flex analysis software was used for the pre-processing of acquired raw spectra for further analysis. N. gonorrhoeae cells grown in the media with HL showed a significant spectral shift, indicating enough incorporation of HL into the newly dividing cells (Fig. 1). Ten significant peak shifts were selected for resistance analysis between mass range of 2000–10,000 Da (m/z) and the mass tolerance of ± 3 Da was designated as the same peak. The peak shift for isolates grown in media with HL with respect to NL was 8 to 80 Da. It is well known that single heavy isotope incorporation increases the mass by 8 Da and every shift was found to be the approximate multiple of 8 (Table S3).
Visual resistance profiling and cut-off drug concentration
Flex analysis software was used to visualize the spectra obtained from three different setups, (a) medium with NL, (b) medium with HL, and (c) media with heavy lysine and drug (HL + Cip/HL + Azi.). The direct visual observation of spectra aptly differentiated resistant and susceptible isolates, resistant isolates displayed the incorporation of the heavy isotope into their proteins in the presence of the drug. Owing to the incorporation of heavy isotopes, the spectra were similar to the spectra obtained in the HL setup. In contrast, spectra of the isolates susceptible to the drug were identical to the spectra obtained from the isolates in the presence of NL (Fig. 2). It was observed that 0.12 µg/ml of ciprofloxacin and 2 µg/ml of azithromycin were enough to inhibit the growth of susceptible N. gonorrhoeae isolates to the extent that there was no or very low incorporation of the HL.
Representative figure showing a comparison of spectra from susceptible (Cip. = 0.12 μg/mL, Azi. = 0.5 μg/mL) and resistant (Cip. = 1 μg/mL Azi. = 2 μg/mL) isolates by direct visual inspection. A, E In susceptible isolate, the spectra of HL + Cip./HL + Azi. are similar to NL. B, F The overlapped view of spectra also showed similarity between NL and HL + Cip/HL + Azi. in susceptible isolate. C, G In susceptible isolate, the spectra of HL + Cip./HL + Azi. are similar to HL. D, H The overlapped view of spectra also showed similarity between HL and HL + Cip. in resistant isolate
Validation of results by virtual gel and PCA dendrogram analysis
The results obtained by the visual inspection were further confirmed by constructing virtual gel and PCA dendrogram, both virtual gel analysis, and PCA dendrogram confirmed our findings. It was noted that HL and HL + Drug spectra generated highly similar banding patterns in virtual gel analysis and generated a single clade in the PCA dendrogram; NL produced a different banding pattern and was placed in a separate clade (Fig. 3) in the case of resistant isolates. When looking for susceptible isolates, the isolates grown in NL and HL + Drug produced a similar banding pattern was generated, whereas the susceptible isolates grown in HL produced a different banding pattern (Fig. 4).
The representative PCA dendrogram derived from three setups depicts separate clustering according to the drug susceptibility status. A The spectra obtained from ciprofloxacin susceptible isolate show clustering of HL + Cip. and NL spectra. B The spectra obtained from ciprofloxacin resistant isolate show clustering of HL + Cip. and HL spectra. C The spectra obtained from azithromycin susceptible isolate show clustering of HL + Azi. and NL spectra. D The spectra obtained from azithromycin resistant isolate show clustering of HL + Azi. and HL spectra
Virtual gel analysis of representative ciprofloxacin and azithromycin resistant and susceptible isolates in three different setups. A The banding pattern of HL + Cip. and NL was similar in susceptible isolate. B The banding pattern of HL + Cip. was similar to the bands in HL in resistant isolate. C The banding pattern of HL + Azi. and NL was similar in susceptible isolate. D The banding pattern of HL + Azi. was similar to the bands in HL in resistant isolate
Discussion
MALDI-TOF MS has revolutionized the identification and antimicrobial susceptibility of bacteria and fungi. It is also an innovative technique employed for the detection and quantification of metabolites, peptides, proteins, recombinant proteins, and degraded products for precise detection of susceptibility profiles [21]. MALDI-TOF MS has earlier been employed for the detection of β-lactamases [22] and has the potential for the detection of drug susceptibility in bacteria and fungi [16,17,18, 23].
The present study demonstrates the development of a drug resistance detection method for N. gonorrhoeae based on stable isotope labeling, involving the incorporation of 13C-labeled amino acids. In this study, we used a modified GW medium, which was deficient in L-lysine, and subsequently, the concentration of lysine was also standardized for the optimal growth of N. gonorrhoeae, and L-lysine was opted in owing to its high incorporation in ribosomal proteins. The L-Lysine at a concentration of 100 mg/L was able to support the growth of N. gonorrhoeae isolates still 200 mg/L of lysine was used for better reproducibility of the results [24]. The cell count and drug concentrations were standardized for the detection of drug resistance using the MALDI-TOF MS. Several cell divisions are necessary to facilitate the optimum incorporation of the heavy isotopes and their detection. In the present study, the time for incubation was standardized to be 6 h compared to previous studies that demonstrated results in approximately 3 h [17]. This delay in time could be attributed to the fastidious nature of N. gonorrhoeae. The classical detection methods of drug resistance in N. gonorrhoeae take 18–24 h and thus, this assay may be helpful in reducing the turnaround time for antimicrobial susceptibility testing. The differentiation of susceptible and resistant isolates was made by assessing their ability to uptake HL in presence of the drug. The differences in spectra acquired from susceptible and resistant isolates grown in the presence of HL + Drug were visually observed to be non-identical. The spectra of susceptible N. gonorrhoeae isolates in presence of HL + Drug were similar to the spectra observed in the isolates grown in a medium with NL, whereas the spectra of resistant isolates grown in the presence of HL + Drug were completely different from the spectra of the isolates grown in the presence of NL and similar to HL. The data acquisition was performed using the default settings of the instrument for the species identification, which demonstrates the ease of use of the same method. This method can be employed in the laboratories using the MALDI-TOF MS for bacterial identification for antimicrobial susceptibility testing without much hassle. The results of the findings were validated by the Virtual 2-D gel analysis and PCA dendrogram. The results thus predicted were completely concordant to the results of the E-tests, which might be due to the availability of the low number of N. gonorrhoeae isolates susceptible to ciprofloxacin and resistant to azithromycin. The time needed to produce results was observed to be 6 h, which is in turn very less compared to the time required to perform routine the E-test or agar dilution methods and the same was noted to be in congruence to the previous studies [18, 23]. The current method is a phenotypic method that exploits changes in the metabolic profile of the organism to decipher antimicrobial susceptibility. Moreover, being a phenotypic method, it can precisely predict antimicrobial susceptibility irrespective of mutations and other mechanisms imparting resistance in the N. gonorrhoeae isolates.
The protocol standardized in the present study can be exploited for the detection of drug susceptibility profiles, after evaluating and standardizing the breakpoints even for other drugs in N. gonorrhoeae. The availability of a smaller number of isolates can be considered as the limitation of the study. The method can thus be used for testing the susceptibility profile of N. gonorrhoeae after evaluation of the breakpoints using a larger number of isolates in a lesser turnaround time.
References
Fair RJ, Tor Y (2014) Antibiotics and bacterial resistance in the 21st century. Perspect Medicinal Chem 6:25–64. https://doi.org/10.4137/PMC.S14459
Unemo M, Shafer WM (2014) Antimicrobial resistance in Neisseria gonorrhoeae in the 21st century: past, evolution, and future. Clin Microbiol Rev 27:587–613. https://doi.org/10.1128/CMR.00010-14
Wi T, Lahra MM, Ndowa F et al (2017) Antimicrobial resistance in Neisseria gonorrhoeae: global surveillance and a call for international collaborative action. PLOS Med 14:e1002344. https://doi.org/10.1371/journal.pmed.1002344
Unemo M, Seifert HS, Hook EW et al (2019) Gonorrhoea. Nat Rev Dis Primers 5:1–23. https://doi.org/10.1038/s41572-019-0128-6
Fletcher-Lartey S, Dronavalli M, Alexander K et al (2019) Trends in antimicrobial resistance patterns in neisseria gonorrhoeae in Australia and New Zealand: a meta-analysis and systematic review. Antibiotics 8. https://doi.org/10.3390/antibiotics8040191
Tapsall J (2001) Antimicrobial resistance in Neisseria gonorrhoeae. Who 1:1–58.http://www.who.int/csr/resources/publications/drugresist/Neisseria_gonorrhoeae.pdf (accessed 7 Dec 2020)
Alcala L, Garcia-Garrote F, Cercenado E et al (1998) Comparison of broth microdilution method using haemophilus test medium and agar dilution method for susceptibility testing of Eikenella corrodens. J Clin Microbiol 36:2386–2388. https://doi.org/10.1128/jcm.36.8.2386-2388.1998
Fluit AC, Visser MR, Schmitz F-J (2001) Molecular detection of antimicrobial resistance. Clin Microbiol Rev 14:836–837. https://doi.org/10.1128/CMR.14.4.836-871.2001
Singhal N, Kumar M, Kanaujia PK et al (2015) MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Front Microbiol 6:791. https://doi.org/10.3389/fmicb.2015.00791
Majcherczyk PA, McKenna T, Moreillon P et al (2006) The discriminatory power of MALDI-TOF mass spectrometry to differentiate between isogenic teicoplanin-susceptible and teicoplanin-resistant strains of methicillin-resistant Staphylococcus aureus. FEMS Microbiol Lett 255:233–239. https://doi.org/10.1111/j.1574-6968.2005.00060.x
Burckhardt I, Zimmermann S (2011) Using matrix-assisted laser desorption ionization-time of flight mass spectrometry to detect carbapenem resistance within 1 to 2.5 hours. J Clin Microbiol 49:3321–4. https://doi.org/10.1128/JCM.00287-11
Edwards-Jones V, Claydon MA, Evason DJ et al (2000) Rapid discrimination between methicillin-sensitive and methicillin-resistant Staphylococcus aureus by intact cell mass spectrometry. J Med Microbiol 49:295–300. https://doi.org/10.1099/0022-1317-49-3-295
Vereshchagin VA, Ilina EN, Zubkov MM et al (2005) Detection of fluoroquinolone resistance single-nucleotide polymorphisms in Neisseria gonorrhoeae gyrA and parC using MALDI-TOF mass spectrometry. Mol Biol 39:806–814. https://doi.org/10.1007/s11008-005-0099-4
de Carolis E, Vella A, Florio AR et al (2012) Use of matrix-assisted laser desorption ionization-time of flight mass spectrometry for caspofungin susceptibility testing of Candida and Aspergillus species. J Clin Microbiol 50:2479–2483. https://doi.org/10.1128/JCM.00224-12
Paul S, Singh P, Shamanth AS et al (2018) Rapid detection of fluconazole resistance in Candida tropicalis by MALDI-TOF MS. Med Mycol 56:234–241. https://doi.org/10.1093/mmy/myx042
Sparbier K, Lange C, Jung J et al (2013) Maldi biotyper-based rapid resistance detection by stable-isotope labeling. J Clin Microbiol 51:3741–3748. https://doi.org/10.1128/JCM.01536-13
Jung JS, Eberl T, Sparbier K et al (2014) Rapid detection of antibiotic resistance based on mass spectrometry and stable isotopes. Eur J Clin Microbiol Infect Dis 33:949–955. https://doi.org/10.1007/s10096-013-2031-5
Paul S, Singh S, Chakrabarti A et al (2019) Stable isotope labelling: an approach for MALDI-TOF MS-based rapid detection of fluconazole resistance in Candida tropicalis. J Antimicrob Chemother 74:1269–1276. https://doi.org/10.1093/jac/dkz019
Wade JJ, Graver MA (2007) A fully defined, clear and protein-free liquid medium permitting dense growth of Neisseria gonorrhoeae from very low inocula. FEMS Microbiol Lett 273:35–37. https://doi.org/10.1111/j.1574-6968.2007.00776.x
Vella A, de Carolis E, Vaccaro L et al (2013) Rapid antifungal susceptibility testing by matrix-assisted laser desorption ionization-time of flight mass spectrometry analysis. J Clin Microbiol 51:2964–2969. https://doi.org/10.1128/JCM.00903-13
Bucknall M, Fung KYC, Duncan MW (2002) Practical quantitative biomedical applications of MALDI-TOF mass spectrometry. J Am Soc Mass Spectrom 13:1015–1027. https://doi.org/10.1016/S1044-0305(02)00426-9
Sparbier K, Schubert S, Weller U et al (2012) Matrix-assisted laser desorption ionization-time of flight mass spectrometry-based functional assay for rapid detection of resistance against β-lactam antibiotics. J Clin Microbiol 50:927–937. https://doi.org/10.1128/JCM.05737-11
Idelevich EA, Sparbier K, Kostrzewa M et al (2018) Rapid detection of antibiotic resistance by MALDI-TOF mass spectrometry using a novel direct-on-target microdroplet growth assay. Clin Microbiol Infect 24:738–743. https://doi.org/10.1016/j.cmi.2017.10.016
Han J, Yi S, Zhao X et al (2019) Improved SILAC method for double labeling of bacterial proteome. J Proteomics 194:89–98. https://doi.org/10.1016/j.jprot.2018.12.011
Acknowledgements
The authors duly acknowledge the Department of Medical Microbiology, PGIMER, Chandigarh for providing all the necessary facilities.
Author information
Authors and Affiliations
Contributions
RD: performing experiments, conceptualization, data analysis, and manuscript writing; SP: data analysis and manuscript writing/correction; PG: manuscript writing/correction; RY: manuscript correction; SSood: supervision; AG: conceptualization and supervision; MRS: conceptualization and supervision; SG: conceptualization and supervision; SS: conceptualization, supervision, and manuscript correction.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Responsible Editor: Tânia A. Tardelli Gomes
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Dadwal, R., Paul, S., Gupta, P. et al. Stable isotope labeling as a promising tool for rapid drug susceptibility testing in Neisseria gonorrhoeae. Braz J Microbiol 54, 1819–1825 (2023). https://doi.org/10.1007/s42770-023-00996-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-023-00996-2